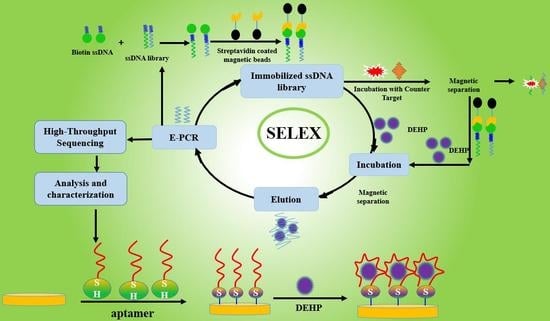
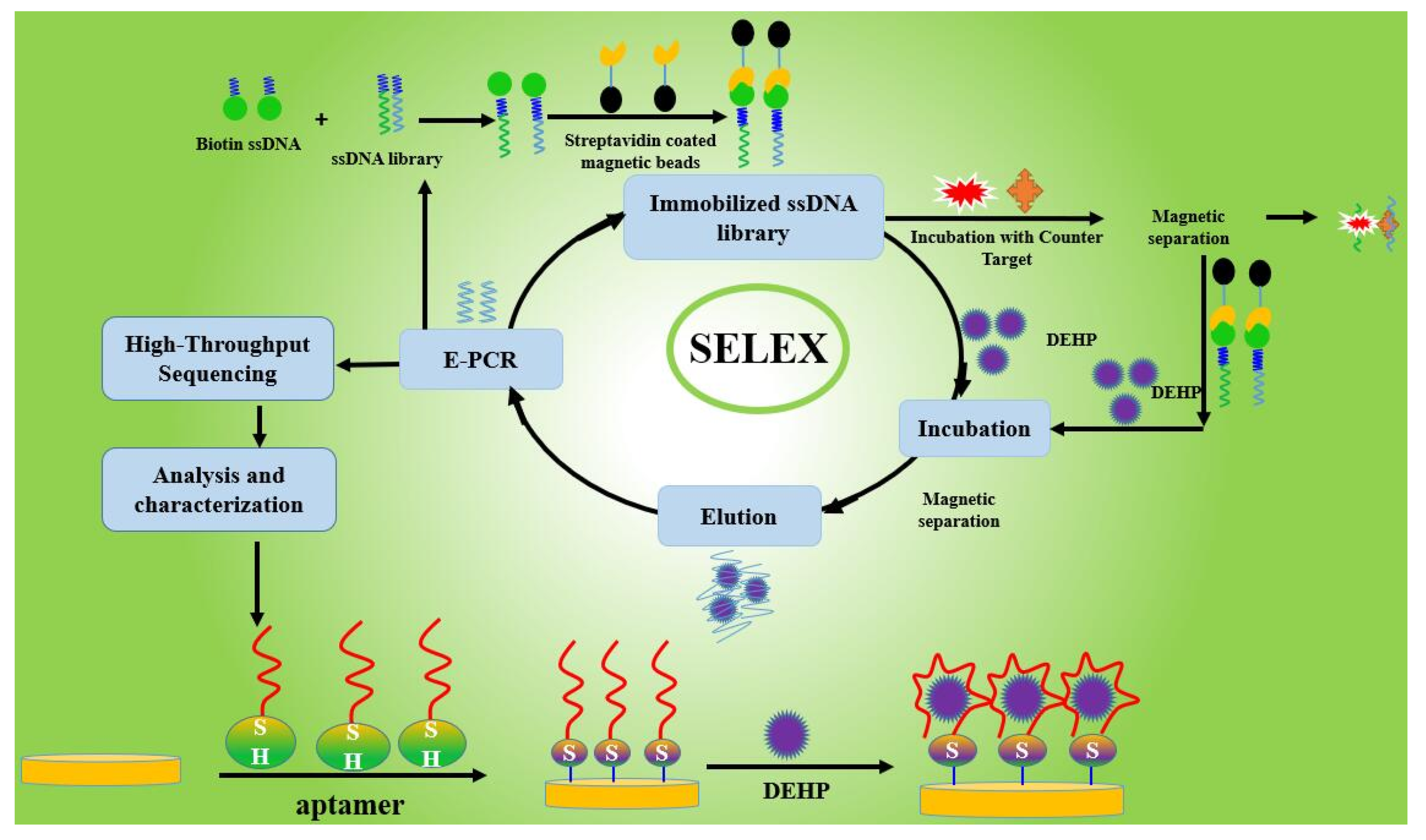
Selection of Aptamers Specific for DEHP Based on ssDNA Library Immobilized SELEX and Development of Electrochemical Impedance Spectroscopy Aptasensor
Abstract
1. Introduction
2. Results and Discussion
2.1. Selection of Aptamers Specific for DEHP Based on ssDNA Immobilized SELEX
2.2. High-Throughput Sequencing and Sequence Analysis of the Enriched Library
2.3. Establishment of Gold Nanoparticles Colomertric Assay to Identify Active Aptamers
2.4. Characterization of Active Aptamers
2.5. Fabrication of Electrochemical Impedance Spectroscope Aptasensor
2.6. Analytical Application of Sulfhydryl Modified Aptamer 31 in DEHP Detection Using EIS Aptasensor
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Selection of Aptamers Specific for DEHP Based on ssDNA library immobilized SELEX
3.3. Establishment of a Real-Time Quantitative PCR Method for the Monitoring Selection Process
3.4. Preparation of Secondary Libraries
3.5. High-Throughput Sequencing and Sequence Analysis of the Enriched Library
3.6. Determination of Binding Activity Between Aptamers and DEHP Using a Gold Nanoparticles Colomertric Assay
3.7. Affinity Analysis of Active Aptamers Using LSPR
3.8. Fabrication of EIS Aptasensor
3.9. Specificity and Sensitivity Analysis of Aptamer 31
3.10. Application of the EIS Aptasensor in Spiked Water Samples
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Melnick, R.L. Is peroxisome proliferation an obligatory precursor step in the carcinogenicity of Di (2-ethylhexyl) phthalate (DEHP)? Environ. Health Perspect. 2001, 109, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Saillenfait, A.M.; Langonne, I.; Leheup, B. Effects of mono-n-butyl phthalate on the development of rat embryos: In vivo and in vitro observations. Toxicol. Appl. Pharm. 2001, 89, 104–112. [Google Scholar] [CrossRef] [PubMed]
- How, C.M.; Yen, P.L.; Wei, C.C.; Li, S.W.; Liao, V.H.C. Early life exposure to di (2-ethylhexyl) phthalate causes age-related declines associated with insulin/IGF-1-like signaling pathway and SKN-1 in Caenorhabditis elegans. Environ. Pollut. 2019, 251, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, R.; Oliveira, N.; Maia, C.; Verde, I. Effects of di(2-etilhexil) phthalate on human umbilical artery. Chemosphere 2019, 228, 278–286. [Google Scholar] [CrossRef]
- Aignasse, M.F.; Prognon, P.; Stachowicz, M.; Gheyouche, R.; Pradeau, D. A new simple and rapid HPLC method for determination of DEHP in PVC packaging and releasing studies. Int. J. Pharm. 1995, 113, 241–246. [Google Scholar] [CrossRef]
- Deng, L.; Wu, F.; Deng, N.S.; Yang, Y. Determination of trace DEHP in aqueous solution by solid phase microextraction coupled with high performance liquid chromatography. Fresen. Environ. Bull. 2005, 14, 494–497. [Google Scholar]
- Li, B.P.; Lin, Q.B.; Song, H.; Li, L.L. Determination of DEHP and DNOP in PVC film by ASE-RP-HPLC. Chin. J. Appl. Chem. 2008, 25, 63–66. [Google Scholar]
- Pinguet, J.; Kerckhove, N.; Eljezi, T.; Lambert, C.; Moreau, E.; Bernard, L.; Boeuf, B.; Decaudin, B.; Genay, S.; Masse, M.; et al. New SPE-LC-MS/MS method for the simultaneous determination in urine of 22 metabolites of DEHP and alternative plasticizers from PVC medical devices. Talanta 2019, 198, 377–389. [Google Scholar] [CrossRef]
- Cai, Z.M.; Zhang, Q.L.; Zhao, W.H.; Yang, K.F.; Wang, L.P.; Luo, N.; Tan, L.T. GC-MS determination of phthalates (DBP and DEHP) dissolved from plastic bags. Phys. Test. Chem. Anal. B 2003, 39, 529–530. [Google Scholar]
- Liang, D.P.; Fang, Y.P.; Liu, W.J.; Zhang, H.; Qiu, Y.T.; Dong, Y.M.; Ning, Y. Direct determination of bis (2-ethylhexyl) phthalate in environmental water samples by electrospray extraction ionization mass spectrometry. Chin. J. Anal. Chem. 2018, 46, 883–888. [Google Scholar]
- Zhang, M.; Hong, W.; Wu, X.; Zhang, Y.; Li, F.; Zhao, S.Q. A highly sensitive and direct competitive enzyme-linked immunosorbent assay for the detection of di-(2-ethylhexyl) phthalate (DEHP) in infant supplies. Anal. Methods UK 2015, 7, 5441–5446. [Google Scholar] [CrossRef]
- Han, Y.; Diao, D.L.; Lu, Z.W.; Li, X.N.; Guo, Q.; Huo, Y.M.; Xu, Q.; Li, Y.S.; Cao, S.L.; Wang, J.C.; et al. Selection of group-specific PAE-binding DNA aptamers via rational designed target immobilization and applications for ultrasensitive and highly selective detection of PAEs. Anal. Chem. 2017, 89, 5270–5277. [Google Scholar] [CrossRef]
- Lim, H.J.; Kim, A.R.; Yoon, M.Y.; You, Y.; Chua, B.; Son, A. Development of quantum dot aptasensor and its portable analyzer for the detection of di-2-ethylhexyl phthalate. Biosens. Bioelectron. 2018, 121, 1–9. [Google Scholar] [CrossRef]
- Tu, D.D.; Garza, T.J.; Coté, L.G. A SERS aptasensor for sensitive and selective detection of bis(2-ethylhexyl) phthalate. Rsc Adv. 2019, 9, 2618–2625. [Google Scholar] [CrossRef] [PubMed]
- Bird, R.E.; Walker, B.W. Single chain antibody variable regions. Trends Biotechnol. 1991, 9, 132–137. [Google Scholar] [CrossRef]
- Zhong, W.; Pu, Y.; Tan, W.H.; Liu, J.; Liao, J.; Liu, B.; Chen, K.; Yu, B.; Hu, Y.L.; Deng, Y.Y. Identification and application of an aptamer targeting papillary thyroid carcinoma using tissue-SELEX. Anal. Chem. 2019, 91, 8289–8297. [Google Scholar] [CrossRef] [PubMed]
- Shao, N.S.; Li, S.H.; Huang, Y.P. New development of SELEX technology and aptamer research. Prog. Biochem. Biophys. 2006, 33, 329–335. [Google Scholar]
- Gong, S.; Wang, Y.L.; Wang, Z.; Sun, Y.Y.; Zhang, W.B. Folding behaviors of purine riboswitch aptamers. Wuhan Univ. J. Nat. Sci. 2018, 23, 43–50. [Google Scholar] [CrossRef]
- Shen, R.; Tang, J.J.; Zhang, Z.Y.; Guo, L.; Xie, J.W. New magnetic beads-based enzyme linked aptamer colorimetric assay for trace amount protein detection. Chem. Res. Chin. Univ. 2009, 4, 701–705. [Google Scholar]
- Duan, N.; Gong, W.H.; Wu, S.J.; Wang, Z.P. An ssDNA library immobilized SELEX technique for selection of an aptamer against ractopamine. Anal. Chim. Acta 2017, 961, 100–105. [Google Scholar] [CrossRef]
- Beheshti-Marnani, A.; Hatefi-Mehrjardi, A.; Es’haghi, Z. A sensitive biosensing method for detecting of ultra-trace amounts of AFB1 based on “Aptamer/reduced graphene oxide” nano-bio interaction. Colloids Surf. B 2019, 175, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.H.; Tan, S.Z.; Zhu, Y.Q.; Li, W.; Chen, T.X.; Wang, Y.; Liu, C. Fluorescent aptamer functionalized graphene oxide biosensor for rapid detection of chloramphenicol. Acta. Chim. Sin. 2019, 77, 253–256. [Google Scholar] [CrossRef]
- Bianco, M.; Sonato, A.; De Girolamo, A.; Pascale, M.; Romanato, F.; Rinaldi, R.; Arima, V. An aptamer-based SPR-polarization platform for high sensitive OTA detection. Sens. Actuat. B Chem. 2017, 241, 314–320. [Google Scholar] [CrossRef]
- Luo, Z.F.; He, L.; Wang, J.J.; Fang, X.N.; Zhang, L.Y. Developing a combined strategy for monitoring the progress of aptamer selection. Analyst 2017, 142, 3136–3139. [Google Scholar] [CrossRef]
- Lee, A.Y.; Ha, N.R.; Jung, I.P.; Kim, S.H.; Kim, A.R.; Yoon, M.Y. Development of a ssDNA aptamer for detection of residual benzylpenicillin. Anal. Biochem. 2017, 531, 1–7. [Google Scholar] [CrossRef]
- Challier, L.; Mavre, F.; Moreau, J.; Fave, C.; Schollhorn, B.; Marchal, D.; Peyrin, E.; Noel, V.; Limoges, B. Simple and highly enantioselective electrochemical aptamer-based binding assay for trace detection of chiral compounds. Anal. Chem. 2012, 84, 5415–5420. [Google Scholar] [CrossRef]
- Li, J.; Zhong, X.; Zhang, H.; Le, X.C.; Zhu, J.J. Binding-induced fluorescence turn-on assay using aptamer-functionalized silver nanocluster DNA probes. Anal. Chem. 2012, 84, 5170–5174. [Google Scholar]
- Gopinath, S.C.B.; Lakshmipriya, T.; Awazu, K. Colorimetric detection of controlled assembly and disassembly of aptamers on unmodified gold nanoparticles. Biosens. Bioelectron. 2014, 51, 115–123. [Google Scholar] [CrossRef]
- Yoo, S.M.; Kim, D.K.; Lee, S.Y. Aptamer-functionalized localized surface plasmon resonance sensor for the multiplexed detection of different bacterial species. Talanta 2015, 132, 112–117. [Google Scholar] [CrossRef]
- Li, S.Y.; Chen, D.Y.; Zhou, Q.T.; Wang, W.; Gao, L.F.; Jiang, J.; Cui, H. A general chemiluminescence strategy for measuring aptamer-target binding and target concentration. Anal. Chem. 2014, 86, 5559–5566. [Google Scholar]
- Rangel, A.E.; Chen, Z.; Ayele, T.M.; Heemstra, J.M. In vitro selection of an XNA aptamer capable of small-molecule recognition. Nucleic Acids Res. 2018, 46, 8057–8068. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.G.; Zhan, S.S.; Wang, L.M.; Zhou, P. Selection of a DNA aptamer for cadmium detection based on cationic polymer mediated aggregation of gold nanoparticles. Analyst 2014, 139, 1550–1561. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.H.; Yang, S.; Zhou, Q.; Yang, L.K.; Wang, P.J.; Fang, Y. The suitable condition of using LSPR model in SERS: LSPR effect versus chemical effect on microparticles surface-modified with nanostructures. Plasmonics 2017, 12, 77–81. [Google Scholar] [CrossRef]
- Kharitonov, A.B.; Alfonta, L.; Katz, E.; Willner, I. Probing of bioaffinity interactions at interfaces using impedance spectroscopy and chronopotentiometry. Electroanal. Chem. 2000, 487, 133–141. [Google Scholar] [CrossRef]
- Zhou, N.D.; Luo, J.B.; Zhang, J.; You, Y.D.; Tian, Y.P. A label-free electrochemical aptasensor for the detection of kanamycin in milk. Anal. Methods 2015, 7, 1991. [Google Scholar] [CrossRef]
- Liu, X.X.; Lu, Q.; Chen, S.R.; Wang, F.; Hou, J.J.; Xu, Z.L.; Meng, C.; Hu, T.Y.; Hou, Y.Y. Selection and identification of novel aptamers specific for clenbuterol based on ssDNA library Immobilized SELEX and gold nanoparticles colomertric assay. Molecules 2018, 23, 2337. [Google Scholar] [CrossRef]
- Yao, X.; Ma, X.D.; Ding, C.; Jia, L. Colorimetric determination of lysozyme based on the aggregation of gold nanoparticles controlled by a cationic polymer and an aptamer. Microchim. Acta 2016, 183, 2353–2359. [Google Scholar] [CrossRef]
- Haiss, W.; Thanh, N.T.K.; Aveyard, J.; Fernig, D.G. Determination of size and concentration of nanoparticles from UV-Vis spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef]
- Lu, Y.; Xu, X.; Jiang, T.; Jin, L.; Zhao, X.D.; Cheng, J.H.; Jin, X.J.; Ma, J.; Piao, H.N.; Piao, L.X. Sertraline ameliorates inflammation in CUMS mice and inhibits TNF-alpha-induced inflammation in microglia cells. Int. Immunopharmacol. 2019, 67, 119–128. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |







| Aptamer | Sequence (5′-3′) | Kd(nM) |
|---|---|---|
| 31 | ACGCATAGGGTGCGACCACATACGCCCCATGTATGTCCCTTGGTTGTGCCCTATGCGT | 2.26 ± 0.06 |
| 123 | ACGCATAGGGCAACCAGACCAGCCCCATCCCCATGTGACTTCTGTTTGGCCTATGCGT | 5.33 ± 0.01 |
| 203 | ACGCATAGGGCAAGACAAACTGCGCCATTCAGCATGCTGTTCGGGTTGGCCTATGCGT | 2.68 ± 0.2 |
| 281 | ACGCATAGGGTGTGCATCAGCAGTACCAACGACGTTGTGGTGTGCTCATCCTATGCGT | 43 ± 0.7 |
| Methods | Linear Range | Limit of Detection (LOD) | References |
|---|---|---|---|
| High-performance liquid chromatography | 50–100,000 ng/mL | / | 5–7 |
| High-performance liquid chromatography–mass spectrometry | 0.01–0.1 ng/mL | 1 pg/mL | 8 |
| Raman spectroscopy | 0.008–182nM | 3.12 pg/mL | 14 |
| Mass spectrometry | 5–1000 ng/mL | 210 pg/mL | 10 |
| Enzyme-linked immunosorbent assay | 10−3–103 ng/mL | 4.2 pg/mL | 11 |
| Multiresidue detection method based on aptamer | 0.5–30 ng/mL | 3.9 pg/mL | 12 |
| An ultrasensitive electrochemical method | 7.629 pg/mL-2 µg/mL | 0.103 pg/mL | This work |
| Samples | Spiked Concentration (μg·L−1) | Detected Concentration (μg·L−1) | RSD % | Recovery % |
|---|---|---|---|---|
| Tap water | 0.03 | 0.031 | 2.74 | 101.77 |
| 1.95 | 1.71 | 1.44 | 87.37 | |
| 500 | 462.59 | 1.57 | 92.52 | |
| Water from YB | 0.03 | 0.043 | 1.69 | 141.32 |
| 1.95 | 1.73 | 2.50 | 88.50 | |
| 500 | 488.44 | 2.54 | 97.69 | |
| Water from Qingshan Lake | 0.03 | 0.026 | 2.15 | 85.70 |
| 1.95 | 1.49 | 2.19 | 76.07 | |
| 500 | 481.22 | 0.55 | 96.24 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Q.; Liu, X.; Hou, J.; Yuan, Q.; Li, Y.; Chen, S. Selection of Aptamers Specific for DEHP Based on ssDNA Library Immobilized SELEX and Development of Electrochemical Impedance Spectroscopy Aptasensor. Molecules 2020, 25, 747. https://doi.org/10.3390/molecules25030747
Lu Q, Liu X, Hou J, Yuan Q, Li Y, Chen S. Selection of Aptamers Specific for DEHP Based on ssDNA Library Immobilized SELEX and Development of Electrochemical Impedance Spectroscopy Aptasensor. Molecules. 2020; 25(3):747. https://doi.org/10.3390/molecules25030747
Chicago/Turabian StyleLu, Qi, Xixia Liu, Jianjun Hou, Qiuxue Yuan, Yani Li, and Sirui Chen. 2020. "Selection of Aptamers Specific for DEHP Based on ssDNA Library Immobilized SELEX and Development of Electrochemical Impedance Spectroscopy Aptasensor" Molecules 25, no. 3: 747. https://doi.org/10.3390/molecules25030747
APA StyleLu, Q., Liu, X., Hou, J., Yuan, Q., Li, Y., & Chen, S. (2020). Selection of Aptamers Specific for DEHP Based on ssDNA Library Immobilized SELEX and Development of Electrochemical Impedance Spectroscopy Aptasensor. Molecules, 25(3), 747. https://doi.org/10.3390/molecules25030747