On Column Binding a Real-Time Biosensor for β-lactam Antibiotics Quantification
Abstract
1. Introduction
2. Results
2.1. Biosensor Assays
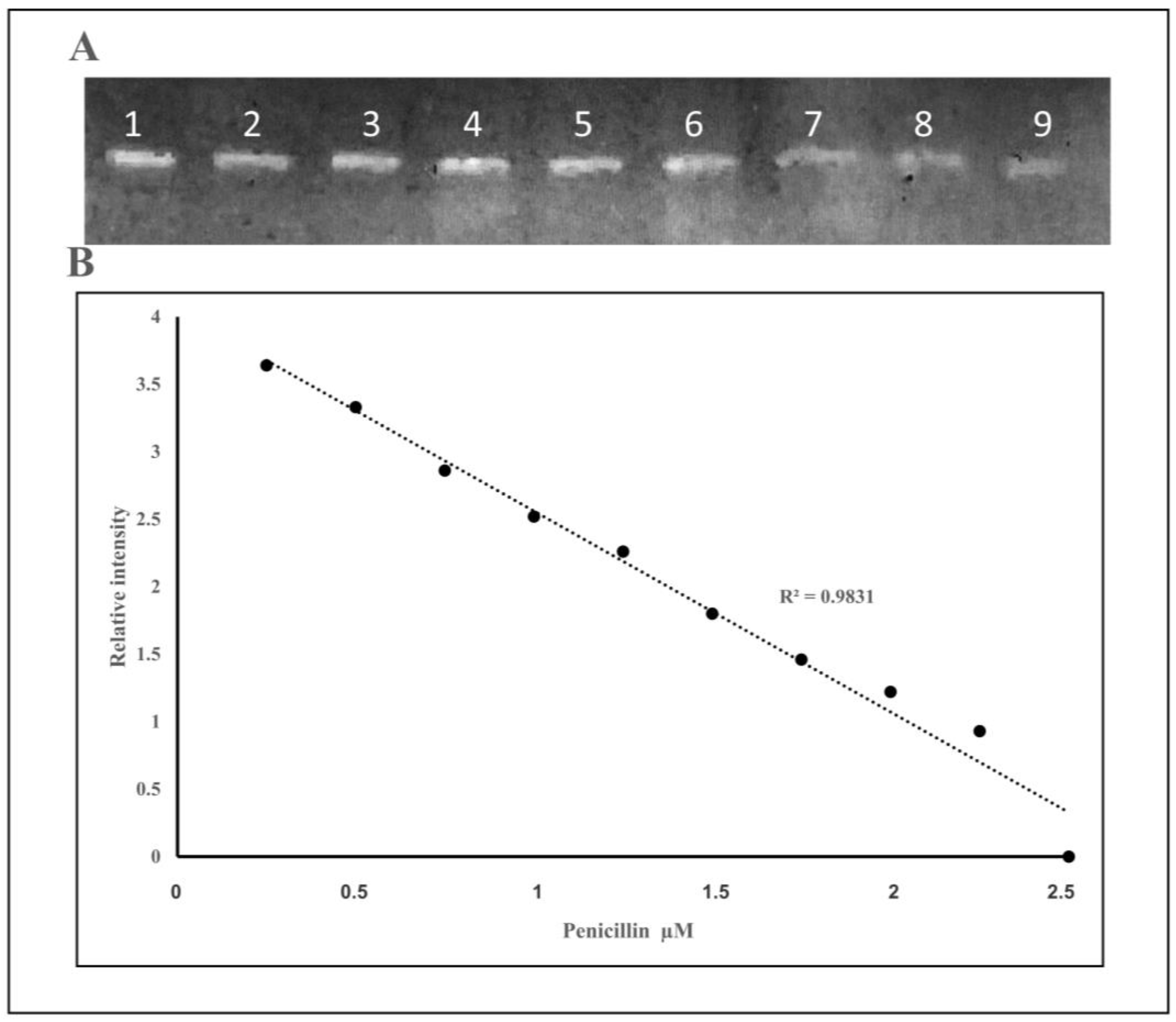
2.1.1. On Column Binding Real-Time Biosensor Assay
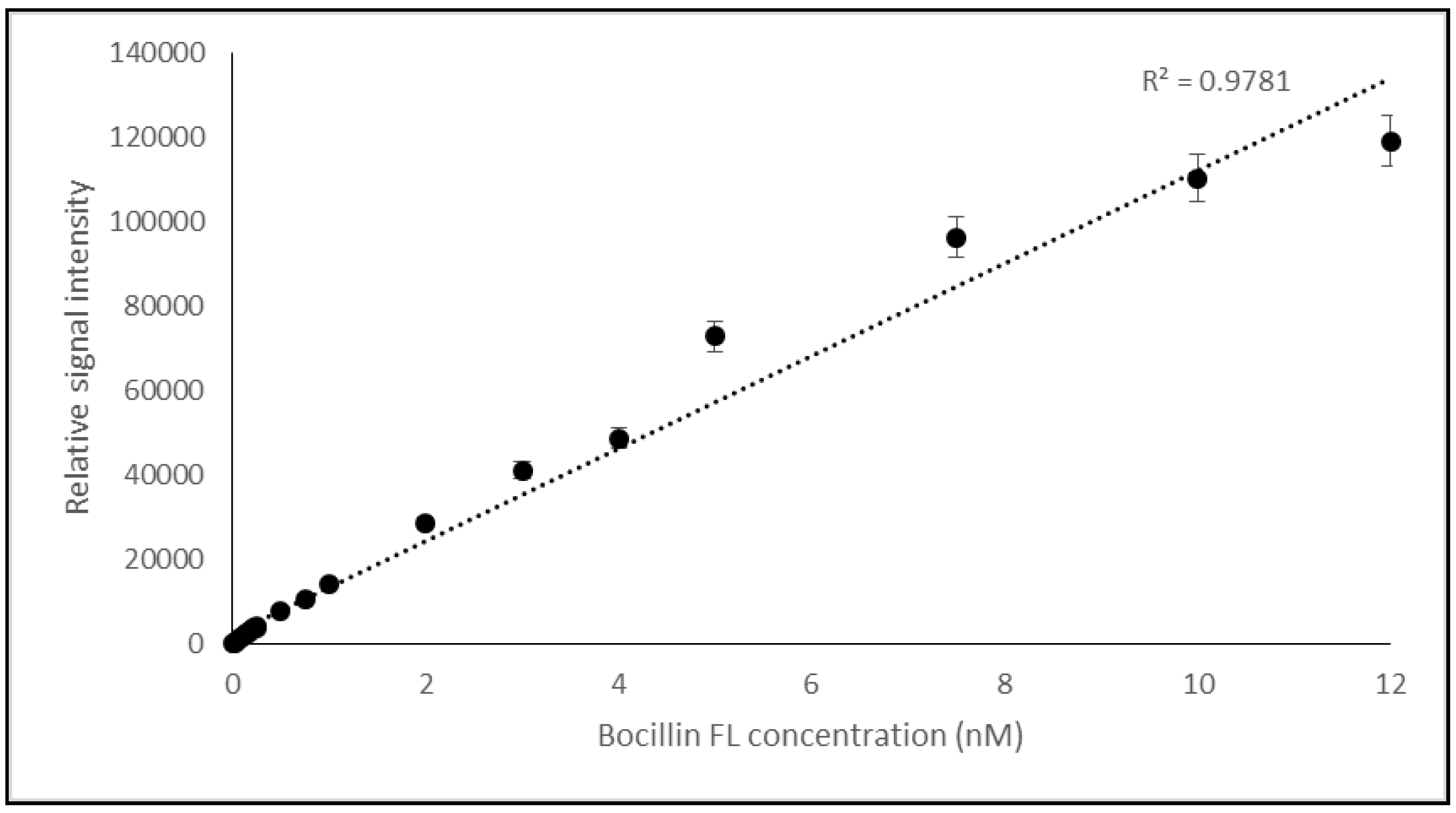
2.1.2. Optimization of Binding of Bocillin FL and PBP2x* for the ChemiDoc-It®2 Imager Based Biosensor
ChemiDoc-It®2 Imager as a Detector of Binding Bocillin FL to PBP2x* of S. pneumoniae
3. Materials and Methods
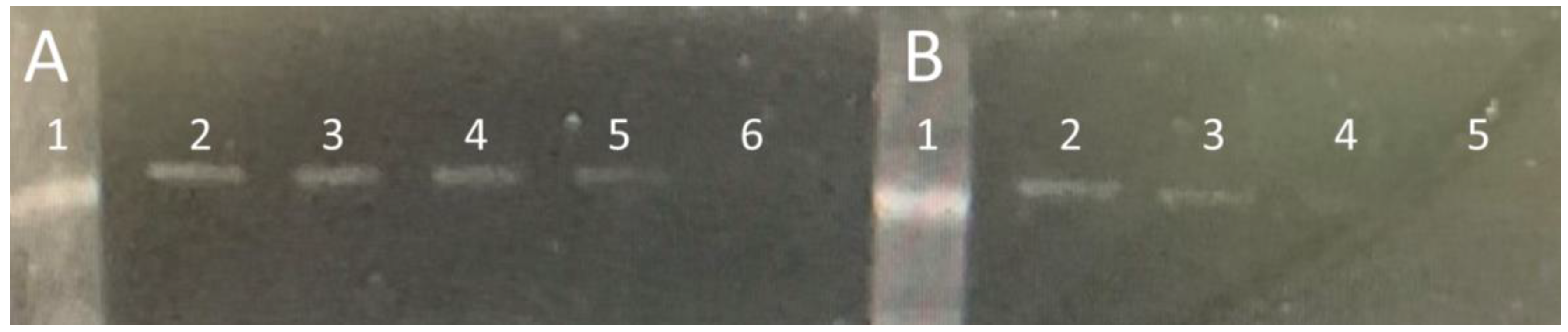
3.1. Expression and Purification of Soluble PBP 2x*
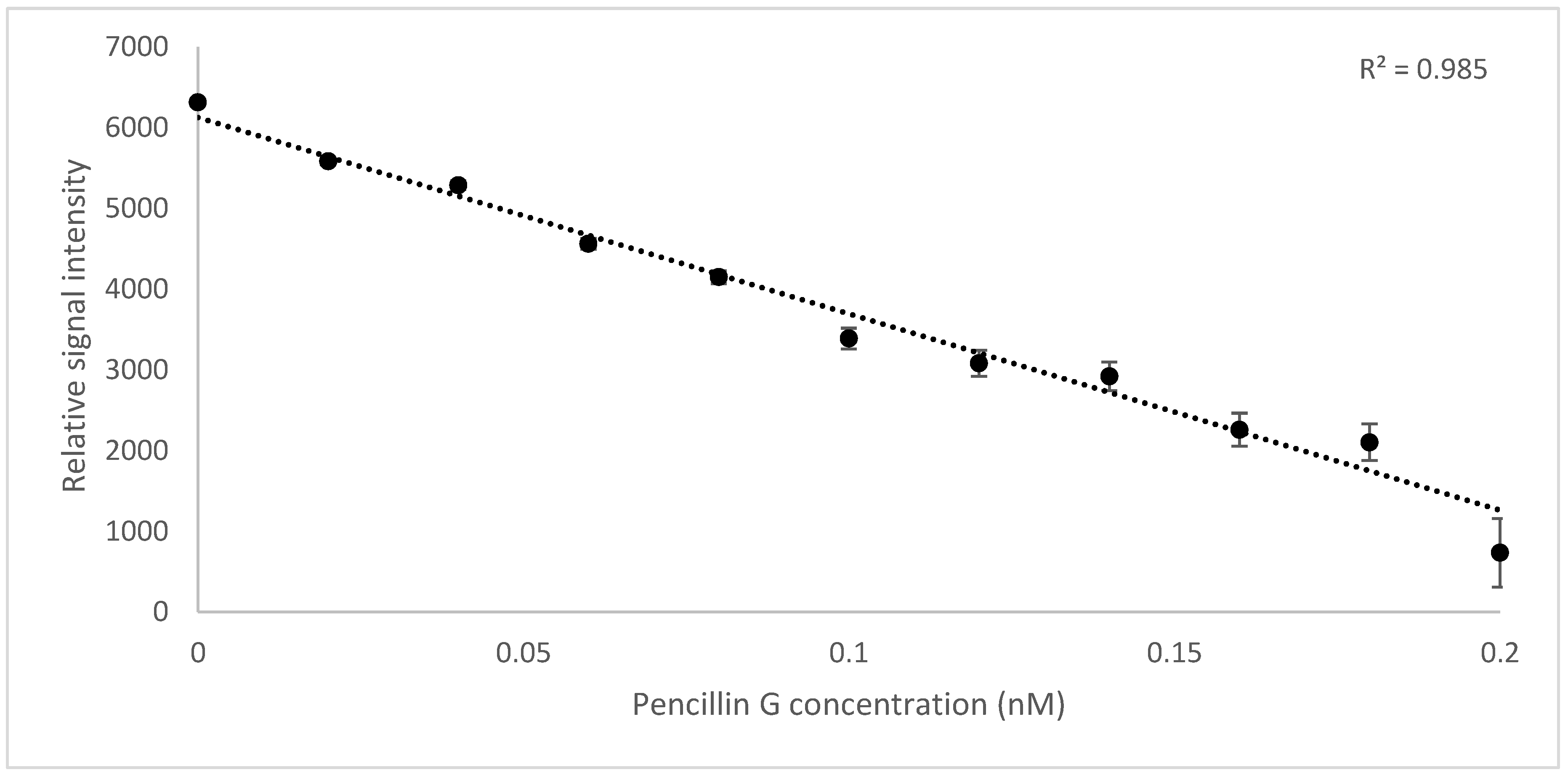
3.2. Biosensor Assay for Penicillin G Detection
3.2.1. On Column Binding Real-Time Biosensor Assay
3.2.2. HPLC Analysis
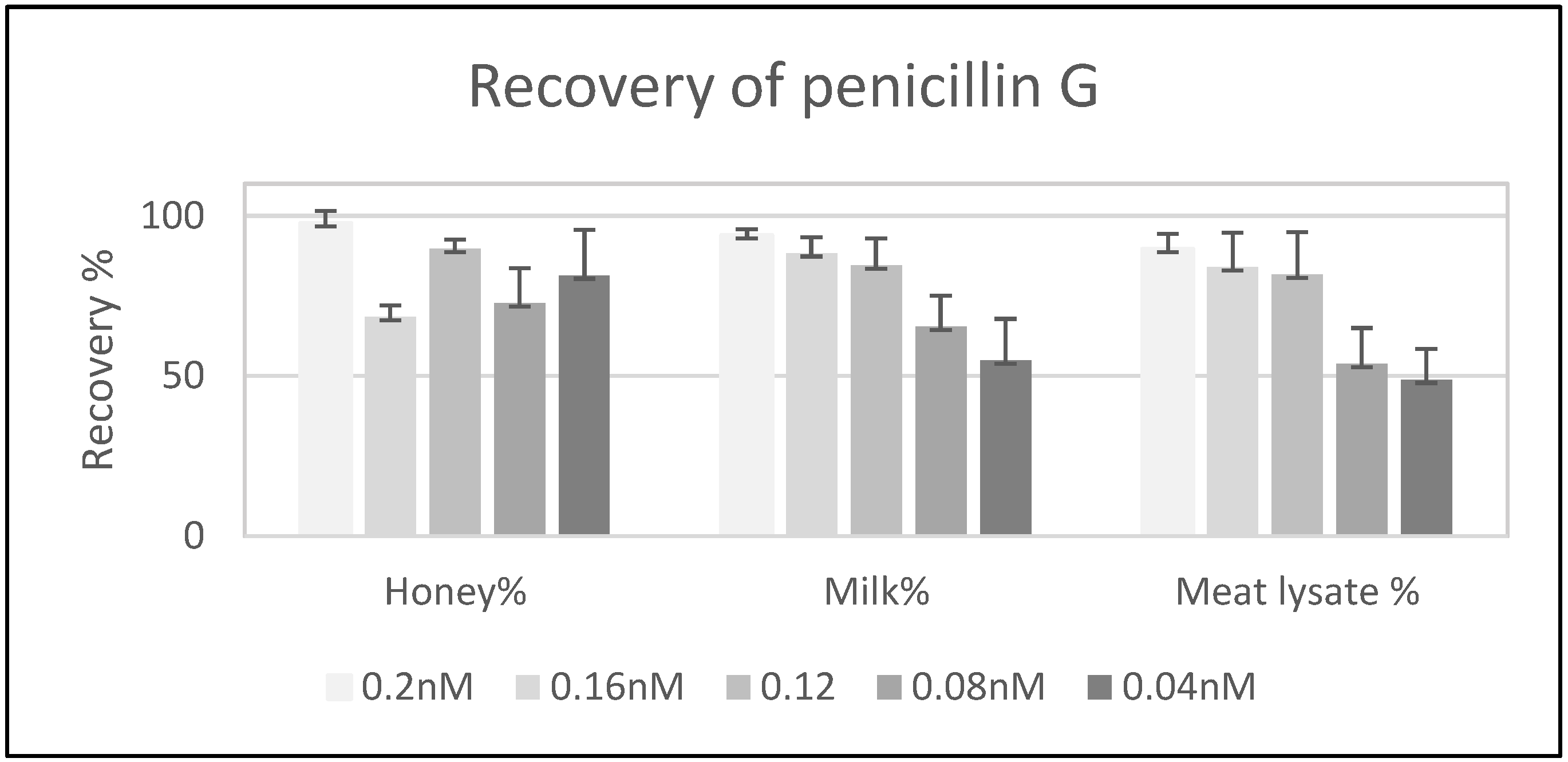
3.2.3. Recovery Test and Method Validation:
3.2.4. Quantification of Penicillin G in the Hospital Environment
3.2.5. ChemiDoc-It®2 Imagers- Based Quantification Assay
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bush, K.; Bradford, P. β-Lactams and β-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247. [Google Scholar] [CrossRef]
- Zeng, K.; Zhang, J.; Wang, Y.; Wang, Z.; Zhang, S.X.; Wu, C.; Shen, J.Z. Development of a rapid multi-residue assay for detecting β-lactams using penicillin binding protein 2x*. Biomed. Environ. Sci. 2013, 26, 100–109. [Google Scholar]
- Fedarovich, A.; Djordjevic, K.A.; Swanson, S.M.; Peterson, Y.K.; Nicholas, R.A.; Davies, C. High-Throughput Screening for Novel Inhibitors of Neisseria gonorrhoeae Penicillin-Binding Protein 2. PLoS ONE 2012, 7, e44918. [Google Scholar] [CrossRef] [PubMed]
- Gaudin, V.; Fontaine, J.; Maris, P. Screening of penicillin residues in milk by a surface plasmon resonance-based biosensor assay: Comparison of chemical and enzymatic sample pre-treatment. Anal. Chim. Acta 2001, 436, 191–198. [Google Scholar] [CrossRef]
- Peng, J.; Cheng, G.; Huang, L.; Wang, Y.; Hao, H.; Peng, D.; Liu, Z.; Yuan, Z. Development of a direct ELISA based on carboxy-terminal of penicillin-binding protein BlaR for the detection of β-lactam antibiotics in foods. Anal. Bioanal. Chem. 2013, 405, 8925–8933. [Google Scholar] [CrossRef] [PubMed]
- Lamar, J.; Petz, M. Development of a receptor-based microplate assay for the detection of beta-lactam antibiotics in different food matrices. Anal. Chim. Acta 2007, 586, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, E.; Bjurling, P.; Sternesjö, Å. Biosensor analysis of penicillin G in milk based on the inhibition of carboxypeptidase activity. Anal. Chim. Acta 2002, 468, 153–159. [Google Scholar] [CrossRef]
- Gustavsson, E.; Degelaen, J.; Bjurling, P.; Sternesjo, A. Determination of β-Lactams in Milk Using a Surface Plasmon Resonance-Based Biosensor. J. Agric. Food Chem. 2004, 52, 2791–2796. [Google Scholar] [CrossRef]
- Xiao, F.; Li, G.; Wu, Y.; Chen, Q.; Wu, Z.; Yu, R. Label-Free Photonic Crystal-Based β-Lactamase Biosensor for β-Lactam Antibiotic and β-Lactamase Inhibitor. Anal. Chem. 2016, 88, 9207–9212. [Google Scholar] [CrossRef]
- Chan, P.-H.; Liu, H.-B.; Chen, Y.W.; Chan, K.-C.; Tsang, C.-W.; Leung, Y.-C.; Wong, K.-Y. Rational Design of a Novel Fluorescent Biosensor for β-Lactam Antibiotics from a Class A β-Lactamase. J. Am. Chem. Soc. 2004, 126, 4074–4075. [Google Scholar] [CrossRef]
- Merola, G.; Martini, E.; Tomassetti, M.; Campanella, L. New immunosensor for β-lactam antibiotics determination in river waste waters. Sensors Actuators B Chem. 2014, 199, 301–313. [Google Scholar] [CrossRef]
- Justino, C.I.L.; Duarte, A.C.; Rocha-Santos, T. Recent Progress in Biosensors for Environmental Monitoring: A Review. Sensors 2017, 17, 2918. [Google Scholar] [CrossRef] [PubMed]
- Preston, D.A.; Wu, C.Y.; Blaszczak, L.C.; Seitz, D.E.; Halligan, N.G. Biological characterization of a new radioactive labeling reagent for bacterial penicillin-binding proteins. Antimicrob. Agents Chemother. 1990, 34, 718–721. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Ning, J.; Cheng, G.; Ahmad, I.; Li, J.; Mingyue, L.; Qu, W.; Iqbal, M.; Shabbir, M.; Yuan, Z. Receptor-based screening assays for the detection of antibiotics residues—A review. Talanta 2017, 166, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Kar, D.; Murugan, R.A.; Mallick, S.; Dutta, M.; Pandey, S.D.; Chowdhury, C.; Ghosh, A.S. A putative low-molecular-mass penicillin-binding protein (PBP) of Mycobacterium smegmatis exhibits prominent physiological characteristics of dd-carboxypeptidase and beta-lactamase. Microbiology 2015, 161, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Goradel, N.H.; Mirzaei, H.; Sahebkar, A.; Poursadeghiyan, M.; Masoudifar, A.; Malekshahi, Z.V.; Negahdari, B. Biosensors for the Detection of Environmental and Urban Pollutions. J. Cell. Biochem. 2017, 119, 207–212. [Google Scholar] [CrossRef]
- Malhotra, S.; Verma, A.; Tyagi, N.; Kumar, V. BioSensors: Principle types and applications. Int. J. Adv. Res. Innov. 2017, 3, 3639–3644. [Google Scholar]
- Srinivasan, B.; Tung, S. Development and Applications of Portable Biosensors. J. Lab. Autom. 2015, 20, 365–389. [Google Scholar] [CrossRef]
- Fronczek, C.F.; Yoon, J.-Y. Biosensors for Monitoring Airborne Pathogens. J. Lab. Autom. 2015, 20, 390–410. [Google Scholar] [CrossRef]
- Pohanka, M. Biosensors Based on Semiconductors, a Review. Int. J. Electrochem. Sci. 2017, 12, 6611–6621. [Google Scholar] [CrossRef]
- Zhao, G.; Meier, T.I.; Kahl, S.; Gee, K.R.; Blaszczak, L.C. BOCILLIN FL, a Sensitive and Commercially Available Reagent for Detection of Penicillin-Binding Proteins. Antimicrob. Agents Chemother. 1999, 43, 1124–1128. [Google Scholar] [CrossRef] [PubMed]
- Garcia, N.B.; Mahasenan, K.V.; Batuecas, M.T.; Lee, M.; Hesek, D.; Petráčková, D.; Doubravová, L.; Branny, P.; Mobashery, S.; Hermoso, J. Allostery, Recognition of Nascent Peptidoglycan, and Cross-linking of the Cell Wall by the Essential Penicillin-Binding Protein 2x of Streptococcus pneumoniae. ACS Chem. Boil. 2018, 13, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Cacciatore, G.; Petz, M.; Rachid, S.; Hakenbeck, R.; Bergwerff, A.A. Development of an optical biosensor assay for detection of β-lactam antibiotics in milk using the penicillin-binding protein 2x*. Anal. Chim. Acta 2004, 520, 105–115. [Google Scholar] [CrossRef]
- Kocaoglu, O.; Tsui, H.-C.T.; Winkler, M.E.; Carlson, E. Profiling of β-Lactam Selectivity for Penicillin-Binding Proteins in Streptococcus pneumoniae D39. Antimicrob. Agents Chemother. 2015, 59, 3548–3555. [Google Scholar] [CrossRef] [PubMed]
- Zerfaß, I.; Hakenbeck, R.; Denapaite, D. An Important Site in PBP2x of Penicillin-Resistant Clinical Isolates of Streptococcus pneumoniae: Mutational Analysis of Thr338v. Antimicrob. Agents Chemother. 2008, 53, 1107–1115. [Google Scholar] [CrossRef]
- Sauvage, E.; Terrak, M. Glycosyltransferases and Transpeptidases/Penicillin-Binding Proteins: Valuable Targets for New Antibacterials. Antibiotics 2016, 5, 12. [Google Scholar] [CrossRef]
- Stone, M.R.L.; Butler, M.S.; Phetsang, W.; Cooper, M.A.; Blaskovich, M.A.T. Fluorescent Antibiotics: New Research Tools to Fight Antibiotic Resistance. Trends Biotechnol. 2018, 36, 523–536. [Google Scholar] [CrossRef]
- Poltronieri, P.; De Blasi, M.; Of, D.U. Detection of Listeria monocytogenes through real-time PCR and biosensor methods. Plant. Soil Environ. 2009, 55, 363–369. [Google Scholar] [CrossRef]
- Vidic, J.; Manzano, M.; Chang, C.M.; Jaffrezic-Renault, N. Advanced biosensors for detection of pathogens related to livestock and poultry. Vet. Res. 2017, 48, 11. [Google Scholar] [CrossRef]
- Karageorgou, E.; Christoforidou, S.; Ioannidou, M.; Psomas, E.; Samouris, G. Detection of β-Lactams and Chloramphenicol Residues in Raw Milk—Development and Application of an HPLC-DAD Method in Comparison with Microbial Inhibition Assays. Foods 2018, 7, 82. [Google Scholar] [CrossRef]
- Worthington, R.J.; Melander, C. Overcoming Resistance to β-Lactam Antibiotics. J. Org. Chem. 2013, 78, 4207–4213. [Google Scholar] [CrossRef]
- Mungroo, N.; Neethirajan, S. Biosensors for the Detection of Antibiotics in Poultry Industry—A Review. Biosens. 2014, 4, 472–493. [Google Scholar] [CrossRef]
- Livermore, D.M. Minimising antibiotic resistance. Lancet Infect. Dis. 2005, 5, 450–459. [Google Scholar] [CrossRef]
- Bouki, C.; Venieri, D.; Diamadopoulos, E. Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: A review. Ecotoxicol. Environ. Saf. 2013, 91, 1–9. [Google Scholar] [CrossRef]
- Wu, X.-L.; Xiang, L.; Yan, Q.-Y.; Jiang, Y.-N.; Li, Y.-W.; Huang, X.-P.; Li, H.; Cai, Q.-Y.; Mo, C.-H. Distribution and risk assessment of quinolone antibiotics in the soils from organic vegetable farms of a subtropical city, Southern China. Sci. Total. Environ. 2014, 487, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Meek, R.W.; Vyas, H.; Piddock, L.J. Nonmedical Uses of Antibiotics: Time to Restrict Their Use? PLoS Boil. 2015, 13, e1002266. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Jaiswal, S.; Sodhi, K.K.; Shree, P.; Singh, D.K.; Agrawal, P.K.; Shukla, P. Antibiotics bioremediation: Perspectives on its ecotoxicity and resistance. Environ. Int. 2019, 124, 448–461. [Google Scholar] [CrossRef] [PubMed]
- Grenni, P.; Ancona, V.; Caracciolo, A.B. Ecological effects of antibiotics on natural ecosystems: A review. Microchem. J. 2018, 136, 25–39. [Google Scholar] [CrossRef]
- Gee, K.R.; Kang, H.C.; Meier, T.I.; Zhao, G.; Blaszcak, L.C. Fluorescent Bocillins: Synthesis and application in the detection of penicillin-binding proteins. Electrophoresis 2001, 22, 960–965. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdullah, S.M.; Rachid, S. On Column Binding a Real-Time Biosensor for β-lactam Antibiotics Quantification. Molecules 2020, 25, 1248. https://doi.org/10.3390/molecules25051248
Abdullah SM, Rachid S. On Column Binding a Real-Time Biosensor for β-lactam Antibiotics Quantification. Molecules. 2020; 25(5):1248. https://doi.org/10.3390/molecules25051248
Chicago/Turabian StyleAbdullah, Shahla M., and Shwan Rachid. 2020. "On Column Binding a Real-Time Biosensor for β-lactam Antibiotics Quantification" Molecules 25, no. 5: 1248. https://doi.org/10.3390/molecules25051248
APA StyleAbdullah, S. M., & Rachid, S. (2020). On Column Binding a Real-Time Biosensor for β-lactam Antibiotics Quantification. Molecules, 25(5), 1248. https://doi.org/10.3390/molecules25051248




