A Comparative Study on Processed Panax ginseng Products Using HR-MAS NMR-Based Metabolomics
Abstract
:1. Introduction
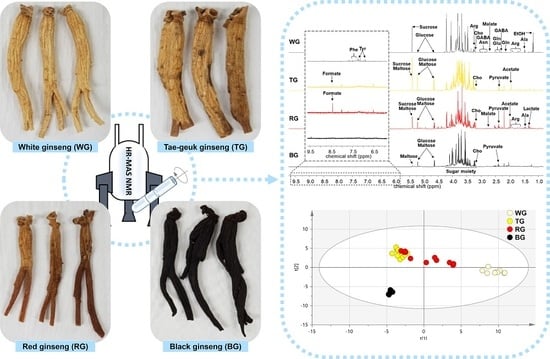
2. Results and Discussion
3. Materials and Methods
3.1. Processed Panax Ginseng Products
3.2. Sample Preparation
3.3. NMR Measurement
3.4. Data Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wang, Y.; Pan, J.Y.; Xiao, X.Y.; Lin, R.C.; Cheng, Y.Y. Simultaneous determination of ginsenosides in Panax ginseng with di erent growth ages using high-performance liquid chromatography–mass spectrometry. Phytochem. Anal. 2006, 17, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-M.; Li, S.-L.; Zhang, H.; Wang, Y.; Zhao, Z.-L.; Chen, S.-L.; Xu, H.-X. Holistic quality evaluation of commercial white and red ginseng using a UPLC-QTOF-MS/MS-based metabolomics approach. J. Pharm. Biomed. Anal. 2012, 62, 258–273. [Google Scholar] [CrossRef] [PubMed]
- Ning, X.; Han, C. Drying characteristics and quality of taegeuk ginseng (Panax ginseng CA Meyer) using far-infrared rays. Int. J. Food Sci. Technol. 2013, 48, 477–483. [Google Scholar] [CrossRef]
- Sun, B.-S.; Gu, L.-J.; Fang, Z.-M.; Wang, C.-Y.; Wang, Z.; Lee, M.-R.; Li, Z.; Li, J.-J.; Sung, C.-K. Simultaneous quantification of 19 ginsenosides in black ginseng developed from Panax ginseng by HPLC–ELSD. J. Pharm. Biomed. Anal. 2009, 50, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Ban, Y.J.; Yang, B.W.; Baik, M.Y.; Hahm, Y.T.; Kim, B.Y. Optimization of the manufacturing process for black ginseng. J. Korean Soc. Appl. Bi. 2010, 53, 71–77. [Google Scholar] [CrossRef]
- Lee, J.W.; Ji, S.-H.; Choi, B.-R.; Choi, D.J.; Lee, Y.-G.; Kim, H.-G.; Kim, G.-S.; Kim, K.; Lee, Y.-H.; Baek, N.-I.; et al. UPLC-QTOF/MS-Based Metabolomics Applied for the Quality Evaluation of Four Processed Panax ginseng Products. Molecules 2018, 23, 2062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, W.C.; Jung, J.; Na, H.S.; Hwang Bo, J.; Kim, H.-G.; Yoon, D.; Choi, B.-R.; Lee, Y.-S.; Kim, G.-S.; Baek, N.-I.; et al. Identification and quantification of major malonyl ginsenosides isolated from Panax ginseng C.A. Meyer. J. Appl. Biol. Chem. 2019, 62, 375–384. [Google Scholar] [CrossRef]
- Xie, Y.; Luo, D.; Cheng, Y.; Ma, J.-F.; Wang, Y.; Liang, Q.; Luo, G. Steaming-induced chemical transformations and holistic quality assessment of red ginseng derived from Panax ginseng by means of HPLC-ESI-MS/MSn-based multicomponent quantification fingerprint. J. Agr. Food Chem. 2012, 60, 8213–8224. [Google Scholar] [CrossRef] [PubMed]
- Du, X.W.; Wills, R.B.H.; Stuart, D.L. Changes in neutral and malonyl ginsenosides in American ginseng (Panax quinquefolium) during drying, storage and ethanolic extraction. Food Chem. 2004, 86, 155–159. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.-J.; Shaykhutdinov, R.; Weljie, A.M.; Vogel, H.J.; Facchini, P.J.; Park, S.-U.; Kim, Y.-K.; Yang, T.-J. Quality assessment of ginseng by 1H NMR metabolite fingerprinting and profiling analysis. J. Agr. Food Chem. 2009, 57, 7513–7522. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Park, H.; He, N.; Lee, H.Y.; Yoon, S. QCanvas: an advanced tool for data clustering and visualization of genomics data. Genomics Inform. 2012, 10, 263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maillard, L.C. Action of amino acids on sugars. Formation of melanoidins in a methodical way. Compt. Rend. 1912, 154, 66–68. [Google Scholar]
- Nam, K.H.; Kim, H.J.; Pack, I.S.; Kim, H.J.; Chung, Y.S.; Kim, S.Y.; Kim, C.G. Global metabolite profiling based on GC–MS and LC–MS/MS analyses in ABF3-overexpressing soybean with enhanced drought tolerance. Appl. Biol. Chem. 2019, 62, 15. [Google Scholar] [CrossRef] [Green Version]
- Yoon, D.; Kim, S.; Lee, M.; Yoon, C.; Kim, S. 1H-NMR-based metabolomic study on toxicity of methomyl and methidathion in fish. J. Environ. Sci. Heal. B 2016, 51, 824–831. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, L.; Johansson, E.; Kettaneh-Wold, N.; Trygg, J.; Wikström, C.; Wold, S. Multi-and Megavariate Data Analysis. Basic Principles and Applications I, 2nd ed.; Umetrics Academy: Umeå, Sweden, 2001; pp. 39–103. [Google Scholar]
- Heussen, P.C.; Janssen, H.G.; Samwel, I.B.; Van Duynhoven, J.P. The use of multivariate modelling of near infra-red spectra to predict the butter fat content of spreads. Anal. Chim. Acta 2007, 595, 176–181. [Google Scholar] [CrossRef] [PubMed]





| Compound | Chemical Shifts (Multiplicities) (ppm) | Wight Ginseng (%) | Tae-geuk Ginseng (%) | Red Ginseng (%) | Black Ginseng (%) |
|---|---|---|---|---|---|
| 4-Aminobutyrate a | 1.90 (m), 2.30 (t), 3.00 (m) | 1.011 ± 0.405 | N.D. | 0.296 ± 0.234 | N.D. |
| Acetate a | 1.94 (s) | N.D. | 3.680 ± 1.080 | 2.565 ± 0.879 | N.D. |
| Alanine b | 1.46 (d), 3.76 (q) | 1.511 ± 1.048 | 0.448 ± 0.190 | 0.753 ± 0.448 | 0.412 ± 0.271 |
| Arginine b | 1.60−1.75 (m), 1.85−1.94 (m), 3.23 (t) | 8.201 ± 2.103 | 3.520 ± 1.673 | 6.132 ± 4.268 | N.D. |
| Asparagine b | 2.84 (dd), 2.94 (dd) | 1.259 ± 1.034 | N.D. | 0.626 ± 0.790 | N.D. |
| Aspartate a | 2.67 (dd), 2.81 (dd) | 0.362 ± 0.254 | N.D. | 0.537 ± 0.276 | N.D. |
| Choline a | 3.18 (s), 3.51 (dd), 4.06 (ddd) | 0.244 ± 0.070 | 0.539 ± 0.132 | 0.400 ± 0.208 | 0.253 ± 0.076 |
| Ethanol a | 1.17 (t), 3.64 (q) | 7.502 ± 4.422 | N.D. | N.D. | N.D. |
| Ethanolamine a | 3.13 (t), 3.81 (t) | 0.161 ± 0.048 | N.D. | N.D. | N.D. |
| Formate c | 8.43 (s) | N.D. | 0.712 ± 0.217 | 1.305 ± 0.360 | N.D. |
| Fructose a | 3.53−3.59 (m), 3.64−3.70 (m), 3.77−3.82 (m), 3.88 (dd), 3.98 (m), 4.01 (dd), 4.09 (m) | 1.407 ± 0.800 | 5.869 ± 2.257 | 4.604 ± 1.351 | 58.747 ± 3.795 |
| Galactose a | 3.48 (dd), 3.64 (dd), 3.68−3.85 (m), 3.92 (d), 3.98 (dd), 4.07 (m), 4.57 (d), 5.25 (d) | 0.510 ± 0.228 | N.D. | N.D. | N.D. |
| Glucose a | 3.25 (m), 3.38−3.50 (m), 3.53 (dd), 3.72−3.91 (m), 4.63 (d), 5.23 (d) | 1.703 ± 1.108 | 4.430 ± 0.801 | 4.658 ± 1.861 | 27.724 ± 6.125 |
| Glutamate a | 2.02−2.16 (m), 2.34−2.37 (m) | 0.524 ± 0.179 | N.D. | N.D. | N.D. |
| Glutamine a | 2.08−2.17 (m), 2.40−2.48 (m), 3.76 (t) | 1.029 ± 0.601 | N.D. | N.D. | N.D. |
| Isoleucine b | 0.92 (t), 0.99 (d), 1.25 (m), 1.46 (m), 1.97 (m), 3.67 (d) | 0.174 ± 0.145 | 0.109 ± 0.079 | 0.118 ± 0.065 | 0.075 ± 0.063 |
| Lactate a | 1.31 (d), 4.10 (q) | N.D. | N.D. | 0.180 ± 0.081 | 0.411 ± 0.296 |
| Leucine a | 0.95 (t), 1.65−1.76 (m), 3.72 (m) | 0.2547 ± 0.1556 | N.D. | N.D. | N.D. |
| Malate a | 2.44 (dd), 2.71 (dd), 4.31 (dd) | 2.0104 ± 0.8500 | N.D. | 3.7763 ± 1.8885 | N.D. |
| Maltose a | 3.26 (dd), 3.41 (m), 3.54−3.97 (m), 4.64 (d), 5.22 (d), 5.40 (d) | N.D. | 49.9092 ± 9.4369 | 30.3911 ± 24.5901 | 1.3598 ± 0.3765 |
| O-Phosphocholine d | 3.21 (s), 4.16 (m) | 0.0165 ± 0.0079 | N.D. | N.D. | N.D. |
| Phenylalanine a | 7.32 (dd), 7.37 (t), 7.42 (m) | 0.1643 ± 0.1712 | N.D. | N.D. | N.D. |
| Proline a | 1.99 (m), 2.06 (m), 2.34 (m), 3.33 (q), 3.41 (q), 4.12 (dd) | 0.3917 ± 0.1456 | N.D. | N.D. | N.D. |
| Pyroglutamate a | 2.02 (m), 2.39 (m), 2.49 (m), 4.16 (dd) | N.D. | 1.8525 ± 1.2858 | 2.9228 ± 1.9959 | 4.2622 ± 1.1718 |
| Pyruvate a | 2.35 (s) | N.D. | 2.1931 ± 0.6792 | 0.6797 ± 0.2923 | 0.3517 ± 0.0567 |
| Succinate a | 2.38 (s) | 0.0762 ± 0.0300 | 1.4194 ± 0.4420 | 0.9192 ± 0.3350 | 0.1590 ± 0.0473 |
| Sucrose e | 3.45 (t), 3.54 (dd), 3.66 (s), 3.74 (t), 3.78−3.89 (m), 4.03 (t), 4.21 (d), 5.40 (d) | 68.5672 ± 6.2442 | 22.1740 ± 6.4451 | 36.3888 ± 19.7104 | 2.3077 ± 1.1327 |
| Threonine a | 1.32 (d), 3.58 (d), 4.25 (m) | 0.2717 ± 0.1349 | N.D. | N.D. | N.D. |
| Tyrosine d | 3.02 (dd), 3.17 (dd), 3.92 (dd), 6.88 (m), 7.18 (m) | 0.2184 ± 0.1642 | N.D. | 0.0869 ± 0.1335 | N.D. |
| Uridine a | 3.79 (dd), 3.90 (dd), 4.11 (m), 4.21 (t), 4.34 (dd), 5.88 (d), 5.90 (d), 7.86 (d) | N.D. | 0.1493 ± 0.0306 | 0.1905 ± 0.0533 | 0.1323 ± 0.0278 |
| Valine b | 0.97 (d), 1.03 (d), 2.26 (m), 3.60 (d) | 0.2409 ± 0.2046 | 0.0805 ± 0.0515 | 0.1361 ± 0.0885 | 0.0822 ± 0.0572 |
| myo-Inositol a | 3.26 (t), 3.51 (dd), 3.60 (t), 4.05 (t) | 2.0610 ± 0.7682 | 2.9154 ± 0.4926 | 2.3364 ± 0.8677 | 3.7231 ± 1.3078 |
| sn-Glycero-3-phosphocholine a | 3.22 (s), 3.64 (m), 3.90 (m), 4.31 (m) | 0.1297 ± 0.0219 | N.D. | N.D. | N.D. |
| OPLS Model | RMSEE | RMSEP | Y Intercept of R2 | Y Intercept of Q2 |
|---|---|---|---|---|
| WG vs. TG | 0.036 | 0.062 | 0.256 | −0.554 |
| WG vs. RG | 0.308 | 0.223 | 0.267 | −0.710 |
| WG vs. BG | 0.080 | 0.192 | 0.317 | −0.584 |
| TG vs. RG | 0.143 | 0.314 | 0.302 | −0.621 |
| TG vs. BG | 0.060 | 0.160 | 0.322 | −0.603 |
| RG vs. BG | 0.041 | 0.061 | 0.302 | −0.543 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoon, D.; Shin, W.C.; Lee, Y.-S.; Kim, S.; Baek, N.-I.; Lee, D.Y. A Comparative Study on Processed Panax ginseng Products Using HR-MAS NMR-Based Metabolomics. Molecules 2020, 25, 1390. https://doi.org/10.3390/molecules25061390
Yoon D, Shin WC, Lee Y-S, Kim S, Baek N-I, Lee DY. A Comparative Study on Processed Panax ginseng Products Using HR-MAS NMR-Based Metabolomics. Molecules. 2020; 25(6):1390. https://doi.org/10.3390/molecules25061390
Chicago/Turabian StyleYoon, Dahye, Woo Cheol Shin, Young-Seob Lee, Suhkmann Kim, Nam-In Baek, and Dae Young Lee. 2020. "A Comparative Study on Processed Panax ginseng Products Using HR-MAS NMR-Based Metabolomics" Molecules 25, no. 6: 1390. https://doi.org/10.3390/molecules25061390
APA StyleYoon, D., Shin, W. C., Lee, Y.-S., Kim, S., Baek, N.-I., & Lee, D. Y. (2020). A Comparative Study on Processed Panax ginseng Products Using HR-MAS NMR-Based Metabolomics. Molecules, 25(6), 1390. https://doi.org/10.3390/molecules25061390