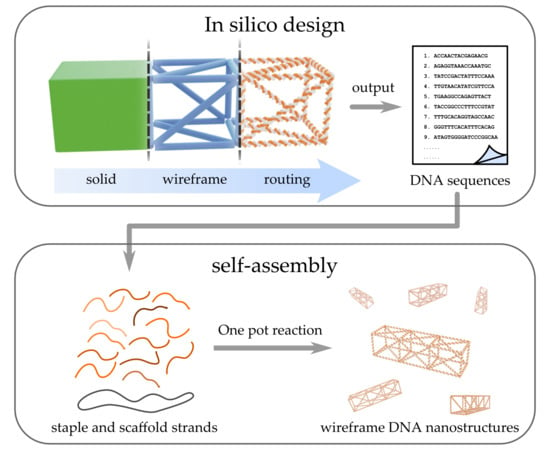
Increasing Complexity in Wireframe DNA Nanostructures
Abstract
Share and Cite
Piskunen, P.; Nummelin, S.; Shen, B.; Kostiainen, M.A.; Linko, V. Increasing Complexity in Wireframe DNA Nanostructures. Molecules 2020, 25, 1823. https://doi.org/10.3390/molecules25081823
Piskunen P, Nummelin S, Shen B, Kostiainen MA, Linko V. Increasing Complexity in Wireframe DNA Nanostructures. Molecules. 2020; 25(8):1823. https://doi.org/10.3390/molecules25081823
Chicago/Turabian StylePiskunen, Petteri, Sami Nummelin, Boxuan Shen, Mauri A. Kostiainen, and Veikko Linko. 2020. "Increasing Complexity in Wireframe DNA Nanostructures" Molecules 25, no. 8: 1823. https://doi.org/10.3390/molecules25081823
APA StylePiskunen, P., Nummelin, S., Shen, B., Kostiainen, M. A., & Linko, V. (2020). Increasing Complexity in Wireframe DNA Nanostructures. Molecules, 25(8), 1823. https://doi.org/10.3390/molecules25081823