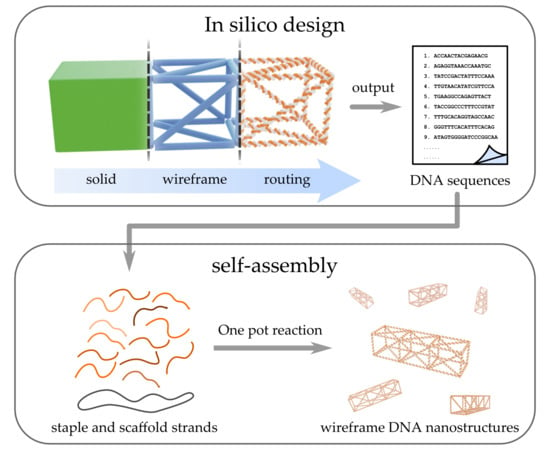
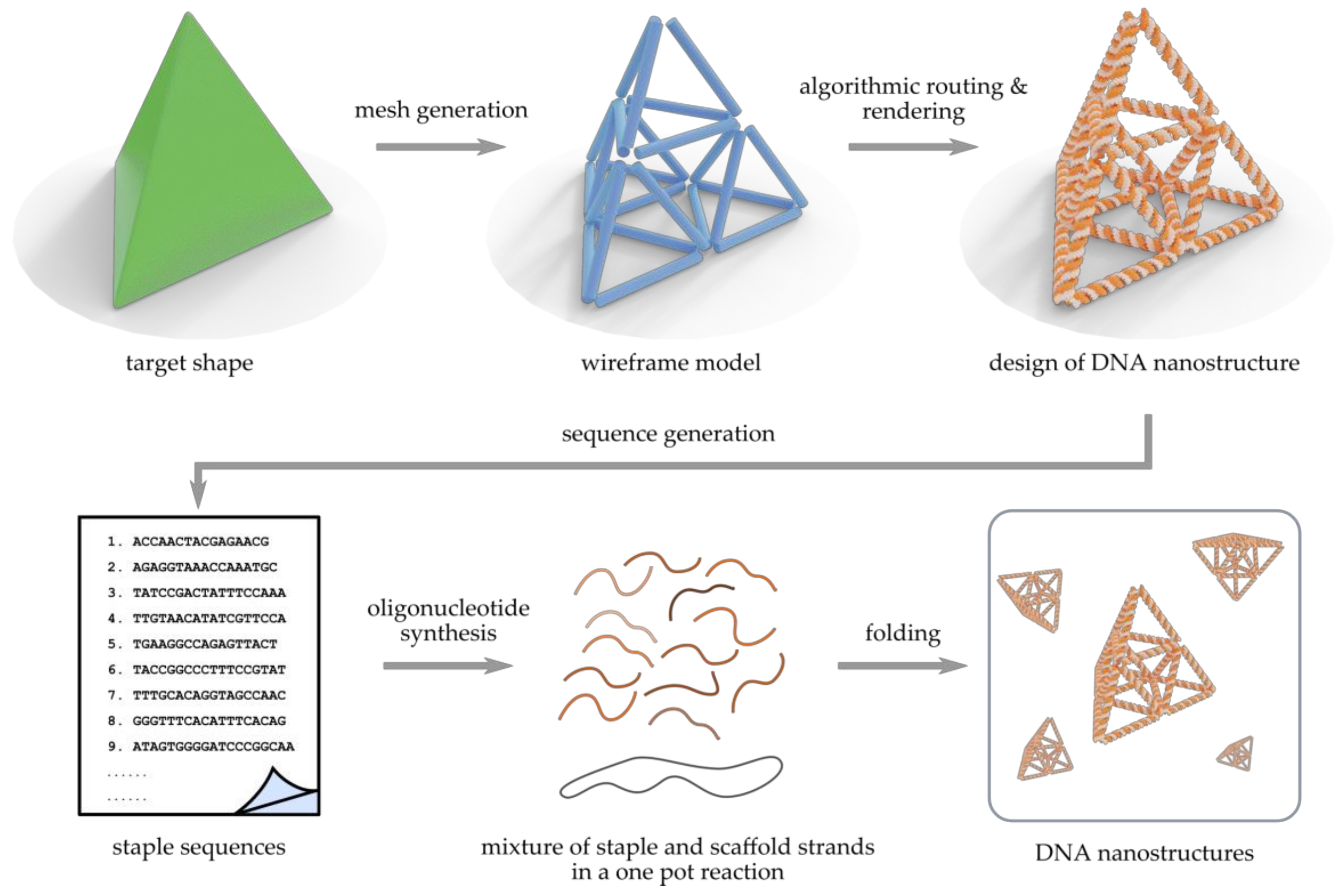
Increasing Complexity in Wireframe DNA Nanostructures
Abstract
:1. Introduction
2. Wireframe Design Principles
3. Shape Space, Design Strategies, and Software
3.1. Gridiron and Simple Meshes
3.2. Semi-Automatic Top-Down Polyhedral DNA Rendering with vHelix
3.3. Automatic Top-Down Fabrication with DAEDALUS, PERDIX, TALOS, METIS, and ATHENA
3.4. Scaffold-Free Approaches
4. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Seeman, N.C. Nucleic acid junctions and lattices. J. Theor. Biol. 1982, 99, 237–247. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H.C. Molecular structure of nucleic acids. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Kallenbach, N.R.; Ma, R.-I.; Seeman, N.C. An immobile nucleic acid junction constructed from oligonucleotides. Nature 1983, 305, 829–831. [Google Scholar] [CrossRef]
- Jones, M.R.; Seeman, N.C.; Mirkin, C.A. Programmable materials and the nature of the DNA bond. Science 2015, 347, 1260901. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C. DNA in a material world. Nature 2003, 421, 427–431. [Google Scholar] [CrossRef]
- Linko, V.; Dietz, H. The enabled state of DNA nanotechnology. Curr. Opin. Biotechnol. 2013, 24, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Dunn, K.E. The business of DNA nanotechnology: Commercialization of origami and other technologies. Molecules 2020, 25, 377. [Google Scholar] [CrossRef] [Green Version]
- Nummelin, S.; Kommeri, J.; Kostiainen, M.A.; Linko, V. Evolution of structural DNA nanotechnology. Adv. Mater. 2018, 30, 1703721. [Google Scholar] [CrossRef] [Green Version]
- Praetorius, F.; Kick, B.; Behler, K.L.; Honemann, M.N.; Weuster-Botz, D.; Dietz, H. Biotechnological mass production of DNA origami. Nature 2017, 552, 84–87. [Google Scholar] [CrossRef]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [Green Version]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Högberg, B.; Graf, F.; Shih, W.M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Andersen, E.S.; Dong, M.; Nielsen, M.M.; Jahn, K.; Subramani, R.; Mamdouh, W.; Golas, M.M.; Sander, B.; Stark, H.; Oliveira, C.L.P.; et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 2009, 459, 73–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, Y.; Sharma, J.; Liu, M.; Jahn, K.; Liu, Y.; Yan, H. Scaffolded DNA origami of a DNA tetrahedron molecular container. Nano Lett. 2009, 9, 2445–2447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, S.M.; Marblestone, A.H.; Teerapittayanon, S.; Vazquez, A.; Church, G.M.; Shih, W.M. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009, 37, 5001–5006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- caDNAno Software. Available online: https://cadnano.org/ (accessed on 25 March 2020).
- de Llano, E.; Miao, H.; Ahmadi, Y.; Wilson, A.J.; Beeby, M.; Viola, I.; Barišić, I. Adenita: Interactive 3D modeling and visualization of DNA nanostructures. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Castro, C.E.; Kilchherr, F.; Kim, D.-N.; Shiao, E.L.; Wauer, T.; Wortmann, P.; Bathe, M.; Dietz, H. A primer to scaffolded DNA origami. Nat. Methods 2011, 8, 221–229. [Google Scholar] [CrossRef]
- Kim, D.-N.; Kilchherr, F.; Dietz, H.; Bathe, M. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 2012, 40, 2862–2868. [Google Scholar] [CrossRef]
- CanDo Software. Available online: https://cando-dna-origami.org/ (accessed on 25 March 2020).
- Maffeo, C.; Yoo, J.; Aksimentiev, A. De novo reconstruction of DNA origami structures through atomistic molecular dynamics simulation. Nucleic Acids Res. 2016, 44, 3013–3019. [Google Scholar] [CrossRef] [Green Version]
- Sharma, R.; Schreck, J.S.; Romano, F.; Louis, A.A.; Doye, J.P.K. Characterizing the motion of jointed DNA nanostructures using a coarse-grained model. ACS Nano 2017, 11, 12426–12435. [Google Scholar] [CrossRef]
- Shi, Z.; Castro, C.E.; Arya, G. Conformational dynamics of mechanically compliant DNA nanostructures from coarse-grained molecular dynamics simulations. ACS Nano 2017, 11, 4617–4630. [Google Scholar] [CrossRef]
- Bathe, M.; Rothemund, P.W.K. DNA nanotechnology: A foundation for programmable nanoscale materials. MRS Bull. 2017, 42, 882–888. [Google Scholar] [CrossRef] [Green Version]
- Hong, F.; Zhang, F.; Liu, Y.; Yan, H. DNA origami: Scaffolds for creating higher order structures. Chem. Rev. 2017, 117, 12584–12640. [Google Scholar] [CrossRef] [PubMed]
- Linko, V. At the dawn of applied DNA nanotechnology. Molecules 2020, 25, 639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzyk, A.; Schreiber, R.; Fan, Z.; Pardatscher, G.; Roller, E.-M.; Högele, A.; Simmel, F.C.; Govorov, A.O.; Liedl, T. DNA-Based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 2012, 483, 311–314. [Google Scholar] [CrossRef] [Green Version]
- Gopinath, A.; Miyazono, F.; Faraon, A.; Rothemund, P.W.K. Engineering and mapping nanocavity emission via precision placement of DNA origami. Nature 2016, 535, 401–405. [Google Scholar] [CrossRef]
- Schnitzbauer, J.; Strauss, M.T.; Schlichthaerle, T.; Schueder, F.; Jungmann, R. Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 2017, 12, 1198–1228. [Google Scholar] [CrossRef]
- Graugnard, E.; Hughes, W.L.; Jungmann, R.; Kostiainen, M.A.; Linko, V. Nanometrology and super-resolution imaging with DNA. MRS Bull. 2017, 42, 951–959. [Google Scholar] [CrossRef] [Green Version]
- Shen, B.; Linko, V.; Tapio, K.; Pikker, S.; Lemma, T.; Gopinath, A.; Gothelf, K.V.; Kostiainen, M.A.; Toppari, J.J. Plasmonic nanostructures through DNA-assisted lithography. Sci. Adv. 2018, 4, eaap8978. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Zhang, G.; Liu, H. DNA-templated nanofabrication. Curr. Opin. Colloid Interface Sci. 2018, 38, 88–99. [Google Scholar] [CrossRef]
- Shen, B.; Kostiainen, M.A.; Linko, V. DNA origami nanophotonics and plasmonics at interfaces. Langmuir 2018, 34, 14911–14920. [Google Scholar] [CrossRef] [Green Version]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Thubagere, A.J.; Li, W.; Johnson, R.F.; Chen, Z.; Doroudi, S.; Lee, Y.L.; Izatt, G.; Wittman, S.; Srinivas, N.; Woods, D.; et al. A cargo-sorting DNA robot. Science 2017, 357, eaan6558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopperger, E.; List, J.; Madhira, S.; Rothfischer, F.; Lamb, D.C.; Simmel, F.C. A self-assembled nanoscale robotic arm controlled by electric fields. Science 2018, 359, 296–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ijäs, H.; Hakaste, I.; Shen, B.; Kostiainen, M.A.; Linko, V. Reconfigurable DNA origami nanocapsule for pH-controlled encapsulation and display of cargo. ACS Nano 2019, 13, 5959–5967. [Google Scholar] [CrossRef] [Green Version]
- Surana, S.; Shenoy, A.R.; Krishnan, Y. Designing DNA nanodevices for compatibility with the immune system of higher organisms. Nat. Nanotechnol. 2015, 10, 741–747. [Google Scholar] [CrossRef] [Green Version]
- Keller, A.; Linko, V. Challenges and perspectives of DNA nanostructures in biomedicine. Angew. Chem. Int. Ed. 2020, 59. [Google Scholar] [CrossRef] [Green Version]
- Dietz, H.; Douglas, S.M.; Shih, W.M. Folding DNA into twisted and curved nanoscale shapes. Science 2009, 325, 725–730. [Google Scholar] [CrossRef] [Green Version]
- Han, D.; Pal, S.; Nangreave, J.; Deng, Z.; Liu, Y.; Yan, H. DNA origami with complex curvatures in three-dimensional space. Science 2011, 332, 342–346. [Google Scholar] [CrossRef] [Green Version]
- Šulc, P.; Romano, F.; Ouldridge, T.E.; Rovigatti, L.; Doye, J.P.; Louis, A.A. Sequence-Dependent thermodynamics of a coarse-grained DNA model. J. Chem. Phys. 2012, 137, 135101. [Google Scholar] [CrossRef]
- DNA Origami Structure Prediction Tool (Coarse-Grained Models). Available online: http://bionano.physics.illinois.edu/origami-structure-prediction (accessed on 25 March 2020).
- Wagenbauer, K.F.; Sigl, C.; Dietz, H. Gigadalton-scale shape-programmable DNA assemblies. Nature 2017, 552, 78–83. [Google Scholar] [CrossRef]
- Tikhomirov, G.; Petersen, P.; Qian, L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Nature 2017, 552, 67–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ijäs, H.; Nummelin, S.; Shen, B.; Kostiainen, M.A.; Linko, V. Dynamic DNA origami devices: From strand-displacement reactions to external-stimuli responsive systems. Int. J. Mol. Sci. 2018, 19, 2114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLuca, M.; Shi, Z.; Castro, C.E.; Arya, G. Dynamic DNA nanotechnology: Toward functional nanoscale devices. Nanoscale Horiz. 2020, 5, 182–201. [Google Scholar] [CrossRef]
- Kielar, C.; Xin, Y.; Shen, B.; Kostiainen, M.A.; Grundmeier, G.; Linko, V.; Keller, A. On the stability of DNA origami nanostructures in low-magnesium buffers. Angew. Chem. Int. Ed. 2018, 57, 9470–9474. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, S.; Ijäs, H.; Linko, V.; Keller, A. Structural stability of DNA origami nanostructures under application-specific conditions. Comput. Struct. Biotechnol. J. 2018, 16, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Bila, H.; Kurinsikal, E.E.; Bastings, M.M.C. Engineering a stable future for DNA-origami as a biomaterial. Biomater. Sci. 2019, 7, 532–541. [Google Scholar] [CrossRef]
- Linko, V.; Kostiainen, M.A. Automated design of DNA origami. Nat. Biotechnol. 2016, 34, 826–827. [Google Scholar] [CrossRef] [Green Version]
- Shih, W.M.; Quispe, J.D.; Joyce, G.F. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature 2004, 427, 618–621. [Google Scholar] [CrossRef]
- Chen, J.; Seeman, N.C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 1991, 350, 631–633. [Google Scholar] [CrossRef]
- Goodman, R.P.; Schaap, I.A.T.; Tardin, C.F.; Erben, C.M.; Berry, R.M.; Schmidt, C.F.; Turberfield, A.J. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science 2005, 310, 1661–1665. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Ye, T.; Su, M.; Zhang, C.; Ribbe, A.E.; Jiang, W.; Mao, C. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature 2008, 452, 198–201. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Park, S.H.; Finkelstein, G.; Reif, J.H.; LaBean, T.H. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science 2003, 301, 1882–1884. [Google Scholar] [CrossRef] [PubMed]
- Iinuma, R.; Ke, Y.; Jungmann, R.; Schlichthaerle, T.; Woehrstein, J.B.; Yin, P. Polyhedra self-assembled from DNA tripods and characterized with 3D DNA-PAINT. Science 2014, 344, 65–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simmel, S.S.; Nickels, P.C.; Liedl, T. Wireframe and tensegrity DNA nanostructures. Acc. Chem. Res. 2014, 47, 1691–1697. [Google Scholar] [CrossRef]
- Orponen, P. Design methods for 3D wireframe DNA nanostructures. Nat. Comput. 2018, 17, 147–160. [Google Scholar] [CrossRef] [Green Version]
- Ellis-Monaghan, J.A.; McDowell, A.; Moffatt, I.; Pangborn, G. DNA origami and the complexity of Eulerian circuits with turning costs. Nat. Comput. 2014, 14, 491–503. [Google Scholar] [CrossRef] [Green Version]
- Han, D.; Pal, S.; Yang, Y.; Jiang, S.; Nangreave, J.; Liu, Y.; Yan, H. DNA gridiron nanostructures based on four-arm junctions. Science 2013, 339, 1412–1415. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Jiang, S.; Wu, S.; Li, Y.; Mao, C.; Liu, Y.; Yan, H. Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nat. Nanotechnol. 2015, 10, 779–784. [Google Scholar] [CrossRef]
- Andersen, E.S. DNA origami rewired. Nat. Nanotechnol. 2015, 10, 733–734. [Google Scholar] [CrossRef]
- Hong, F.; Jiang, S.; Wang, T.; Liu, Y.; Yan, H. 3D framework DNA origami with layered crossovers. Angew. Chem. Int. Ed. 2016, 55, 12832–12835. [Google Scholar] [CrossRef]
- Williams, S.; Lund, K.; Lin, C.; Wonka, P.; Lindsay, S.; Yan, H. Tiamat: A three-dimensional editing tool for complex DNA structures. In DNA Computing, DNA 2008: Lecture Notes in Computer Science; Goel, A., Simmel, F.C., Sosík, P., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; Volume 5347, pp. 90–101. [Google Scholar]
- Tiamat Software. Available online: http://yanlab.asu.edu/Resources.html (accessed on 25 March 2020).
- Pan, K.; Kim, D.-N.; Zhang, F.; Adendorff, M.R.; Yan, H.; Bathe, M. Lattice-free prediction of three-dimensional structure of programmed DNA assemblies. Nat. Commun. 2014, 5, 5578. [Google Scholar] [CrossRef] [PubMed]
- Matthies, M.; Agarwal, N.P.; Schmidt, T.L. Design and synthesis of triangulated DNA origami trusses. Nano Lett. 2016, 16, 2108–2113. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, N.P.; Matthies, M.; Joffroy, B.; Schmidt, T.L. Structural transformation of wireframe DNA origami via DNA polymerase assisted gap-filling. ACS Nano 2018, 12, 2546–2553. [Google Scholar] [CrossRef] [PubMed]
- Benson, E.; Mohammed, A.; Gardell, J.; Masich, S.; Czeizler, E.; Orponen, P.; Högberg, B. DNA rendering of polyhedral meshes at the nanoscale. Nature 2015, 523, 441–444. [Google Scholar] [CrossRef] [Green Version]
- vHelix Software. Available online: http://vhelix.net/ (accessed on 25 March 2020).
- Liedl, T. Pathfinder for DNA constructs. Nature 2015, 523, 412–413. [Google Scholar] [CrossRef]
- Benson, E.; Mohammed, A.; Rayneau-Kirkhope, D.; Gådin, A.; Orponen, P.; Högberg, B. Effects of design choices on the stiffness of wireframe DNA origami structures. ACS Nano 2018, 12, 9291–9299. [Google Scholar] [CrossRef] [Green Version]
- Benson, E.; Mohammed, A.; Bosco, A.; Teixeira, A.I.; Orponen, P.; Högberg, B. Computer-aided production of scaffolded DNA nanostructures from flat sheet meshes. Angew. Chem. Int. Ed. 2016, 56, 8869–8872. [Google Scholar] [CrossRef]
- Veneziano, R.; Ratanalert, S.; Zhang, K.; Zhang, F.; Yan, H.; Chiu, W.; Bathe, M. Designer nanoscale DNA assemblies programmed from the top down. Science 2016, 352, 1534. [Google Scholar] [CrossRef] [Green Version]
- DAEDALUS Software. Available online: https://daedalus-dna-origami.org/ (accessed on 25 March 2020).
- Jun, H.; Zhang, F.; Shepherd, T.; Ratanalert, S.; Qi, X.; Yan, H.; Bathe, M. Autonomously designed free-form 2D DNA origami. Sci. Adv. 2019, 5, eaav0655. [Google Scholar] [CrossRef] [Green Version]
- PERDIX Software. Available online: http://perdix-dna-origami.org/ (accessed on 25 March 2020).
- Jun, H.; Shepherd, T.R.; Zhang, K.; Bricker, W.P.; Li, S.; Chiu, W.; Bathe, M. Automated sequence design of 3D polyhedral wireframe DNA origami with honeycomb edges. ACS Nano 2019, 13, 2083–2093. [Google Scholar] [CrossRef]
- TALOS Software. Available online: http://talos-dna-origami.org/ (accessed on 25 March 2020).
- Hahn, J.; Wickham, S.F.; Shih, W.M.; Perrault, S.D. Addressing the instability of DNA nanostructures in tissue culture. ACS Nano 2014, 8, 8765–8775. [Google Scholar] [CrossRef] [PubMed]
- Ponnuswamy, N.; Bastings, M.M.C.; Nathwani, B.; Ryu, J.H.; Chou, L.Y.T.; Vinther, M.; Li, W.A.; Anastassacos, F.M.; Mooney, D.J.; Shih, W.M. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun. 2017, 8, 15654. [Google Scholar] [CrossRef] [PubMed]
- Jun, H.; Wang, X.; Bricker, W.P.; Bathe, M. Automated sequence design of 2D wireframe DNA origami with honeycomb edges. Nat. Commun. 2019, 10, 5419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- METIS Software. Available online: https://metis-dna-origami.org/ (accessed on 25 March 2020).
- Jun, H.; Wang, X.; Bricker, W.P.; Jackson, S.; Bathe, M. Rapid prototyping of wireframe scaffolded DNA origami using ATHENA. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- ATHENA Software. Available online: https://github.com/lcbb/athena (accessed on 25 March 2020).
- Wei, B.; Dai, M.; Yin, P. Complex shapes self-assembled from single-stranded DNA tiles. Nature 2012, 485, 623–626. [Google Scholar] [CrossRef] [Green Version]
- Ke, Y.; Ong, L.L.; Shih, W.M.; Yin, P. Three-dimensional structures self-assembled from DNA bricks. Science 2012, 338, 1177–1183. [Google Scholar] [CrossRef] [Green Version]
- Gothelf, K.V. LEGO-like DNA Structures. Science 2012, 338, 1159–1160. [Google Scholar] [CrossRef]
- Slone, S.M.L.; Maffeo, C.; Sobh, A.R.N.; Aksimentiev, A. LegoGen Software. 2016. Available online: https://nanohub.org/resources/legogen (accessed on 25 March 2020).
- Ong, L.L.; Hanikel, N.; Yaghi, O.K.; Grun, C.; Strauss, M.T.; Bron, P.; Lai-Kee-Him, J.; Schueder, F.; Wang, B.; Wang, P.; et al. Programmable self-assembly of three-dimensional nanostructures from 10,000 unique components. Nature 2017, 552, 72–77. [Google Scholar] [CrossRef]
- NanoBricks Software. Available online: http://molecular.systems/software (accessed on 25 March 2020).
- Wang, W.; Chen, S.; An, B.; Huang, K.; Bai, T.; Xu, M.; Bellot, G.; Ke, Y.; Xiang, Y.; Wei, B. Complex wireframe DNA nanostructures from simple building blocks. Nat. Commun. 2019, 10, 1067. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.; Yang, D.; Tan, Z.; Chen, S.; Xiang, Y.; Mao, C.; Wei, B. Self-assembly of wireframe DNA nanostructures from junction motifs. Angew. Chem. Int. Ed. 2019, 58, 12123–12127. [Google Scholar] [CrossRef]
- Veneziano, R.; Moyer, T.J.; Stone, M.B.; Shepherd, T.R.; Schief, W.R.; Irvine, D.J.; Bathe, M. Role of nanoscale antigen organization on B-cell activation probed using DNA origami. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Hartl, C.; Fischer, S.; Frank, K.; Nickels, P.; Heuer-Jungemann, A.; Nickel, B.; Liedl, T. 3D DNA origami crystals. Adv. Mater. 2018, 30, 1800273. [Google Scholar] [CrossRef] [PubMed]
- Julin, S.; Nummelin, S.; Kostiainen, M.A.; Linko, V. DNA nanostructure-directed assembly of metal nanoparticle superlattices. J. Nanopart. Res. 2018, 20, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heuer-Jungemann, A.; Liedl, T. From DNA tiles to functional DNA materials. Trends Chem. 2019, 1, 799–814. [Google Scholar] [CrossRef]
- Tian, Y.; Lhermitte, J.R.; Bai, L.; Vo, T.; Xin, H.L.; Li, H.; Li, R.; Fukuto, M.; Yager, K.G.; Kahn, J.S.; et al. Ordered three-dimensional nanomaterials using DNA-prescribed and valence-controlled material voxels. Nat. Mater. 2020, 19. [Google Scholar] [CrossRef] [PubMed]
- Bae, W.; Kocabey, S.; Liedl, T. DNA nanostructures in vitro, in vivo and on membranes. Nano Today 2019, 26, 98–107. [Google Scholar] [CrossRef]
- Li, S.; Jiang, Q.; Liu, S.; Zhang, Y.; Tian, Y.; Song, C.; Wang, J.; Zou, Y.; Anderson, G.J.; Han, J.Y.; et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 2018, 36, 258–264. [Google Scholar] [CrossRef]
- Jiang, D.; Ge, Z.; Im, H.-J.; England, C.G.; Ni, D.; Hou, J.; Zhang, L.; Kutyreff, C.J.; Yan, Y.; Liu, Y.; et al. DNA origami nanostructures can exhibit preferential renal uptake and alleviate acute kidney injury. Nat. Biomed. Eng. 2018, 2, 865–877. [Google Scholar] [CrossRef]
- Kollmann, F.; Ramakrishnan, S.; Shen, B.; Grundmeier, G.; Kostiainen, M.A.; Linko, V.; Keller, A. Superstructure-dependent loading of DNA origami nanostructures with a groove-binding drug. ACS Omega 2018, 3, 9441–9448. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Shen, B.; Kostiainen, M.A.; Grundmeier, G.; Keller, A.; Linko, V. Real-Time observation of superstructure-dependent DNA origami digestion by DNase I using high-speed atomic force microscopy. ChemBioChem 2019, 20, 2818–2823. [Google Scholar] [CrossRef]
- Suma, A.; Stopar, A.; Nicholson, A.W.; Castronovo, M.; Carnevale, V. Global and local mechanical properties control endonuclease reactivity of a DNA origami nanostructure. Nucleic Acids Res. 2020, gkaa080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiviaho, J.K.; Linko, V.; Ora, A.; Tiainen, T.; Järvihaavisto, E.; Mikkilä, J.; Tenhu, H.; Nonappa; Kostiainen, M.A. Cationic polymers for DNA origami coating—Examining their binding efficiency and tuning the enzymatic reaction rates. Nanoscale 2016, 8, 11674–11680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, N.P.; Matthies, M.; Gür, F.N.; Osada, K.; Schmidt, T.L. Block copolymer micellization as a protection strategy for DNA origami. Angew. Chem. Int. Ed. 2017, 56, 5460–5464. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, Y.; de Llano, E.; Barišić, I. (Poly)cation-induced protection of conventional and wireframe DNA origami nanostructures. Nanoscale 2018, 10, 7494–7504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrault, S.D.; Shih, W.M. Virus-Inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano 2014, 8, 5132–5140. [Google Scholar] [CrossRef] [PubMed]
- Mikkilä, J.; Eskelinen, A.-P.; Niemelä, E.H.; Linko, V.; Frilander, M.J.; Törmä, P.; Kostiainen, M.A. Virus-Encapsulated DNA origami nanostructures for cellular delivery. Nano Lett. 2014, 14, 2196–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auvinen, H.; Zhang, H.; Nonappa; Kopilow, A.; Niemelä, E.H.; Nummelin, S.; Correia, A.; Santos, H.A.; Linko, V.; Kostiainen, M.A. Protein coating of DNA nanostructures for enhanced stability and immunocompatibility. Adv. Healthc. Mater. 2017, 6, 1700692. [Google Scholar] [CrossRef] [Green Version]
- Lacroix, A.; Edwardson, T.G.W.; Hancock, M.A.; Dore, M.D.; Sleiman, H.F. Development of DNA nanostructures for high-affinity binding to human serum albumin. J. Am. Chem. Soc. 2017, 139, 7355–7362. [Google Scholar] [CrossRef]
- Wang, S.-T.; Gray, M.A.; Xuan, S.; Lin, Y.; Byrnes, J.; Nguyen, A.I.; Todorova, N.; Stevens, M.M.; Bertozzi, C.R.; Zuckermann, R.N.; et al. DNA origami protection and molecular interfacing through engineered sequence-defined peptoids. Proc. Natl. Acad. Sci. USA 2020, 117, 6339–6348. [Google Scholar] [CrossRef] [Green Version]
- Gerling, T.; Kube, M.; Kick, B.; Dietz, H. Sequence-Programmable covalent bonding of designed DNA assemblies. Sci. Adv. 2018, 4, eaau1157. [Google Scholar] [CrossRef] [Green Version]
- Anastassacos, F.M.; Zhao, Z.; Zeng, Y.; Shih, W.M. Glutaraldehyde cross-linking of oligolysines coating DNA origami greatly reduces susceptibility to nuclease degradation. J. Am. Chem. Soc. 2020, 142, 3311–3315. [Google Scholar] [CrossRef] [PubMed]
- Linko, V.; Shen, B.; Tapio, K.; Toppari, J.J.; Kostiainen, M.A.; Tuukkanen, S. One-step large-scale deposition of salt-free DNA origami nanostructures. Sci. Rep. 2015, 5, 15634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kielar, C.; Ramakrishnan, S.; Fricke, S.; Grundmeier, G.; Keller, A. Dynamics of DNA origami lattice formation at solid-liquid interfaces. ACS Appl. Mater. Interfaces 2018, 10, 44844–44853. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sun, W.; Yang, C.; Zhu, Z. Scaling up DNA self-assembly. ACS Appl. Bio Mater. 2020, 3. [Google Scholar] [CrossRef]
- Engelhardt, F.A.S.; Praetorius, F.; Wachauf, C.H.; Brüggenthies, G.; Kohler, F.; Kick, B.; Kadletz, K.L.; Nhi Pham, P.; Behler, K.L.; Gerling, T.; et al. Custom-size, functional, and durable DNA origami with design-specific scaffolds. ACS Nano 2019, 13, 5015–5027. [Google Scholar] [CrossRef]
- Tilibit Nanosystems. Available online: https://www.tilibit.com/ (accessed on 25 March 2020).




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piskunen, P.; Nummelin, S.; Shen, B.; Kostiainen, M.A.; Linko, V. Increasing Complexity in Wireframe DNA Nanostructures. Molecules 2020, 25, 1823. https://doi.org/10.3390/molecules25081823
Piskunen P, Nummelin S, Shen B, Kostiainen MA, Linko V. Increasing Complexity in Wireframe DNA Nanostructures. Molecules. 2020; 25(8):1823. https://doi.org/10.3390/molecules25081823
Chicago/Turabian StylePiskunen, Petteri, Sami Nummelin, Boxuan Shen, Mauri A. Kostiainen, and Veikko Linko. 2020. "Increasing Complexity in Wireframe DNA Nanostructures" Molecules 25, no. 8: 1823. https://doi.org/10.3390/molecules25081823
APA StylePiskunen, P., Nummelin, S., Shen, B., Kostiainen, M. A., & Linko, V. (2020). Increasing Complexity in Wireframe DNA Nanostructures. Molecules, 25(8), 1823. https://doi.org/10.3390/molecules25081823