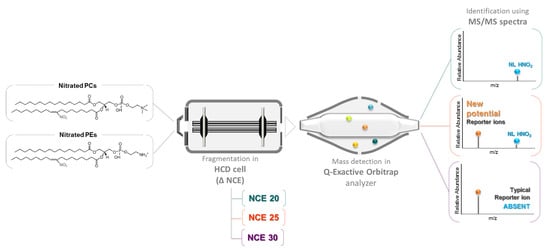
Advancing Target Identification of Nitrated Phospholipids in Biological Systems by HCD Specific Fragmentation Fingerprinting in Orbitrap Platforms
Abstract
:1. Introduction
2. Results
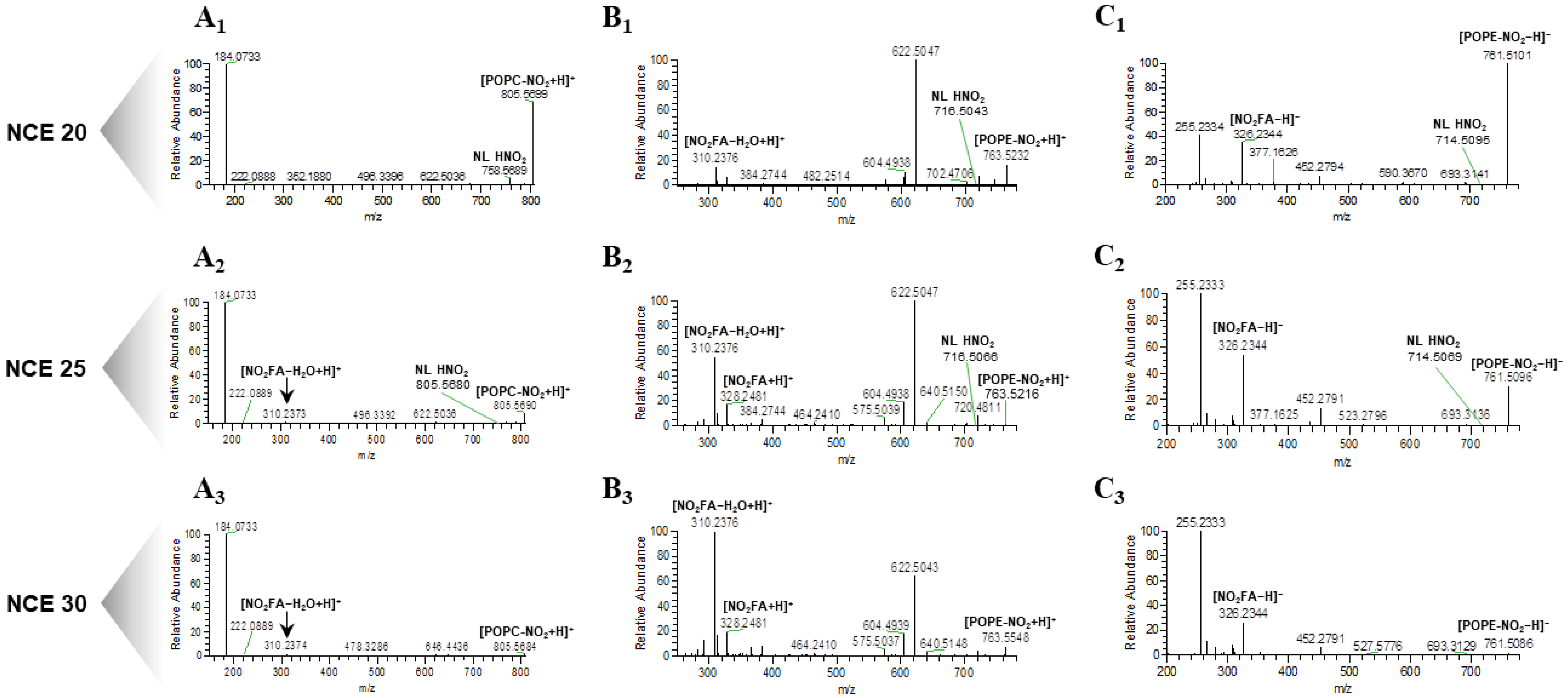
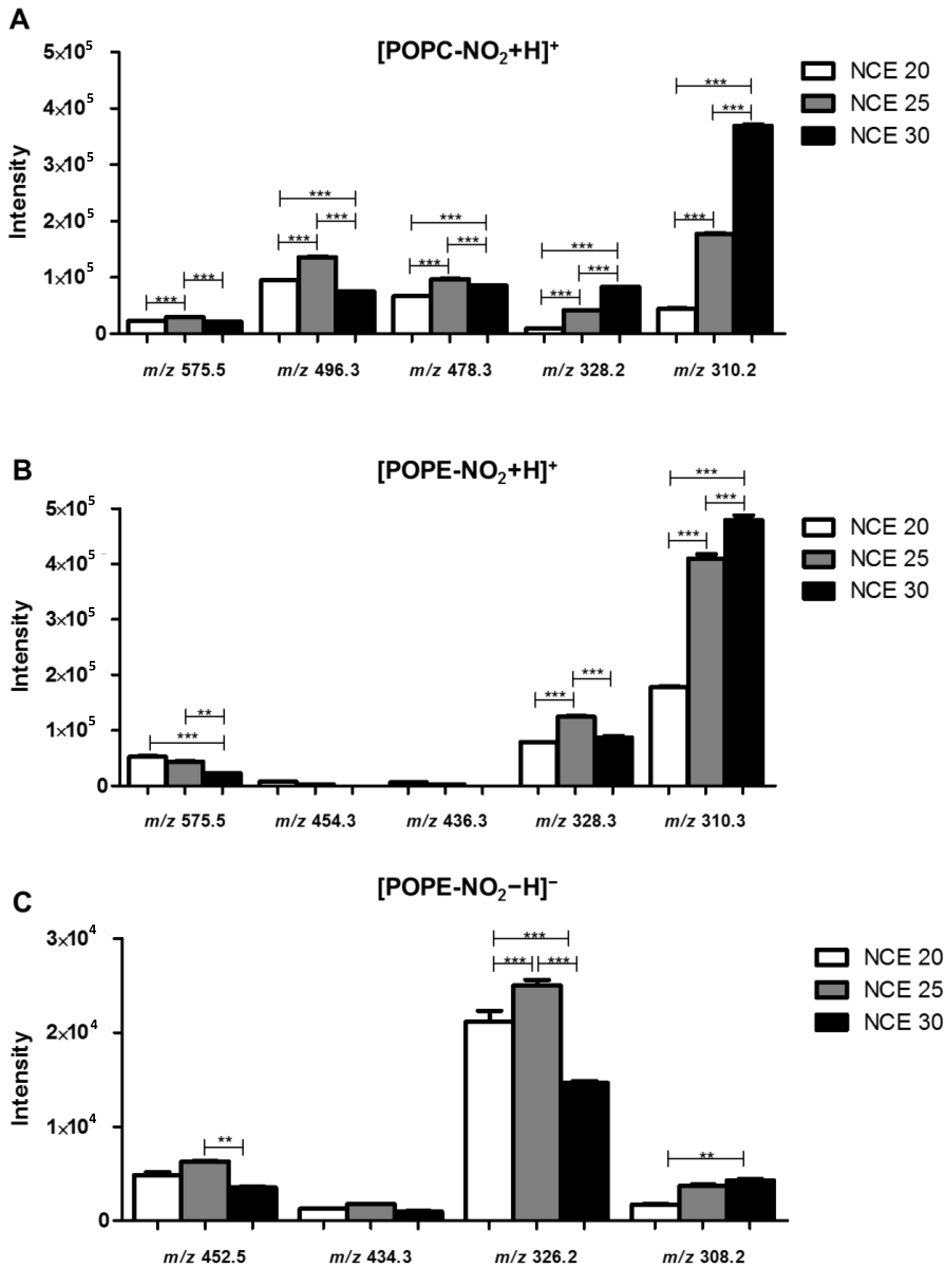
2.1. Optimization of the Normalized Collision Energy for the Study of Nitrated PL Standards
2.2. Identification of Nitrated PL in Cell Lipid Extracts
3. Discussion
4. Materials and Methods
4.1. Reagents/Chemicals
4.2. Nitration of Phospholipids Using In Vitro Model Systems
4.3. Phospholipid Quantification by Phosphorous Measurement Assay
4.4. HCD-Orbitrap Instrumental Conditions
4.5. Cell Culture and Treatments of Adrenal Carcinoma SW13/cl.2 Cells
4.6. Lipid Extraction from SW13/cl.2 Cells
4.7. HPLC-ESI-MS and MS/MS Analysis
4.8. Data and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Spickett, C.M.; Pitt, A.R. Oxidative lipidomics coming of age: Advances in analysis of oxidized phospholipids in physiology and pathology. Antioxid. Redox Signal. 2015, 22, 1646–1666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maciel, E.; Domingues, P.; Domingues, M.R.M. Liquid chromatography/tandem mass spectrometry analysis of long-chain oxidation products of cardiolipin induced by the hydroxyl radical. Rapid Commun. Mass Spectrom. 2011, 25, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Tyurina, Y.Y.; Domingues, R.M.; Tyurin, V.A.; Maciel, E.; Domingues, P.; Amoscato, A.A.; Bayir, H.; Kagan, V.E. Characterization of cardiolipins and their oxidation products by LC-MS analysis. Chem. Phys. Lipids 2014, 179, 3–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freeman, B.A.; Baker, P.R.S.; Schopfer, F.J.; Woodcock, S.R.; Napolitano, A.; d’Ischia, M. Nitro-fatty Acid Formation and Signaling. J. Biol. Chem. 2008, 283, 15515–15519. [Google Scholar] [CrossRef] [Green Version]
- Rubbo, H.; Trostchansky, A.; O’Donnell, V.B. Peroxynitrite-mediated lipid oxidation and nitration: Mechanisms and consequences. Arch. Biochem. Biophys. 2009, 484, 167–172. [Google Scholar] [CrossRef]
- Milic, I.; Griesser, E.; Vemula, V.; Ieda, N.; Nakagawa, H.; Miyata, N.; Galano, J.-M.; Oger, C.; Durand, T.; Fedorova, M. Profiling and relative quantification of multiply nitrated and oxidized fatty acids. Anal. Bioanal. Chem. 2015, 407, 5587–5602. [Google Scholar] [CrossRef]
- Melo, T.; Montero-Bullón, J.-F.; Domingues, P.; Domingues, M.R.M. Discovery of bioactive nitrated lipids and nitro-lipid-protein adducts using mass spectrometry-based approaches. Redox Biol. 2019, 23, 1–16. [Google Scholar] [CrossRef]
- Buchan, G.J.; Bonacci, G.; Fazzari, M.; Salvatore, S.R.; Gelhaus Wendell, S. Nitro-fatty acid formation and metabolism. Nitric Oxide 2018, 79, 38–44. [Google Scholar] [CrossRef] [Green Version]
- Schopfer, F.J.; Khoo, N.K.H. Nitro-fatty acid logistics: Formation, biodistribution, signaling, and pharmacology. Trends Endocrinol. Metab. 2019, 30, 505–519. [Google Scholar] [CrossRef]
- Melo, T.; Domingues, P.; Ferreira, R.; Milic, I.; Fedorova, M.; Santos, S.M.; Segundo, M.A.; Domingues, M.R.M. Recent advances on mass spectrometry analysis of nitrated phospholipids. Anal. Chem. 2016, 88, 2622–2629. [Google Scholar] [CrossRef]
- Neves, B.; Domingues, P.; Oliveira, M.M.; Domingues, M.R.M.; Melo, T. Profile of phosphatidylserine modifications under nitroxidative stress conditions using a liquid chromatography-mass spectrometry based Approach. Molecules 2018, 24, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melo, T.; Domingues, P.; Ribeiro-Rodrigues, T.M.; Girão, H.; Segundo, M.A.; Domingues, M.R.M. Characterization of phospholipid nitroxidation by LC-MS in biomimetic models and in H9c2 Myoblast using a lipidomic approach. Free Radic. Biol. Med. 2017, 106, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Montero-Bullon, J.-F.; Melo, T.; Domingues, M.R.M.; Domingues, P. Liquid chromatography/tandem mass spectrometry characterization of nitroso, nitrated and nitroxidized cardiolipin products. Free Radic. Biol. Med. 2019, 144, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Colombo, S.; Melo, T.; Martínez-López, M.; Carrasco, M.J.; Domingues, M.R.M.; Pérez-Sala, D.; Domingues, P. Phospholipidome of endothelial cells shows a different adaptation response upon oxidative, glycative and lipoxidative stress. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Anjos, S.; Feiteira, E.; Cerveira, F.; Melo, T.; Reboredo, A.; Colombo, S.; Dantas, R.; Costa, E.; Moreira, A.; Santos, S.; et al. Lipidomics reveals similar changes in serum phospholipid signatures of overweight and obese pediatric subjects. J. Proteome Res. 2019, 18, 3174–3183. [Google Scholar] [CrossRef]
- Wang, M.; Wang, C.; Han, R.H.; Han, X. Novel advances in shotgun lipidomics for biology and medicine. Prog. Lipid Res. 2016, 61, 83–108. [Google Scholar] [CrossRef] [Green Version]
- Almeida, R.; Pauling, J.K.; Sokol, E.; Hannibal-Bach, H.K.; Ejsing, C.S. Comprehensive lipidome analysis by shotgun lipidomics on a hybrid quadrupole-orbitrap-linear ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 2015, 26, 133–148. [Google Scholar] [CrossRef]
- Bayat, P.; Lesage, D.; Cole, R.B. Low-energy collision-induced dissociation (low-energy CID), collision-induced dissociation (CID), and higher energy collision dissociation (HCD) mass spectrometry for structural elucidation of saccharides and clarification of their dissolution mechanism in DMAc/LiCl. J. Mass Spectrom. 2018, 53, 705–716. [Google Scholar] [CrossRef]
- Zuo, M.-Q.; Sun, R.-X.; Fang, R.-Q.; He, S.-M.; Dong, M.-Q. Characterization of collision-induced dissociation of deprotonated peptides of 4–16 amino acids using high-resolution mass spectrometry. Int. J. Mass Spectrom. 2019, 445, 116186. [Google Scholar] [CrossRef]
- Chiva, C.; Sabidó, E. HCD-only fragmentation method balances peptide identification and quantitation of TMT-labeled samples in hybrid linear ion trap/orbitrap mass spectrometers. J. Proteom. 2014, 96, 263–270. [Google Scholar] [CrossRef]
- Yang, K.; Zhao, Z.; Gross, R.W.; Han, X. Identification and quantitation of unsaturated fatty acid isomers by electrospray ionization tandem mass spectrometry: A shotgun lipidomics approach. Anal. Chem. 2011, 83, 4243–4250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, X.; Gross, R.W. Shotgun lipidomics: Multidimensional MS analysis of cellular lipidomes. Expert Rev. Proteom. 2005, 2, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Comstock, K.; Huang, Y. Multiple Fragmentation Methods for Small Molecule Characterization on a Dual Pressure Linear Ion Trap Orbitrap Hybrid Mass Spectrometer; Thermo Scientific: San Jose, CA, USA, 2011; pp. 1–8. [Google Scholar]
- Hinneburg, H.; Stavenhagen, K.; Schweiger-Hufnagel, U.; Pengelley, S.; Jabs, W.; Seeberger, P.H.; Silva, D.V.; Wuhrer, M.; Kolarich, D. The art of destruction: Optimizing collision energies in Quadrupole-Time of Flight (Q-TOF) Instruments for glycopeptide-based glycoproteomics. J. Am. Soc. Mass Spectrom. 2016, 27, 507–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, X.; Gross, R.W. Shotgun lipidomics: Electrospray Ionization Mass Spectrometric Analysis and Quantitation of Cellular Lipidomes directly from Crude Extracts of Biological samples. Mass Spectrom. Rev. 2005, 24, 367–412. [Google Scholar] [CrossRef]
- Villacorta, L.; Minarrieta, L.; Salvatore, S.R.; Khoo, N.K.; Rom, O.; Gao, Z.; Berman, R.C.; Jobbagy, S.; Li, L.; Woodcock, S.R.; et al. In situ generation, metabolism and immunomodulatory signaling actions of nitro-conjugated linoleic acid in a murine model of inflammation. Redox Biol. 2018, 15, 522–531. [Google Scholar] [CrossRef]
- Kansanen, E.; Kuosmanen, S.M.; Ruotsalainen, A.-K.; Hynynen, H.; Levonen, A.-L. Nitro-oleic acid regulates endothelin signaling in human endothelial cells. Mol. Pharmacol. 2017, 92, 481–490. [Google Scholar] [CrossRef]
- Charles, R.L.; Rudyk, O.; Prysyazhna, O.; Kamynina, A.; Yang, J.; Morisseau, C.; Hammock, B.D.; Freeman, B.A.; Eaton, P. Protection from hypertension in mice by the Mediterranean diet is mediated by nitro fatty acid inhibition of soluble epoxide hydrolase. Proc. Natl. Acad. Sci. USA 2014, 111, 8167–8172. [Google Scholar] [CrossRef] [Green Version]
- Baker, P.R.S.; Lin, Y.; Schopfer, F.J.; Woodcock, S.R.; Groeger, A.L.; Batthyany, C.; Sweeney, S.; Long, M.H.; Baker, L.M.S.; Branchaud, B.P.; et al. Fatty Acid Transduction of Nitric Oxide Signaling: Multiple Nitrated Unsaturated Fatty Acid Derivatives exist in Human Blood and Urine and serve as Endogenous Peroxisome Proliferator-Activated Recetor Ligands. J. Biol. Chem. 2005, 280, 42464–42475. [Google Scholar] [CrossRef] [Green Version]
- Jones, L.H. Chemistry and biology of biomolecule nitration. Chem. Biol. 2012, 19, 1086–1092. [Google Scholar] [CrossRef] [Green Version]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Melo, T.; Marques, S.S.; Ferreira, I.; Cruz, M.T.; Domingues, P.; Segundo, M.A.; Domingues, M.R.M. New Insights into the Anti-Inflammatory and Antioxidant Properties of Nitrated Phospholipids. Lipids 2018, 53, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Duarte, S.; Melo, T.; Domingues, M.R.M.; de Dios Alché, J.; Pérez-Sala, D. Insight into the cellular effects of nitrated phospholipids: Evidence for pleiotropic mechanisms of action. Free Radic. Biol. Med. 2019, 144, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Koivusalo, M.; Haimi, P.; Heikinheimo, L.; Kostiainen, R.; Somerharju, P. Quantitative determination of phospholipid compositions by ESI-MS: Effects of acyl chain length, unsaturation, and lipid concentration on instrument response. J. Lipid Res. 2001, 42, 663–672. [Google Scholar] [PubMed]
- Wang, Y.; Armando, A.M.; Quehenberger, O.; Yan, C.; Dennis, E.A. Comprehensive ultra-performance liquid chromatographic separation and mass spectrometric analysis of eicosanoid metabolites in human samples. J. Chromatogr. A 2014, 1359, 60–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buré, C.; Ayciriex, S.; Testet, E.; Schmitter, J.-M. A single run LC-MS/MS method for phospholipidomics. Anal. Bioanal. Chem. 2013, 405, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.V.; Macek, B.; Lange, O.; Makarov, A.; Horning, S.; Mann, M. Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 2007, 4, 709–712. [Google Scholar] [CrossRef]
- Olsen, J.V.; Schwartz, J.C.; Griep-Raming, J.; Nielsen, M.L.; Damoc, E.; Denisov, E.; Lange, O.; Remes, P.; Taylor, D.; Splendore, M.; et al. A Dual Pressure Linear Ion Trap Orbitrap Instrument with Very High Dequencing Speed. Mol. Cell. Proteom. 2009, 8, 2759–2769. [Google Scholar] [CrossRef] [Green Version]
- Bayat, P.; Lesage, D.; Cole, R.B. Tutorial: Ion activation in Tandem Mass Spectrometry using ultra-high resolution instrumentation. Mass Spectrom. Rev. 2020, 1–23. [Google Scholar] [CrossRef]
- Chakravartula, S.V.S.; Balazy, M. Characterization of Nitro Arachidonic Acid and Nitro Linoleic Acid by Mass Spectrometry. Anal. Lett. 2012, 45, 2412–2424. [Google Scholar] [CrossRef]
- Lima, É.S.; Di Mascio, P.; Rubbo, H.; Abdalla, D.S.P. Characterization of Linoleic Acid Nitration in Human Blood Plasma by Mass Spectrometry. Biochemistry 2002, 41, 10717–10722. [Google Scholar] [CrossRef]
- Bartlett, E.M.; Lewis, D.H. Spectrophotometric determination of phosphate esters in the presence and absence of orthophosphate. Anal. Biochem. 1970, 36, 159–167. [Google Scholar] [CrossRef]
- Weinman, E.O.; Chaikoff, I.L.; Entenman, C.; Dauben, W.G. Turnover rates of phosphate and fatty acid moieties of plasma phospholipids. J. Biol. Chem. 1950, 187, 643–649. [Google Scholar]
- Pérez-Sala, D.; Oeste, C.L.; Martínez, A.E.; Carrasco, M.J.; Garzón, B.; Cañada, F.J. Vimentin filament organization and stress sensing depend on its single cysteine residue and zinc binding. Nat. Commun. 2015, 6, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bligh, E.G.; Dyer, W.J. A Rapid Method of Total Lipid Extration and Purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Sample Availability: Samples of the compounds are not available from the authors. |



| Typical Neutral Losses and Product Ions Observed for NO2-PC and NO2-PE in Positive Ion Mode | |||||||
| Neutral Losses | Proposed Identification | (POPC + NO2 + H)+ | (POPE + NO2 + H)+ | (PLPC + NO2 + H)+ | (PLPE + NO2 + H)+ | (PAPC + NO2 + H)+ | (PAPE + NO2 + H)+ |
| 805.8 | 763.6 | 803.6 | 761.5 | 827.8 | 785.6 | ||
| 18 Da | -H2O | 787.6 | 745.5 | 785.5 | 743.5 | 809.6 | 767.5 |
| 43 Da | -C2H5N | --- | 720.4 | -- | 718.4 | -- | 742.5 |
| 47 Da | -HNO2 | 758.7 | 716.5 | 756.5 | 714.6 | 780.6 | 738.5 |
| 59 Da | -C3H9N | 746.5 | -- | 744.34 | -- | 768.7 | -- |
| 90 Da (43 + 47) | -(C2H5N + HNO2) | -- | 673.4 | -- | 671.6 | -- | 695.5 |
| 106 Da (59 + 47) | -(C3H9N + HNO2) | 699.6 | --- | 697.4 | -- | 721.5 | -- |
| 141 Da | -C2H8PO4N | -- | 622.5 | -- | 620.5 | -- | 644.5 |
| 183 Da | -C5H14PO4N | 622.6 | -- | 620.6 | -- | 644.6 | -- |
| 188 Da (141 + 47) | -(C2H8PO4N + HNO2) | --- | 575.4 | -- | 573.4 | -- | 597.6 |
| 230 Da (183 + 47) | -(C5H14PO4N + HNO2) | 575.5 | -- | 573.6 | -- | 597.7 | -- |
| 238 Da | -R1C=C=O | 567.6 | 525.4 | 565.4 | 523.3 | 589.3 | 547.4 |
| 256 Da | -R1COOH | 549.6 | 507.4 | 547.4 | 505.5 | 571.5 | 529.5 |
| Product Ions | |||||||
| Nitro Oleic Acid (NO2-OA) | Nitro Linoleic Acid (NO2-LA) | Nitro Arachidonic Acid (NO2-AA) | |||||
| (NO2-FA + H)+ | 328.3 | 328.3 | 326.4 | 326.4 | 350.3 | 350.3 | |
| (NO2-FA-H2O + H)+ | 310.3 | 310.3 | 308.3 | 308.3 | 332.3 | 332.3 | |
| (NO2-FA-2H2O + H)+ | 292.3 | 292.3 | 290.3 | 290.3 | 314.2 | 314.2 | |
| (NO2-FA-NO2 + H)+ | 281.3 | 281.3 | 279.3 | 279.3 | 303.3 | 303.3 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neves, B.; Duarte, S.; Domingues, P.; Pérez-Sala, D.; Oliveira, M.M.; Domingues, M.d.R.; Melo, T. Advancing Target Identification of Nitrated Phospholipids in Biological Systems by HCD Specific Fragmentation Fingerprinting in Orbitrap Platforms. Molecules 2020, 25, 2120. https://doi.org/10.3390/molecules25092120
Neves B, Duarte S, Domingues P, Pérez-Sala D, Oliveira MM, Domingues MdR, Melo T. Advancing Target Identification of Nitrated Phospholipids in Biological Systems by HCD Specific Fragmentation Fingerprinting in Orbitrap Platforms. Molecules. 2020; 25(9):2120. https://doi.org/10.3390/molecules25092120
Chicago/Turabian StyleNeves, Bruna, Sofia Duarte, Pedro Domingues, Dolores Pérez-Sala, Maria Manuel Oliveira, Maria do Rosário Domingues, and Tânia Melo. 2020. "Advancing Target Identification of Nitrated Phospholipids in Biological Systems by HCD Specific Fragmentation Fingerprinting in Orbitrap Platforms" Molecules 25, no. 9: 2120. https://doi.org/10.3390/molecules25092120
APA StyleNeves, B., Duarte, S., Domingues, P., Pérez-Sala, D., Oliveira, M. M., Domingues, M. d. R., & Melo, T. (2020). Advancing Target Identification of Nitrated Phospholipids in Biological Systems by HCD Specific Fragmentation Fingerprinting in Orbitrap Platforms. Molecules, 25(9), 2120. https://doi.org/10.3390/molecules25092120