Phylogenetic Analysis and In Vitro Bifunctional Nuclease Assay of Arabidopsis BBD1 and BBD2
Abstract
:1. Introduction
2. Results and Discussion
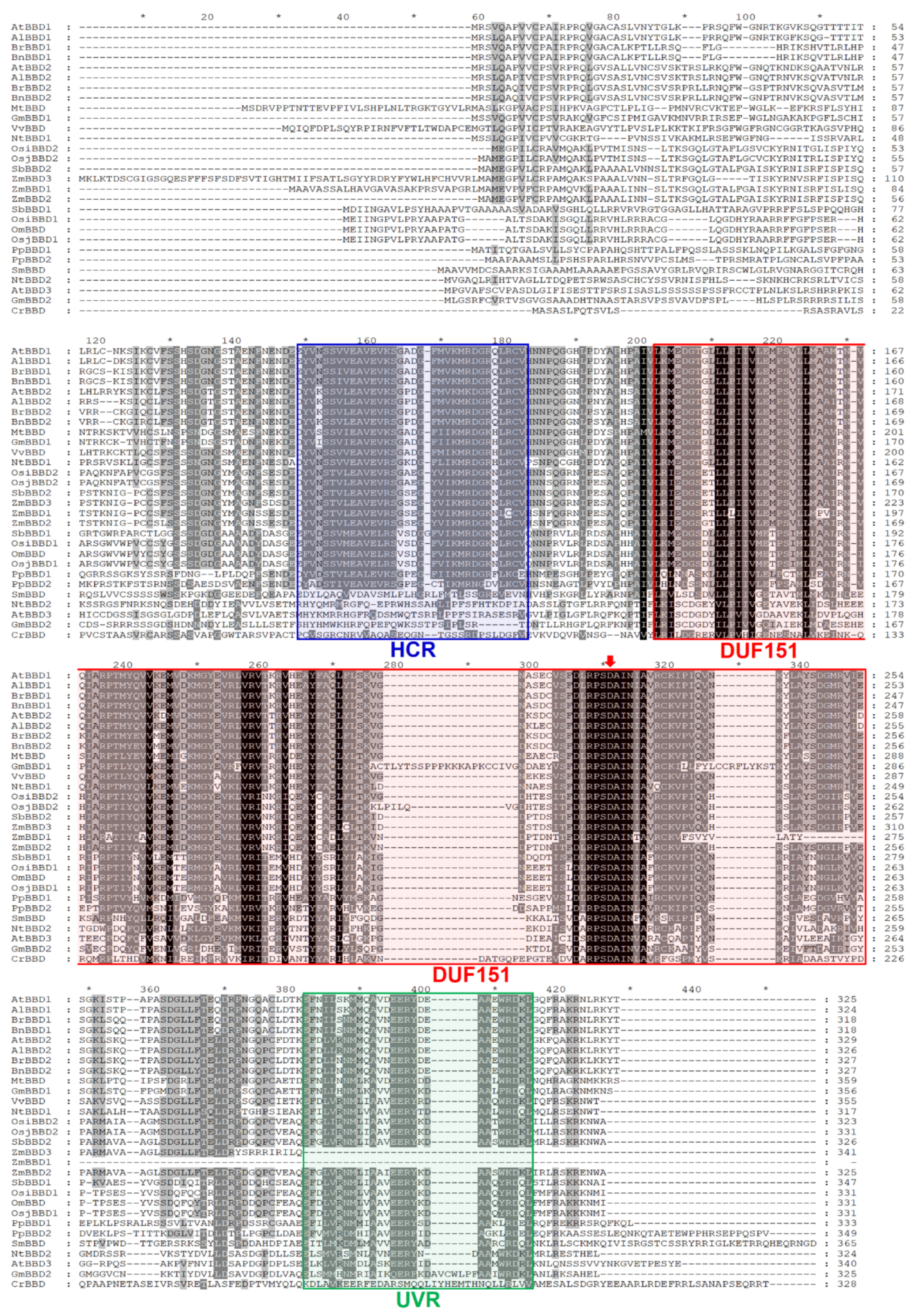
2.1. DUF151-Containing Proteins Are Phylogenetically Divided into Two Major Groups, and Eukaryotic Proteins Are Only Identified in the Viridiplantae
2.2. AtBBD1 and AtBBD2 Contain Three Characteristic Domains and Are the Closest Arabidopsis Homologs of OmBBD
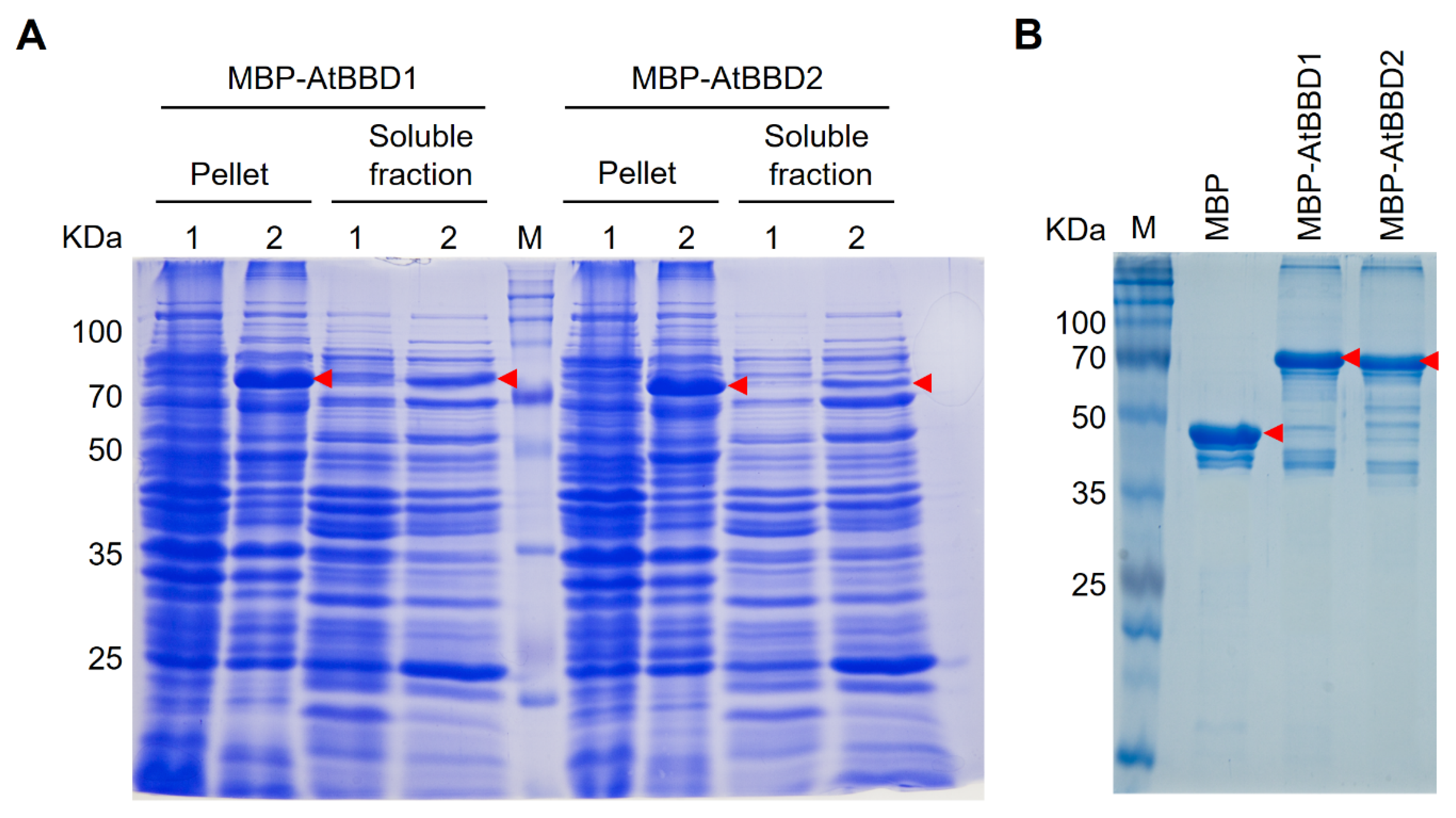
2.3. Recombinant MBP-Tagged AtBBD1 and AtBBD2 Proteins Purified Using Amylose Affinity Chromatography
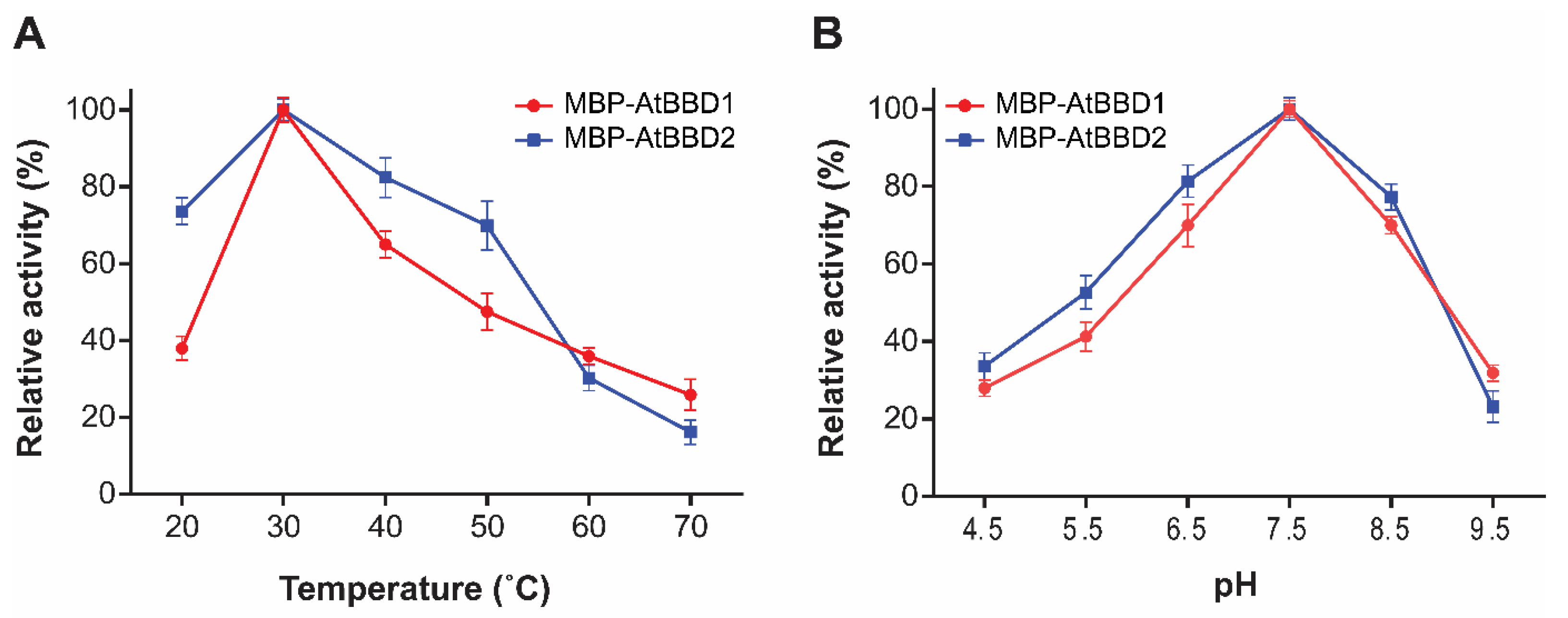
2.4. AtBBD1 and AtBBD2 Possess Non-Substrate-Specific DNase Activity that Is Affected by Temperature and pH But Do Not Require Bivalent Cations
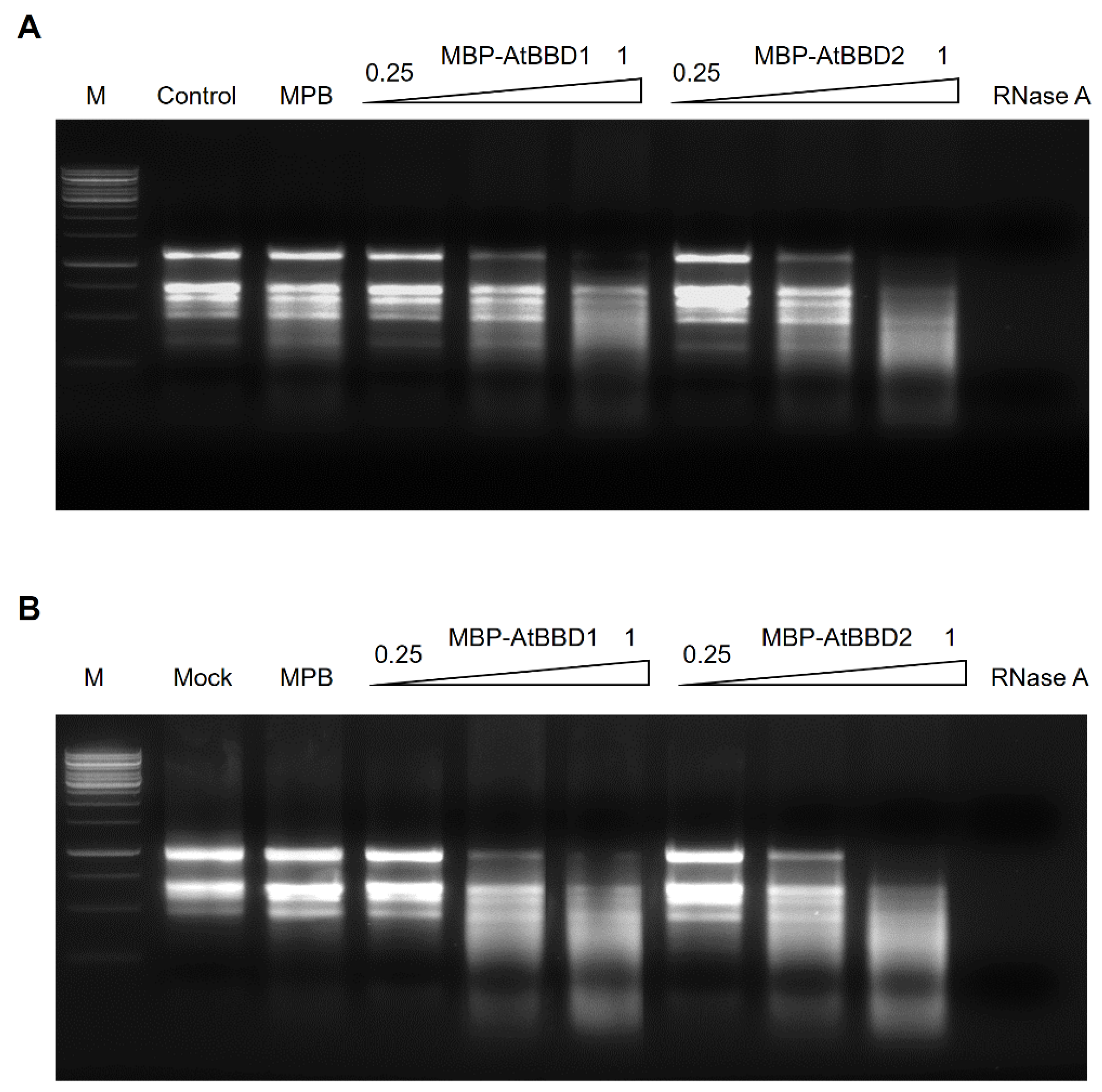
2.5. AtBBD1 and AtBBD2 Have Non-Substrate-Specific RNase Activity
2.6. AtBBD1 and AtBBD2 Each form a Homodimer and the HCR Domain is Possibly Crucial for Dimerization
3. Materials and Methods
3.1. Gene Cloning and Construction of Expression Vector
3.2. Expression and Purification of MBP-BBD1 and MBP-BBD2
3.3. Nuclease Activity of Recombinant BBD1 and BBD2
3.4. Construction of Phylogenetic Tree and Sequence Analysis
3.5. Yeast Two-Hybrid
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yang, W. Nucleases: Diversity of structure, function and mechanism. Q. Rev. Biophys. 2011, 44, 1–93. [Google Scholar] [CrossRef] [PubMed]
- Canetti, L.; Lomaniec, E.; Elkind, Y.; Lers, A. Nuclease activities associated with dark-induced and natural leaf senescence in parsley. Plant Sci. 2002, 163, 873–880. [Google Scholar] [CrossRef]
- Lambert, R.; Quiles, F.A.; Gálvez-Valdivieso, G.; Piedras, P. Nucleases activities during French bean leaf aging and dark-induced senescence. J. Plant Physiol. 2017, 218, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Perez-Amador, M.A.; Abler, M.L.; De Rocher, E.J.; Thompson, D.M.; Van Hoof, A.; LeBrasseur, N.D.; Lers, A.; Green, P.J. Identification of BFN1, a bifunctional nuclease induced during leaf and stem senescence in Arabidopsis. Plant Physiol. 2000, 122, 169–179. [Google Scholar] [CrossRef] [Green Version]
- Matoušek, J.; Kozlová, P.; Orctová, L.; Schmitz, A.; Pešina, K.; Bannach, O.; Diermann, N.; Steger, G.; Riesner, D. Accumulation of viroid-specific small RNAs and increase in nucleolytic activities linked to viroid-caused pathogenesis. Biol. Chem. 2007, 388, 1–13. [Google Scholar] [CrossRef]
- Aoyagi, S.; Sugiyama, M.; Fukuda, H. BEN1 and ZEN1 cDNAs encoding S1-type DNases that are associated with programmed cell death in plants. FEBS Lett. 1998, 429, 134–138. [Google Scholar] [CrossRef] [Green Version]
- Balk, J.; Chew, S.K.; Leaver, C.J.; McCabe, P.F. The intermembrane space of plant mitochondria contains a DNase activity that may be involved in programmed cell death. Plant J. 2003, 34, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Farage-Barhom, S.; Burd, S.; Sonego, L.; Perl-Treves, R.; Lers, A. Expression analysis of the BFN1 nuclease gene promoter during senescence, abscission, and programmed cell death-related processes. J. Exp. Bot. 2008, 59, 3247–3258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lombardi, L.; Ceccarelli, N.; Picciarelli, P.; Lorenzi, R. DNA degradation during programmed cell death in Phaseolus coccineus suspensor. Plant Physiol. Biochem. 2007, 45, 221–227. [Google Scholar] [CrossRef]
- Ito, J.; Fukuda, H. ZEN1 is a key enzyme in the degradation of nuclear DNA during programmed cell death of tracheary elements. Plant Cell 2002, 14, 3201–3211. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.M.; Pang, Y.; Zeng, J.; Ding, Q.; Yin, S.Y.; Liu, C.; Lu, M.Z.; Cui, K.M.; He, X.Q. The Ca2+-dependent DNases are Involved in Secondary Xylem Development in Eucommia ulmoides. J. Integr. Plant Biol. 2012, 54, 456–470. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, R.; Tang, L.Y.; Zhang, L.; Yamada, H.; Twell, D.; Sakamoto, W. A conserved, Mg2+-dependent exonuclease degrades organelle dna during Arabidopsis pollen development. Plant Cell 2011, 23, 1608–1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Hanson, M.R. Programmed cell death during pollination-induced petal senescence in Petunia. Plant Physiol. 2000, 122, 1323–1333. [Google Scholar] [CrossRef] [Green Version]
- Mittler, R.; Lam, E. Identification, characterization, and purification of a tobacco endonuclease activity induced upon hypersensitive response cell death. Plant Cell 1995, 7, 1951–1962. [Google Scholar] [PubMed] [Green Version]
- You, M.K.; Shin, H.Y.; Kim, Y.J.; Ok, S.H.; Cho, S.K.; Jeung, J.U.; Yoo, S.D.; Kim, J.K.; Shin, J.S. Novel bifunctional nucleases, OmBBD and AtBBD1, are involved in abscisic acid-mediated callose deposition in arabidopsis. Plant Physiol. 2010, 152, 1015–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sui, W.; Guo, K.; Li, L.; Liu, S.; Takano, T.; Zhang, X. Arabidopsis Ca2+-dependent nuclease AtCaN2 plays a negative role in plant responses to salt stress. Plant Sci. 2019, 281, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Muramoto, Y.; Watanabe, A.; Nakamura, T.; Takabe, T. Enhanced expression of a nuclease gene in leaves of barley plants under salt stress. Gene 1999, 234, 315–321. [Google Scholar] [CrossRef]
- Plchova, H.; Hartung, F.; Puchta, H. Biochemical Characterization of an Exonuclease from Arabidopsis thaliana Reveals Similarities to the DNA Exonuclease of the Human Werner Syndrome Protein. J. Biol. Chem. 2003, 278, 44128–44138. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, M.; Ito, J.; Aoyagi, S.; Fukuda, H. Endonucleases. In Programmed Cell Death in Higher Plants; Lam, E., Fukuda, H., Greenberg, J., Eds.; Springer: Dordrecht, The Netherlands, 2000; pp. 143–153. [Google Scholar]
- Domínguez, F.; Cejudo, F.J. Identification of a nuclear-localized nuclease from wheat cells undergoing programmed cell death that is able to trigger DNA fragmentation and apoptotic morphology on nuclei from human cells. Biochem. J. 2006, 397, 529–536. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Kermode, A.R. Proteases associated with programmed cell death of megagametophyte cells after germination of white spruce (Picea glauca) seeds. Plant Mol. Biol. 2003, 52, 729–744. [Google Scholar] [CrossRef]
- Langston, B.J.; Bai, S.; Jones, M.L. Increases in DNA fragmentation and induction of a senescence-specific nuclease are delayed during corolla senescence in ethylene-insensitive (etr1-1) transgenic petunias. J. Exp. Bot. 2005, 56, 15–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakamoto, W.; Takami, T. Nucleases in higher plants and their possible involvement in DNA degradation during leaf senescence. J. Exp. Bot. 2014, 65, 3835–3843. [Google Scholar] [CrossRef] [PubMed]
- Triques, K.; Sturbois, B.; Gallais, S.; Dalmais, M.; Chauvin, S.; Clepet, C.; Aubourg, S.; Rameau, C.; Caboche, M.; Bendahmane, A. Characterization of Arabidopsis thaliana mismatch specific endonucleases: Application to mutation discovery by TILLING in pea. Plant J. 2007, 51, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- Lesniewicz, K.; Karlowski, W.M.; Pienkowska, J.R.; Krzywkowski, P.; Poreba, E. The plant s1-like nuclease family has evolved a highly diverse range of catalytic capabilities. Plant Cell Physiol. 2013, 54, 1064–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isono, K.; Satoh, K.; Kobayashi, H. Molecular cloning of a cDNA encoding a novel Ca2+-dependent nuclease of Arabidopsis that is similar to staphylococcal nuclease. Biochim. Biophys. Acta Gene Struct. Expr. 2000, 1491, 267–272. [Google Scholar] [CrossRef]
- Guo, K.; Liu, S.; Takano, T.; Zhang, X. Molecular cloning, expression, and characterization of a Ca 2+-dependent nuclease of Arabidopsis thaliana. Protein Expr. Purif. 2012, 83, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Leśniewicz, K.; Poreba, E.; Smolarkiewicz, M.; Wolff, N.; Stanisławski, S.; Wojtaszek, P. Plant plasma membrane-bound staphylococcal-like DNases as a novel class of eukaryotic nucleases. BMC Plant Biol. 2012, 12, 195. [Google Scholar] [CrossRef] [Green Version]
- Seo, J.S.; Koo, Y.J.; Jung, C.; Yeu, S.Y.; Song, J.T.; Kim, J.K.; Choi, Y.; Lee, J.S.; Do Choi, Y. Identification of a Novel Jasmonate-Responsive Element in the AtJMT Promoter and Its Binding Protein for AtJMT Repression. PLoS ONE 2013, 8, 30. [Google Scholar] [CrossRef] [Green Version]
- Moolenaar, G.F.; Franken, K.L.M.C.; Dijkstra, D.M.; Thomas-Oates, J.E.; Visse, R.; Van De Putte, P.; Goosen, N. The C-terminal region of the UvrB protein of Escherichia coli contains an important determinant for UvrC binding to the preincision complex but not the catalytic site for 3′-incision. J. Biol. Chem. 1995, 270, 30508–30515. [Google Scholar] [CrossRef] [Green Version]
- Zacharia, V.M.; Manzanillo, P.S.; Nair, V.R.; Marciano, D.K.; Kinch, L.N.; Grishin, N.V.; Cox, J.S.; Shiloh, M.U. Cor, a novel carbon monoxide resistance gene, is essential for Mycobacterium tuberculosis pathogenesis. MBio 2013, 4, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Park, D.W.; Kim, S.S.; Nam, M.K.; Kim, G.Y.; Kim, J.; Rhim, H. Improved recovery of active GST-fusion proteins from insoluble aggregates: Solubilization and purification conditions using PKM2 and HtrA2 as model proteins. BMB Rep. 2011, 44, 279–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massiah, M.A.; Wright, K.M.; Du, H. Obtaining soluble folded proteins from inclusion bodies using sarkosyl, triton X-100, and CHAPS: Application to LB and M9 minimal media. Curr. Protoc. Protein Sci. 2016, 84, 6.13.1–6.13.24. [Google Scholar] [CrossRef] [PubMed]
- Sivagnanam, U.; Narayana Murthy, S.; Gummadi, S.N. Identification and characterization of the novel nuclease activity of human phospholipid scramblase 1. BMC Biochem. 2016, 17, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.; Zeng, Y.; Zhuang, X.; Sun, L.; Yao, X.; Pimpl, P.; Jiang, L. Organelle pH in the Arabidopsis Endomembrane System. Mol. Plant 2013, 6, 1419–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podzimek, T.; Matoušek, J.; Lipovová, P.; Poučková, P.; Spiwok, V.; Šantrůček, J. Biochemical properties of three plant nucleases with anticancer potential. Plant Sci. 2011, 180, 343–351. [Google Scholar] [CrossRef]
- Koval’, T.; Lipovová, P.; Podzimek, T.; Matoušek, J.; Dušková, J.; Skálová, T.; Štěpánková, A.; Hašek, J.; Dohnálek, J. Plant multifunctional nuclease TBN1 with unexpected phospholipase activity: Structural study and reaction-mechanism analysis. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 213–226. [Google Scholar] [CrossRef]
- Rayala, S.; Francis, V.G.; Sivagnanam, U.; Gummadi, S.N. N-terminal proline-rich domain is required for scrambling activity of human phospholipid scramblases. J. Biol. Chem. 2014, 289, 13206–13218. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Ben-Efraim, I.; Bigcas, J.L.; Junqueira, D.; Wiedmer, T.; Sims, P.J. Phospholipid scramblase 1 binds to the promoter region of the inositol 1,4,5-triphosphate receptor type 1 gene to enhance its expression. J. Biol. Chem. 2005, 280, 35062–35068. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Talluri, S.; Pal, J.; Yuan, X.; Lu, R.; Nanjappa, P.; Samur, M.K.; Munshi, N.C.; Shammas, M.A. Role of apurinic/apyrimidinic nucleases in the regulation of homologous recombination in myeloma: Mechanisms and translational significance. Blood Cancer J. 2018, 8, 92. [Google Scholar] [CrossRef]
- Kelley, M.R.; Parsons, S.H. Redox regulation of the DNA repair function of the human AP endonuclease Ape1/ref-1. Antioxid. Redox Signal. 2001, 3, 671–683. [Google Scholar] [CrossRef]
- Spraggon, G.; Pantazatos, D.; Klock, H.E.; Wilson, I.A.; Woods, V.L.; Lesley, S.A. On the use of DXMS to produce more crystallizable proteins: Structures of the T. maritima proteins TM0160 and TM1171. Protein Sci. 2009, 13, 3187–3199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunitz, M. Crystalline desoxyribonuclease; isolation and general properties; spectrophotometric method for the measurement of desoxyribonuclease activity. J. Gen. Physiol. 1950, 33, 349–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Sample Availability: All the vector constructs are available from the authors and can be provided upon request. |





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huque, A.K.M.M.; So, W.M.; You, M.K.; Shin, J.S. Phylogenetic Analysis and In Vitro Bifunctional Nuclease Assay of Arabidopsis BBD1 and BBD2. Molecules 2020, 25, 2169. https://doi.org/10.3390/molecules25092169
Huque AKMM, So WM, You MK, Shin JS. Phylogenetic Analysis and In Vitro Bifunctional Nuclease Assay of Arabidopsis BBD1 and BBD2. Molecules. 2020; 25(9):2169. https://doi.org/10.3390/molecules25092169
Chicago/Turabian StyleHuque, A. K. M. Mahmudul, Won Mi So, Min Kyoung You, and Jeong Sheop Shin. 2020. "Phylogenetic Analysis and In Vitro Bifunctional Nuclease Assay of Arabidopsis BBD1 and BBD2" Molecules 25, no. 9: 2169. https://doi.org/10.3390/molecules25092169






