Recent Advances in Organelle-Targeted Fluorescent Probes
Abstract
1. Introduction
2. Design Strategies and Recent Examples
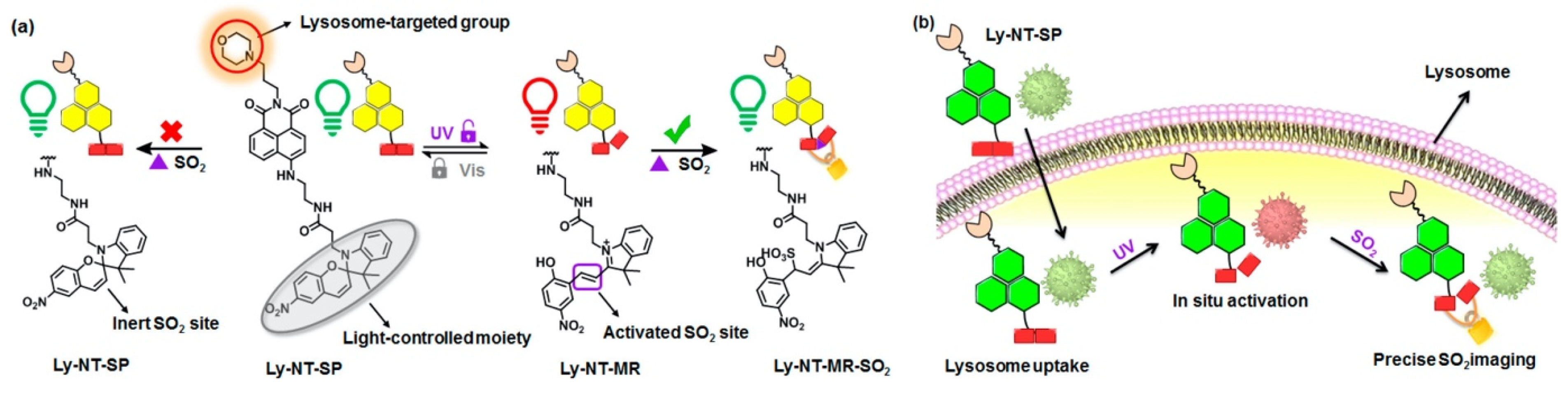
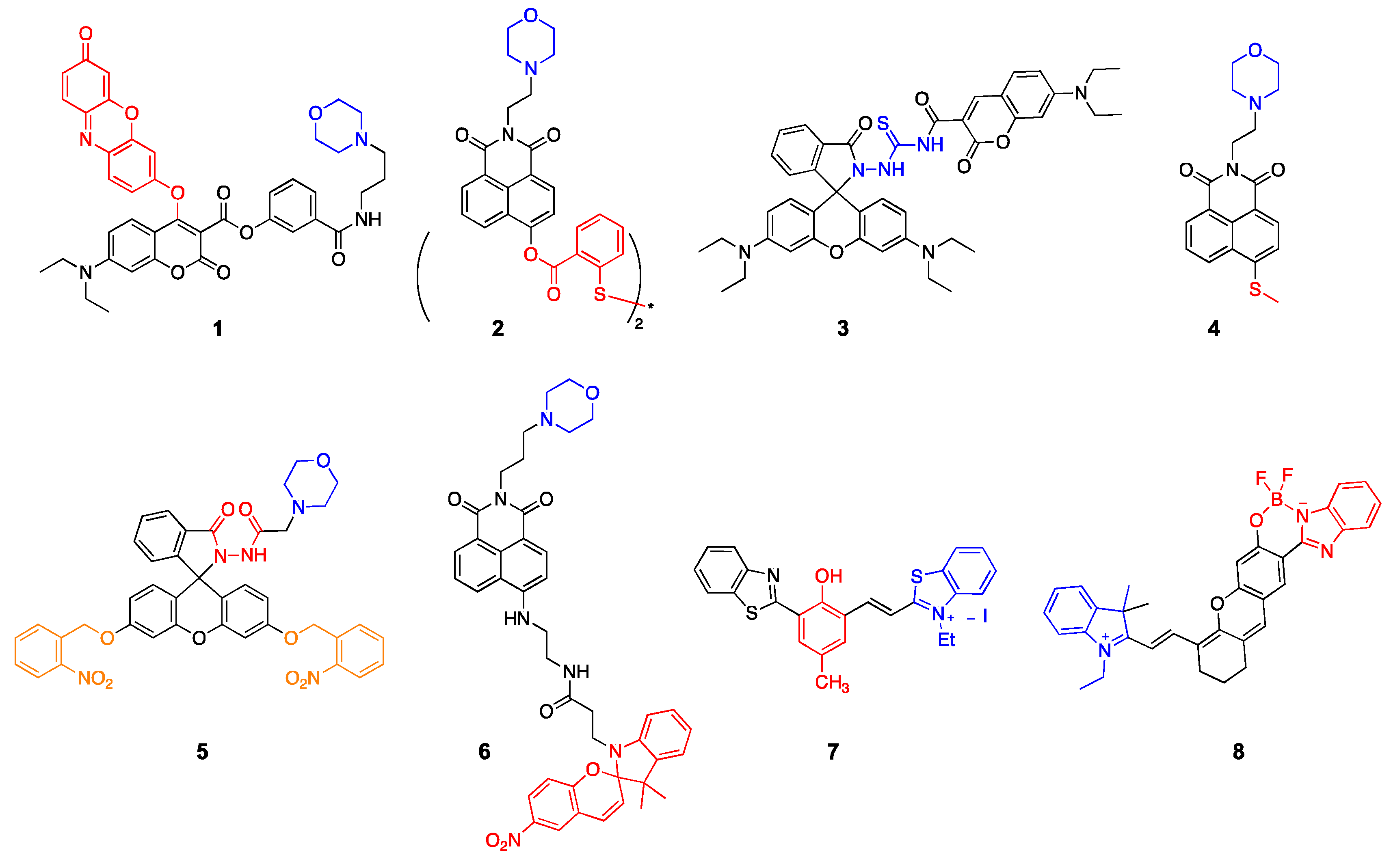
2.1. Lysosome-Targeted Probes
2.2. Nucleus-Targeted Probes
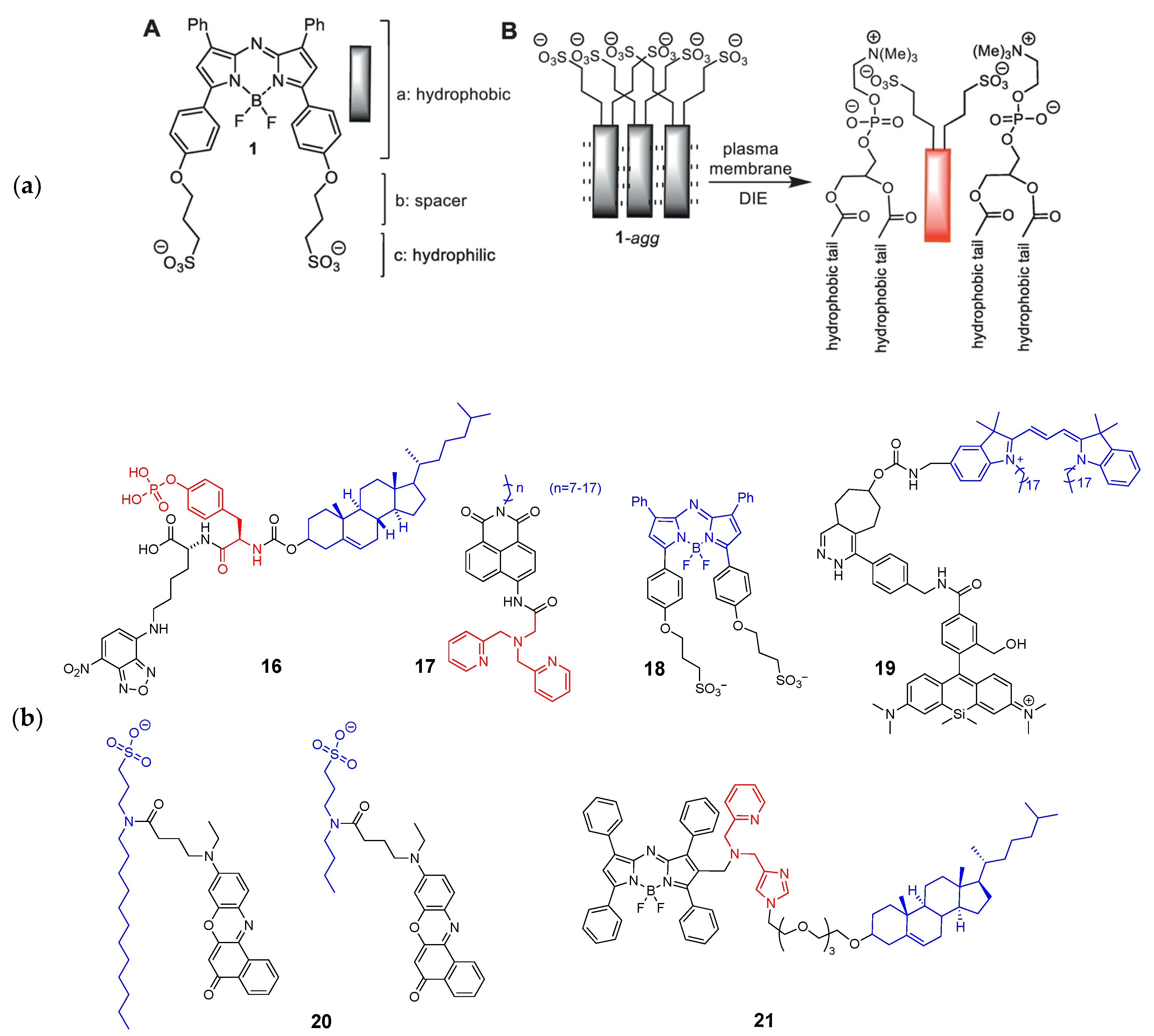
2.3. Membrane-Targeted Probes
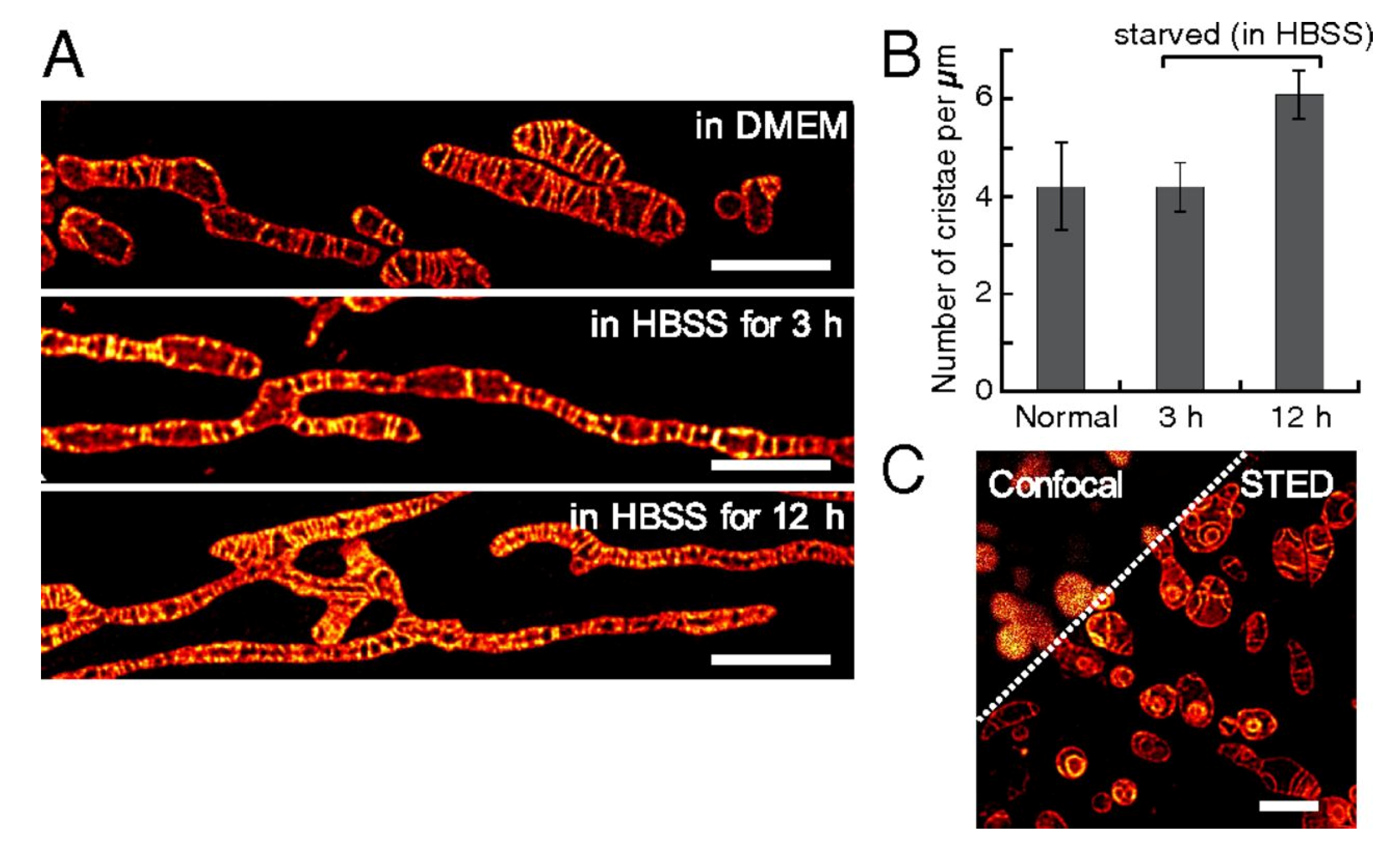
2.4. Mitochondrion-Targeted Probes
2.5. Probes Targeting the Endoplasmic Reticulum (ER) and Golgi Apparatus
2.6. Probes Targeting Other Organelles
3. Conclusions and Outlook
Funding
Conflicts of Interest
References
- Perera, R.M.; Zoncu, R. The Lysosome as a Regulatory Hub. Annu. Rev. Cell Dev. Biol. 2016, 32, 223–253. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Ren, D. Lysosomal physiology. Annu. Rev. Physiol. 2015, 77, 57–80. [Google Scholar] [CrossRef] [PubMed]
- Bonam, S.R.; Wang, F.; Muller, S. Lysosomes as a therapeutic target. Nat. Rev. Drug Discov. 2019, 18, 923–948. [Google Scholar] [CrossRef] [PubMed]
- Deus, C.M.; Yambire, K.F.; Oliveira, P.J.; Raimundo, N. Mitochondria-Lysosome Crosstalk: From Physiology to Neurodegeneration. Trends Mol. Med. 2020, 26, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Hastings, K.T.; Cresswell, P. Disulfide reduction in the endocytic pathway: Immunological functions of gamma-interferon-inducible lysosomal thiol reductase. Antioxid. Redox Signal. 2011, 15, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xu, L.; Chen, W.; Huang, J.; Huang, C.; Sheng, J.; Song, X. A Lysosome-Targetable Fluorescent Probe for Simultaneously Sensing Cys/Hcy, GSH, and H2S from Different Signal Patterns. ACS Sens. 2018, 3, 2513–2517. [Google Scholar] [CrossRef]
- Firestone, R.A.; Pisano, J.M.; Bonney, R.J. Lysosomotropic agents. 1. Synthesis and cytotoxic action of lysosomotropic detergents. J. Med. Chem. 1979, 22, 1130–1133. [Google Scholar] [CrossRef]
- Casey, J.R.; Grinstein, S.; Orlowski, J. Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 2010, 11, 50–61. [Google Scholar] [CrossRef]
- Li, G.; Ma, S.; Tang, J.; Ye, Y. Lysosome-targeted two-photon fluorescent probes for rapid detection of H2S in live cells. New J. Chem. 2019, 43, 1267–1274. [Google Scholar] [CrossRef]
- Yuan, Q.; Zhao, Z.-M.; Zhang, Y.-R.; Su, L.; Miao, J.-Y.; Zhao, B.-X. A lysosome-targeted ratiometric fluorescent probe for detection of hypochlorous acid in living cells. Sens. Actuators B Chem. 2017, 247, 736–741. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, X.; Zhang, R.; Liu, Y.; Ren, X.; Xian, M.; Ye, Y.; Zhao, Y. Lysosomal-Targeted Two-Photon Fluorescent Probe to Sense Hypochlorous Acid in Live Cells. Anal. Chem. 2017, 89, 10384–10390. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Li, Z.; Nie, J.; Wang, L.; Lin, W. A photocaged fluorescent probe for imaging hypochlorous acid in lysosomes. Chem. Commun. 2018, 54, 9238–9241. [Google Scholar] [CrossRef]
- Zhang, W.; Huo, F.; Yue, Y.; Zhang, Y.; Chao, J.; Cheng, F.; Yin, C. Heat Stroke in Cell Tissues Related to Sulfur Dioxide Level Is Precisely Monitored by Light-Controlled Fluorescent Probes. J. Am. Chem. Soc. 2020, 142, 3262–3268. [Google Scholar] [CrossRef] [PubMed]
- Villamil Giraldo, A.M.; Appelqvist, H.; Ederth, T.; Ollinger, K. Lysosomotropic agents: Impact on lysosomal membrane permeabilization and cell death. Biochem. Soc. Trans. 2014, 42, 1460–1464. [Google Scholar] [CrossRef] [PubMed]
- Nadanaciva, S.; Lu, S.; Gebhard, D.F.; Jessen, B.A.; Pennie, W.D.; Will, Y. A high content screening assay for identifying lysosomotropic compounds. Toxicol. In Vitro 2011, 25, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Dahal, D.; McDonald, L.; Bi, X.; Abeywickrama, C.; Gombedza, F.; Konopka, M.; Paruchuri, S.; Pang, Y. An NIR-emitting lysosome-targeting probe with large Stokes shift via coupling cyanine and excited-state intramolecular proton transfer. Chem. Commun. 2017, 53, 3697–3700. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Meng, X.; Yang, H.; Song, L.; Liu, S.; Xu, A.; Chen, Z.; Huang, W.; Zhao, Q. Lysosome-specific sensing and imaging of pH variations in vitro and in vivo utilizing a near-infrared boron complex. J. Mater. Chem. B 2019, 7, 3569–3575. [Google Scholar] [CrossRef]
- Hetzer, M.W. The nuclear envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000539. [Google Scholar] [CrossRef]
- Nano, A.; Boynton, A.N.; Barton, J.K. A rhodium-cyanine fluorescent probe: Detection and signaling of mismatches in DNA. J. Am. Chem. Soc. 2017, 139, 17301–17304. [Google Scholar] [CrossRef]
- Boyle, K.M.; Barton, J.K. A Family of Rhodium Complexes with Selective Toxicity toward Mismatch Repair-Deficient Cancers. J. Am. Chem. Soc. 2018, 140, 5612–5624. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, H.; Chen, H.; Li, Q.; Guan, A.; Wang, L.; Shi, Y.; Xu, S.; Liu, M.; Tang, Y. Direct visualization of nucleolar G-quadruplexes in live cells by using a fluorescent light-up probe. Biochim. Et Biophys. Acta BBA Gen. Subj. 2018, 1862, 1101–1106. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sun, H.; Wang, L.; Liu, Y.; Chen, H.; Li, Q.; Guan, A.; Liu, M.; Tang, Y. Real-time monitoring of DNA G-quadruplexes in living cells with a small-molecule fluorescent probe. Nucleic Acids Res. 2018, 46, 7522–7532. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, J.; Barooah, N.; Dhamodharan, V.; Harikrishna, S.; Pradeepkumar, P.I.; Bhasikuttan, A.C. Thioflavin T as an efficient inducer and selective fluorescent sensor for the human telomeric G-quadruplex DNA. J. Am. Chem. Soc. 2013, 135, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Bucevicius, J.; Keller-Findeisen, J.; Gilat, T.; Hell, S.W.; Lukinavicius, G. Rhodamine-Hoechst positional isomers for highly efficient staining of heterochromatin. Chem. Sci. 2019, 10, 1962–1970. [Google Scholar] [CrossRef]
- Lammle, C.A.; Varady, A.; Muller, T.G.; Sturtzel, C.; Riepl, M.; Mathes, B.; Eichhorst, J.; Sporbert, A.; Lehmann, M.; Krausslich, H.G.; et al. Photocaged Hoechst Enables Subnuclear Visualization and Cell Selective Staining of DNA in vivo. Chembiochem 2020. [Google Scholar] [CrossRef]
- Dervan, P.B.; Edelson, B.S. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr. Opin. Struct. Biol. 2003, 13, 284–299. [Google Scholar] [CrossRef]
- Vaijayanthi, T.; Bando, T.; Pandian, G.N.; Sugiyama, H. Progress and prospects of pyrrole-imidazole polyamide-fluorophore conjugates as sequence-selective DNA probes. Chembiochem 2012, 13, 2170–2185. [Google Scholar] [CrossRef]
- Yang, F.; Nickols, N.G.; Li, B.C.; Marinov, G.K.; Said, J.W.; Dervan, P.B. Antitumor activity of a pyrrole-imidazole polyamide. Proc. Natl. Acad. Sci. USA 2013, 110, 1863–1868. [Google Scholar] [CrossRef]
- Raskatov, J.A.; Meier, J.L.; Puckett, J.W.; Yang, F.; Ramakrishnan, P.; Dervan, P.B. Modulation of NF-kappaB-dependent gene transcription using programmable DNA minor groove binders. Proc. Natl. Acad. Sci. USA 2012, 109, 1023–1028. [Google Scholar] [CrossRef]
- Tsubono, Y.; Kawamoto, Y.; Hidaka, T.; Pandian, G.N.; Hashiya, K.; Bando, T.; Sugiyama, H. A Near-Infrared Fluorogenic Pyrrole-Imidazole Polyamide Probe for Live-Cell Imaging of Telomeres. J. Am. Chem. Soc. 2020, 142, 17356–17363. [Google Scholar] [CrossRef]
- Cheng, Y.; Sun, C.; Ou, X.; Liu, B.; Lou, X.; Xia, F. Dual-targeted peptide-conjugated multifunctional fluorescent probe with AIEgen for efficient nucleus-specific imaging and long-term tracing of cancer cells. Chem. Sci. 2017, 8, 4571–4578. [Google Scholar] [CrossRef]
- Wen, Y.; Huo, F.; Yin, C. A glycine spacer improved peptidyl-nuclear-localized efficiency for fluorescent imaging nuclear H2O2. Sens. Actuators B Chem. 2019, 296, 126624. [Google Scholar] [CrossRef]
- Wen, Y.; Liu, K.; Yang, H.; Li, Y.; Lan, H.; Liu, Y.; Zhang, X.; Yi, T. A highly sensitive ratiometric fluorescent probe for the detection of cytoplasmic and nuclear hydrogen peroxide. Anal. Chem. 2014, 86, 9970–9976. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, P.; Seo, A.Y.; Pasolli, H.A.; Song, Y.E.; Johnson, M.C.; Lippincott-Schwartz, J. A lipid-based partitioning mechanism for selective incorporation of proteins into membranes of HIV particles. Nat. Cell. Biol. 2019, 21, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.T.; Kreutzberger, A.J.B.; Kiessling, V.; Ganser-Pornillos, B.K.; White, J.M.; Tamm, L.K. HIV virions sense plasma membrane heterogeneity for cell entry. Sci. Adv. 2017, 3, e1700338. [Google Scholar] [CrossRef]
- Staubach, S.; Hanisch, F.-G. Lipid rafts: Signaling and sorting platforms of cells and their roles in cancer. Exp. Rev. Proteom. 2011, 8, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Di Paolo, G.; Kim, T.W. Linking lipids to Alzheimer’s disease: Cholesterol and beyond. Nat. Rev. Neurosci. 2011, 12, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Schengrund, C.L. Lipid rafts: Keys to neurodegeneration. Brain Res. Bull. 2010, 82, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Feng, Z.; Del Signore, S.J.; Rodal, A.A.; Xu, B. Active probes for imaging membrane dynamics of live cells with high spatial and temporal resolution over extended time scales and areas. J. Am. Chem. Soc. 2018, 140, 3505–3509. [Google Scholar] [CrossRef]
- Deng, F.; Liu, L.; Qiao, Q.; Huang, C.; Miao, L.; Xu, Z. A general strategy to develop cell membrane fluorescent probes with location- and target-specific fluorogenicities: A case of a Zn(2+) probe with cellular selectivity. Chem. Commun. 2019, 55, 15045–15048. [Google Scholar] [CrossRef]
- Wu, D.; Cheung, S.; Sampedro, G.; Chen, Z.-L.; Cahill, R.A.; O’Shea, D.F. A DIE responsive NIR-fluorescent cell membrane probe. Biochim. et Biophys. Acta BBA Biomembr. 2018, 1860, 2272–2280. [Google Scholar] [CrossRef] [PubMed]
- Takakura, H.; Zhang, Y.; Erdmann, R.S.; Thompson, A.D.; Lin, Y.; McNellis, B.; Rivera-Molina, F.; Uno, S.N.; Kamiya, M.; Urano, Y.; et al. Long time-lapse nanoscopy with spontaneously blinking membrane probes. Nat. Biotechnol. 2017, 35, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Mockl, L.; Moerner, W.E. Super-resolution Microscopy with Single Molecules in Biology and Beyond-Essentials, Current Trends, and Future Challenges. J. Am. Chem. Soc. 2020, 142, 17828–17844. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.; Tyson, J.; Shaw, J.E.; Rivera-Molina, F.; Koleske, A.J.; Schepartz, A.; Toomre, D.K. Two-color nanoscopy of organelles for extended times with HIDE probes. Nat. Commun. 2020, 11, 4271. [Google Scholar] [CrossRef]
- Danylchuk, D.I.; Moon, S.; Xu, K.; Klymchenko, A.S. Switchable Solvatochromic Probes for Live-Cell Super-resolution Imaging of Plasma Membrane Organization. Angew. Chem. Int. Ed. Engl. 2019, 58, 14920–14924. [Google Scholar] [CrossRef]
- Kim, J.J.; Hong, J.; Yu, S.; You, Y. Deep-Red-Fluorescent Zinc Probe with a Membrane-Targeting Cholesterol Unit. Inorg. Chem. 2020, 59, 11562–11576. [Google Scholar] [CrossRef]
- Parton, R.G.; Del Pozo, M.A. Caveolae as plasma membrane sensors, protectors and organizers. Nat. Rev. Mol. Cell Biol. 2013, 14, 98–112. [Google Scholar] [CrossRef]
- Mouritsen, O.G.; Zuckermann, M.J. What’s so special about cholesterol? Lipids 2004, 39, 1101–1113. [Google Scholar] [CrossRef]
- Chow, J.; Rahman, J.; Achermann, J.C.; Dattani, M.T.; Rahman, S. Mitochondrial disease and endocrine dysfunction. Nat. Rev. Endocrinol. 2017, 13, 92–104. [Google Scholar] [CrossRef]
- Vyas, S.; Zaganjor, E.; Haigis, M.C. Mitochondria and Cancer. Cell 2016, 166, 555–566. [Google Scholar] [CrossRef]
- Johri, A.; Beal, M.F. Mitochondrial dysfunction in neurodegenerative diseases. J. Pharm. Exp. 2012, 342, 619–630. [Google Scholar] [CrossRef]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Zielonka, M.; Dranka, B.; Kumar, S.N.; Myers, C.R.; Bennett, B.; Garces, A.M.; Dias Duarte Machado, L.G.; Thiebaut, D.; Ouari, O.; et al. Detection of mitochondria-generated reactive oxygen species in cells using multiple probes and methods: Potentials, pitfalls, and the future. J. Biol. Chem. 2018, 293, 10363–10380. [Google Scholar] [CrossRef] [PubMed]
- Wisnovsky, S.; Lei, E.K.; Jean, S.R.; Kelley, S.O. Mitochondrial Chemical Biology: New Probes Elucidate the Secrets of the Powerhouse of the Cell. Cell Chem. Biol. 2016, 23, 917–927. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Liu, X.; Zhang, Y.; Yang, L.; Fang, Q.; Geng, Y.; Chen, W.; Song, X. A mitochondria-targeting ratiometric fluorescent probe for imaging hydrogen peroxide with long-wavelength emission and large stokes shift. Sens. Actuators B Chem. 2018, 276, 247–253. [Google Scholar] [CrossRef]
- Tang, Y.; Ma, Y.; Xu, A.; Xu, G.; Lin, W. A turn-on fluorescent probe for endogenous formaldehyde in the endoplasmic reticulum of living cells. Methods Appl. Fluoresc. 2017, 5, 024005. [Google Scholar] [CrossRef]
- Hu, Q.; Qin, C.; Huang, L.; Wang, H.; Liu, Q.; Zeng, L. Selective visualization of hypochlorite and its fluctuation in cancer cells by a mitochondria-targeting ratiometric fluorescent probe. Dyes Pigment. 2018, 149, 253–260. [Google Scholar] [CrossRef]
- Zhu, B.; Wu, L.; Zhang, M.; Wang, Y.; Liu, C.; Wang, Z.; Duan, Q.; Jia, P. A highly specific and ultrasensitive near-infrared fluorescent probe for imaging basal hypochlorite in the mitochondria of living cells. Biosens. Bioelectron. 2018, 107, 218–223. [Google Scholar] [CrossRef]
- Lin, C.W.; Shulok, J.R.; Kirley, S.D.; Cincotta, L.; Foley, J.W. Lysosomal localization and mechanism of uptake of Nile blue photosensitizers in tumor cells. Cancer Res. 1991, 51, 2710–2719. [Google Scholar]
- Schermelleh, L.; Ferrand, A.; Huser, T.; Eggeling, C.; Sauer, M.; Biehlmaier, O.; Drummen, G.P.C. Super-resolution microscopy demystified. Nat. Cell Biol. 2019, 21, 72–84. [Google Scholar] [CrossRef]
- Zielonka, J.; Joseph, J.; Sikora, A.; Hardy, M.; Ouari, O.; Vasquez-Vivar, J.; Cheng, G.; Lopez, M.; Kalyanaraman, B. Mitochondria-Targeted Triphenylphosphonium-Based Compounds: Syntheses, Mechanisms of Action, and Therapeutic and Diagnostic Applications. Chem. Rev. 2017, 117, 10043–10120. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Taki, M.; Sato, Y.; Tamura, Y.; Yaginuma, H.; Okada, Y.; Yamaguchi, S. A photostable fluorescent marker for the superresolution live imaging of the dynamic structure of the mitochondrial cristae. Proc. Natl. Acad. Sci. USA 2019, 116, 15817–15822. [Google Scholar] [CrossRef] [PubMed]
- Goujon, A.; Colom, A.; Strakova, K.; Mercier, V.; Mahecic, D.; Manley, S.; Sakai, N.; Roux, A.; Matile, S. Mechanosensitive Fluorescent Probes to Image Membrane Tension in Mitochondria, Endoplasmic Reticulum, and Lysosomes. J. Am. Chem. Soc. 2019, 141, 3380–3384. [Google Scholar] [CrossRef] [PubMed]
- Dal Molin, M.; Verolet, Q.; Colom, A.; Letrun, R.; Derivery, E.; Gonzalez-Gaitan, M.; Vauthey, E.; Roux, A.; Sakai, N.; Matile, S. Fluorescent flippers for mechanosensitive membrane probes. J. Am. Chem. Soc. 2015, 137, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Zhang, X.; Lake, R.J.; Pawel, G.T.; Guo, Z.; Pei, R.; Lu, Y. A photo-regulated aptamer sensor for spatiotemporally controlled monitoring of ATP in the mitochondria of living cells. Chem. Sci. 2020, 11, 713–720. [Google Scholar] [CrossRef]
- Zhou, W.; Huang, P.-J.J.; Ding, J.; Liu, J. Aptamer-based biosensors for biomedical diagnostics. Analyst 2014, 139, 2627–2640. [Google Scholar] [CrossRef]
- Lan, L.; Yao, Y.; Ping, J.; Ying, Y. Recent Progress in Nanomaterial-Based Optical Aptamer Assay for the Detection of Food Chemical Contaminants. ACS Appl. Mater. Interfaces 2017, 9, 23287–23301. [Google Scholar] [CrossRef]
- Wang, T.; Chen, C.; Larcher, L.M.; Barrero, R.A.; Veedu, R.N. Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv. 2019, 37, 28–50. [Google Scholar] [CrossRef]
- Weissig, V. DQAsomes as the prototype of mitochondria-targeted pharmaceutical nanocarriers: Preparation, characterization, and use. In Mitochondrial Medicine; Springer: New York, NY, USA, 2015; pp. 1–11. [Google Scholar]
- Jimenez-Sanchez, A.; Lei, E.K.; Kelley, S.O. A Multifunctional Chemical Probe for the Measurement of Local Micropolarity and Microviscosity in Mitochondria. Angew. Chem. Int. Ed. Engl. 2018, 57, 8891–8895. [Google Scholar] [CrossRef]
- Nam, H.Y.; Song, D.; Eo, J.; Choi, N.E.; Hong, J.A.; Hong, K.T.; Lee, J.S.; Seo, J.; Lee, J. Activity-Based Probes for the High Temperature Requirement A Serine Proteases. ACS Chem. Biol. 2020. [Google Scholar] [CrossRef]
- Brandizzi, F.; Barlowe, C. Organization of the ER-Golgi interface for membrane traffic control. Nat. Rev. Mol. Cell Biol. 2013, 14, 382–392. [Google Scholar] [CrossRef]
- Verissimo, F.; Pepperkok, R. Imaging ER-to-Golgi transport: Towards a systems view. J. Cell Sci. 2013, 126, 5091–5100. [Google Scholar] [CrossRef] [PubMed]
- Kjer-Nielsen, L.; van Vliet, C.; Erlich, R.; Toh, B.-H.; Gleeson, P.A. The Golgi-targeting sequence of the peripheral membrane protein p230. J. Cell Sci. 1999, 112, 1645–1654. [Google Scholar] [PubMed]
- Liu, X.; Zheng, X.F. Endoplasmic reticulum and Golgi localization sequences for mammalian target of rapamycin. Mol. Biol. Cell 2007, 18, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Wlodkowic, D.; Skommer, J.; McGuinness, D.; Hillier, C.; Darzynkiewicz, Z. ER-Golgi network--a future target for anti-cancer therapy. Leuk. Res. 2009, 33, 1440–1447. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekharan, N.V.; Simmons, D.L. The cyclooxygenases. Genome Biol. 2004, 5, 241. [Google Scholar] [CrossRef] [PubMed]
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007, 8, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Machamer, C.E. The Golgi complex in stress and death. Front. Neurosci. 2015, 9, 421. [Google Scholar] [CrossRef] [PubMed]
- Li, S.J.; Zhou, D.Y.; Li, Y.; Liu, H.W.; Wu, P.; Ou-Yang, J.; Jiang, W.L.; Li, C.Y. Efficient Two-Photon Fluorescent Probe for Imaging of Nitric Oxide during Endoplasmic Reticulum Stress. ACS Sens. 2018, 3, 2311–2319. [Google Scholar] [CrossRef]
- Zhu, H.; Liu, C.; Liang, C.; Tian, B.; Zhang, H.; Zhang, X.; Sheng, W.; Yu, Y.; Huang, S.; Zhu, B. A new phenylsulfonamide-based Golgi-targeting fluorescent probe for H2S and its bioimaging applications in living cells and zebrafish. Chem. Commun. 2020, 56, 4086–4089. [Google Scholar] [CrossRef]
- Fan, L.; Wang, X.; Ge, J.; Li, F.; Zhang, C.; Lin, B.; Shuang, S.; Dong, C. A Golgi-targeted off-on fluorescent probe for real-time monitoring of pH changes in vivo. Chem. Commun. 2019, 55, 6685–6688. [Google Scholar] [CrossRef] [PubMed]
- Pagano, R.E.; Martin, O.C.; Kang, H.C.; Haugland, R.P. A novel fluorescent ceramide analogue for studying membrane traffic in animal cells: Accumulation at the Golgi apparatus results in altered spectral properties of the sphingolipid precursor. J. Cell Biol. 1991, 113, 1267–1279. [Google Scholar] [CrossRef] [PubMed]
- Gray-Schopfer, V.; Wellbrock, C.; Marais, R. Melanoma biology and new targeted therapy. Nature 2007, 445, 851–857. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Wang, Y.; Fu, Q.; Sun, F.; Na, N.; Ouyang, J. Melanosome-Targeting Near-Infrared Fluorescent Probe with Large Stokes Shift for in Situ Quantification of Tyrosinase Activity and Assessing Drug Effects on Differently Invasive Melanoma Cells. Anal. Chem. 2018, 90, 6206–6213. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Won, M.; Kim, J.S.; Lee, M.H. Ratiometric fluorescent probe for monitoring tyrosinase activity in melanosomes of melanoma cancer cells. Sens. Actuators B Chem. 2020, 15, 128306. [Google Scholar] [CrossRef]
- Smith, J.J.; Aitchison, J.D. Peroxisomes take shape. Nat. Rev. Mol. Cell Biol. 2013, 14, 803–817. [Google Scholar] [CrossRef]
- Brocard, C.; Hartig, A. Peroxisome targeting signal 1: Is it really a simple tripeptide? Biochim. Biophys. Acta 2006, 1763, 1565–1573. [Google Scholar] [CrossRef]
- Dansen, T.B.; Pap, E.H.W.; Wanders, R.J.; Wirtz, K.W. Targeted fluorescent probes in peroxisome function. Histochem. J. 2001, 33, 65–69. [Google Scholar] [CrossRef]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef]
- Li, X.; Liang, X.; Yin, J.; Lin, W. Organic fluorescent probes for monitoring autophagy in living cells. Chem. Soc. Rev. 2020. [Google Scholar] [CrossRef]
- Lopez, A.; Fleming, A.; Rubinsztein, D.C. Seeing is believing: Methods to monitor vertebrate autophagy in vivo. Open Biol. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Olzmann, J.A.; Carvalho, P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019, 20, 137–155. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.J.; Hsu, C.L.; Yang, J.Y.; Yang, W.Y. Monodansylpentane as a blue-fluorescent lipid-droplet marker for multi-color live-cell imaging. PLoS ONE 2012, 7, e32693. [Google Scholar] [CrossRef] [PubMed]
- Fam, T.K.; Klymchenko, A.S.; Collot, M. Recent Advances in Fluorescent Probes for Lipid Droplets. Materials 2018, 11, 1768. [Google Scholar] [CrossRef] [PubMed]
- Collot, M.; Fam, T.K.; Ashokkumar, P.; Faklaris, O.; Galli, T.; Danglot, L.; Klymchenko, A.S. Ultrabright and Fluorogenic Probes for Multicolor Imaging and Tracking of Lipid Droplets in Cells and Tissues. J. Am. Chem. Soc. 2018, 140, 5401–5411. [Google Scholar] [CrossRef] [PubMed]
- Tatenaka, Y.; Kato, H.; Ishiyama, M.; Sasamoto, K.; Shiga, M.; Nishitoh, H.; Ueno, Y. Monitoring Lipid Droplet Dynamics in Living Cells by Using Fluorescent Probes. Biochemistry 2019, 58, 499–503. [Google Scholar] [CrossRef]
- Xia, T.; Li, N.; Fang, X. Single-molecule fluorescence imaging in living cells. Annu. Rev. Phys. Chem. 2013, 64, 459–480. [Google Scholar] [CrossRef]
- Debie, P.; Hernot, S. Emerging Fluorescent Molecular Tracers to Guide Intra-Operative Surgical Decision-Making. Front. Pharm. 2019, 10, 510. [Google Scholar] [CrossRef]


| Entry | Organelle | Targeting Moiety | Detecting Analytes | λex/λem (nm) | Ref. |
|---|---|---|---|---|---|
| 1 | Lysosome | morpholine | H2S Cys/HCy GSH | 580/602 (H2S) 376/480 (Cys/HCy) 438/540 (GSH) | [6] |
| 2 | Lysosome | morpholine | H2S | 410/550 | [9] |
| 3 | Lysosome | monothio-bishydrazide | HClO | 410/480 | [10] |
| 4 | Lysosome | morpholine | HClO | 405/505 | [11] |
| 5 | Lysosome | morpholine | HClO | 480/525 | [12] |
| 6 | Lysosome | morpholine | SO2 | 450/535 | [13] |
| 7 | Lysosome | benzothiazolium | pH | 415/694 | [16] |
| 8 | Lysosome | hemicyanine | pH | 635/730 | [17] |
| 9 | Nucleus | rhodium complex | mismatched DNA | 520/570 | [19] |
| 10 | Nucleus | benzothiazole | DNA G-quadruplex | 405/635 | [21] |
| 11 | Nucleus | Hoechst | AT-rich region in DNA | 352/455 | [24] |
| 12 | Nucleus | Hoechst | AT-rich region in DNA | 355/455 | [25] |
| 13 | Nucleus | Py-Im polyamide | telomeres | 645/665 | [30] |
| 14 | Nucleus | NLS a | integrin and CD13 | 450/560 | [31] |
| 15 | Nucleus | NLS a | H2O2 | 353/551 | [32] |
| 16 | Membrane | cholesterol | membrane structure | 465/550 | [39] |
| 17 | Membrane | alkyl chain (C = 8–18) | Zn2+ | 405/525 | [40] |
| 18 | Membrane | bis-sulfonic acids | membrane structure | 700/720 | [41] |
| 19 | Membrane | alkyl chain (C = 17) | membrane structure | 642/700 | [42] |
| 20 | Membrane | Nile Red | lipid composition | 540/620 | [45] |
| 21 | Membrane | cholesterol | Zn2+ | 622/663 | [46] |
| 22 | Mitochondria | quinoline | H2O2 | 340/594 | [55] |
| 23 | Mitochondria | N-methylpyridine | H2O2 | 405/669 | [56] |
| 24 | Mitochondria | hemicyanine | ClO− | 455/632 | [57] |
| 25 | Mitochondria | Nile Blue | ClO− | 600/672 | [58] |
| 26 | Mitochondria | TPP a | mitochondrial ultrastructure | 488/(λSTED) 660 | [62] |
| 27 | Mitochondria | TPP a | mitochondrial membrane tension | 430/570 | [64] |
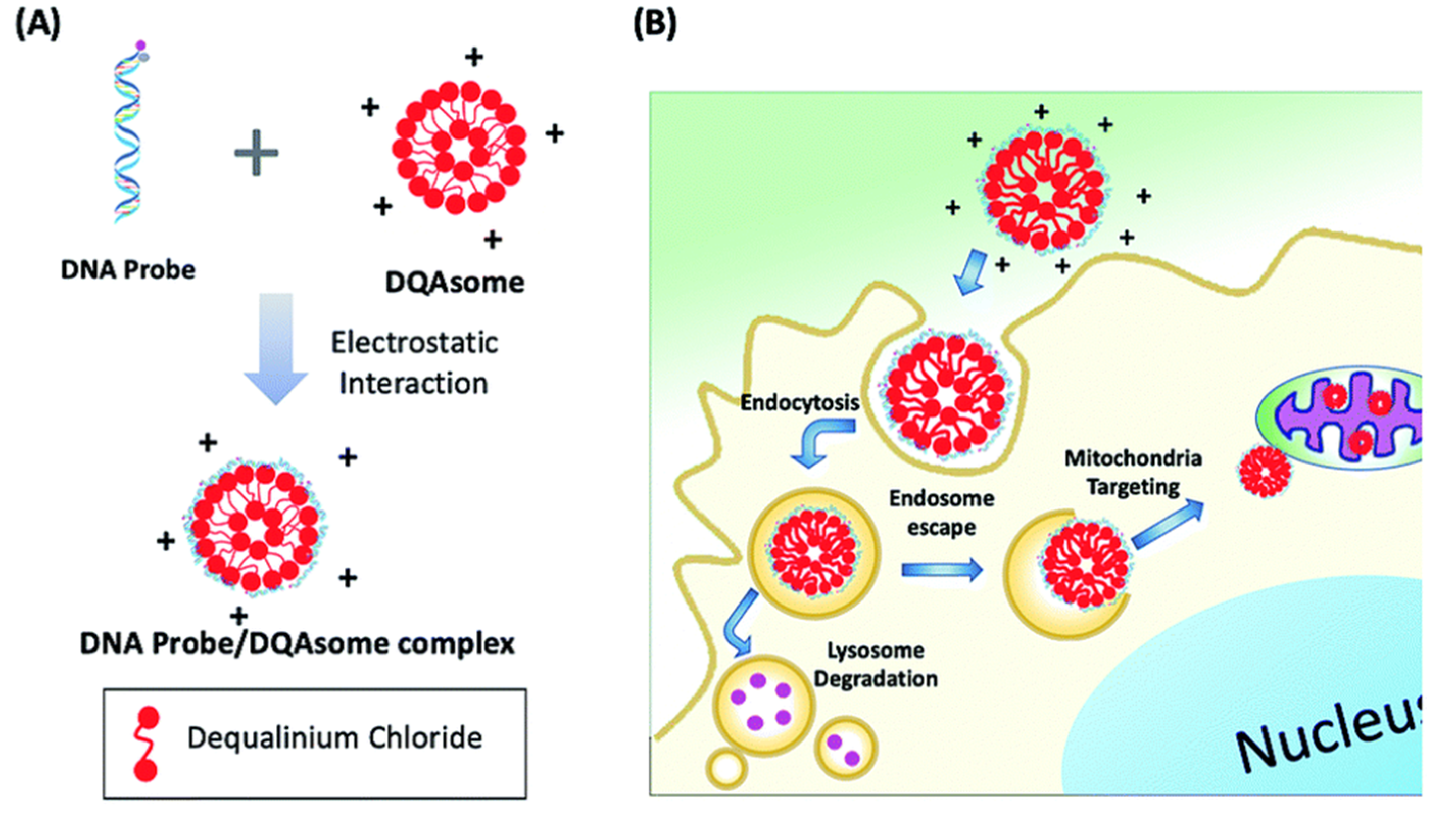
| 28 | Mitochondria | DQAsome | ATP | 530/565 | [65] |
| 29 | Mitochondria | mitochondria-penetrating peptide | mitochondrial polarity and viscosity | 320/485 (polarity) 320/550 (viscosity) | [70] |
| 30 | Mitochondria | mitochondria-targeting peptoid | mitochondrial serine protease activity | 494/512 | [71] |
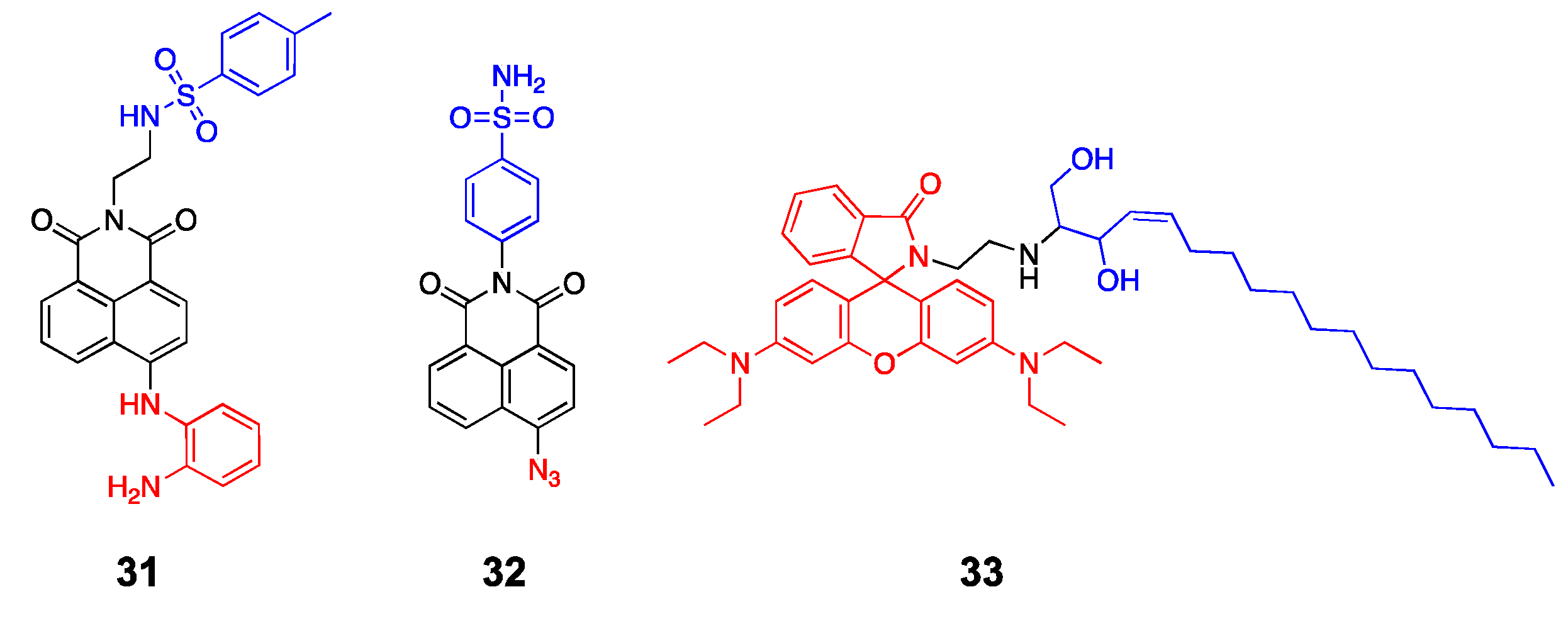
| 31 | ER | p-toluenesulfonamide | NO | 440/538 | [80] |
| 32 | Golgi | phenylsulfonamide | H2S | 440/550 | [81] |
| 33 | Golgi | sphingosine | pH | 577/600 | [82] |
| 34 | Melanosome | m-hydroxybenzyl | tyrosinase activity | 500/675 | [85] |
| 35 | Melanosome | 3-hydroxybenzyl | tyrosinase activity | 405/460 | [86] |
| 36 | Peroxisome | acetyl-CKGGAKL | peroxisome biogenesis | 530/550 | [89] |
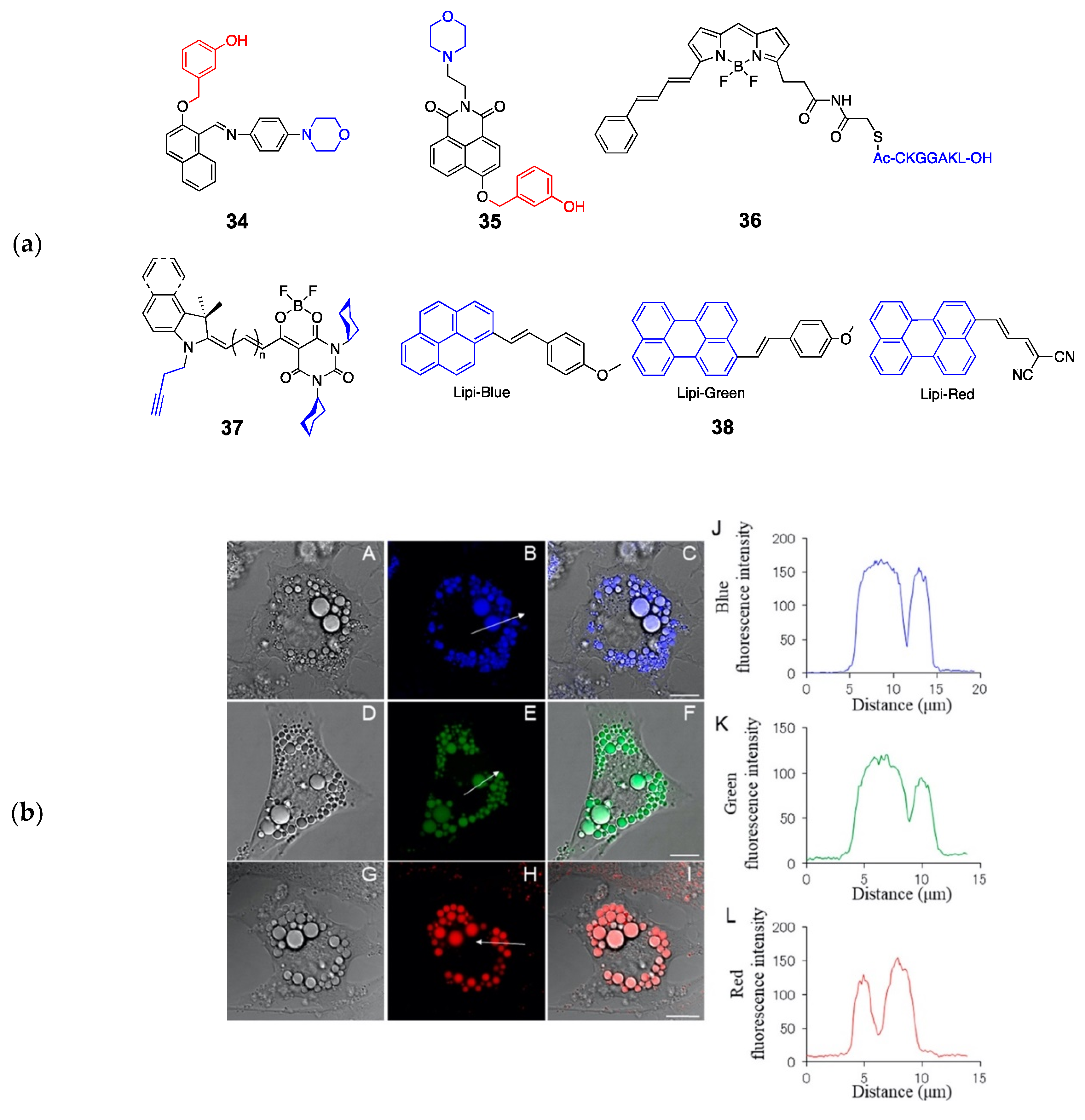
| 37 | LD | cyclohexyl | neutral lipids | 526-770/550-794 (multicolor) | [96] |
| 38 | LD | pyrene, perylene | neutral lipids | 381-520/445-650 (multicolor) | [97] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, N.-E.; Lee, J.-Y.; Park, E.-C.; Lee, J.-H.; Lee, J. Recent Advances in Organelle-Targeted Fluorescent Probes. Molecules 2021, 26, 217. https://doi.org/10.3390/molecules26010217
Choi N-E, Lee J-Y, Park E-C, Lee J-H, Lee J. Recent Advances in Organelle-Targeted Fluorescent Probes. Molecules. 2021; 26(1):217. https://doi.org/10.3390/molecules26010217
Chicago/Turabian StyleChoi, Na-Eun, Ji-Yu Lee, Eun-Chae Park, Ju-Hee Lee, and Jiyoun Lee. 2021. "Recent Advances in Organelle-Targeted Fluorescent Probes" Molecules 26, no. 1: 217. https://doi.org/10.3390/molecules26010217
APA StyleChoi, N.-E., Lee, J.-Y., Park, E.-C., Lee, J.-H., & Lee, J. (2021). Recent Advances in Organelle-Targeted Fluorescent Probes. Molecules, 26(1), 217. https://doi.org/10.3390/molecules26010217






