Discovery of Monoacylglycerol Lipase (MAGL) Inhibitors Based on a Pharmacophore-Guided Virtual Screening Study
Abstract
1. Introduction
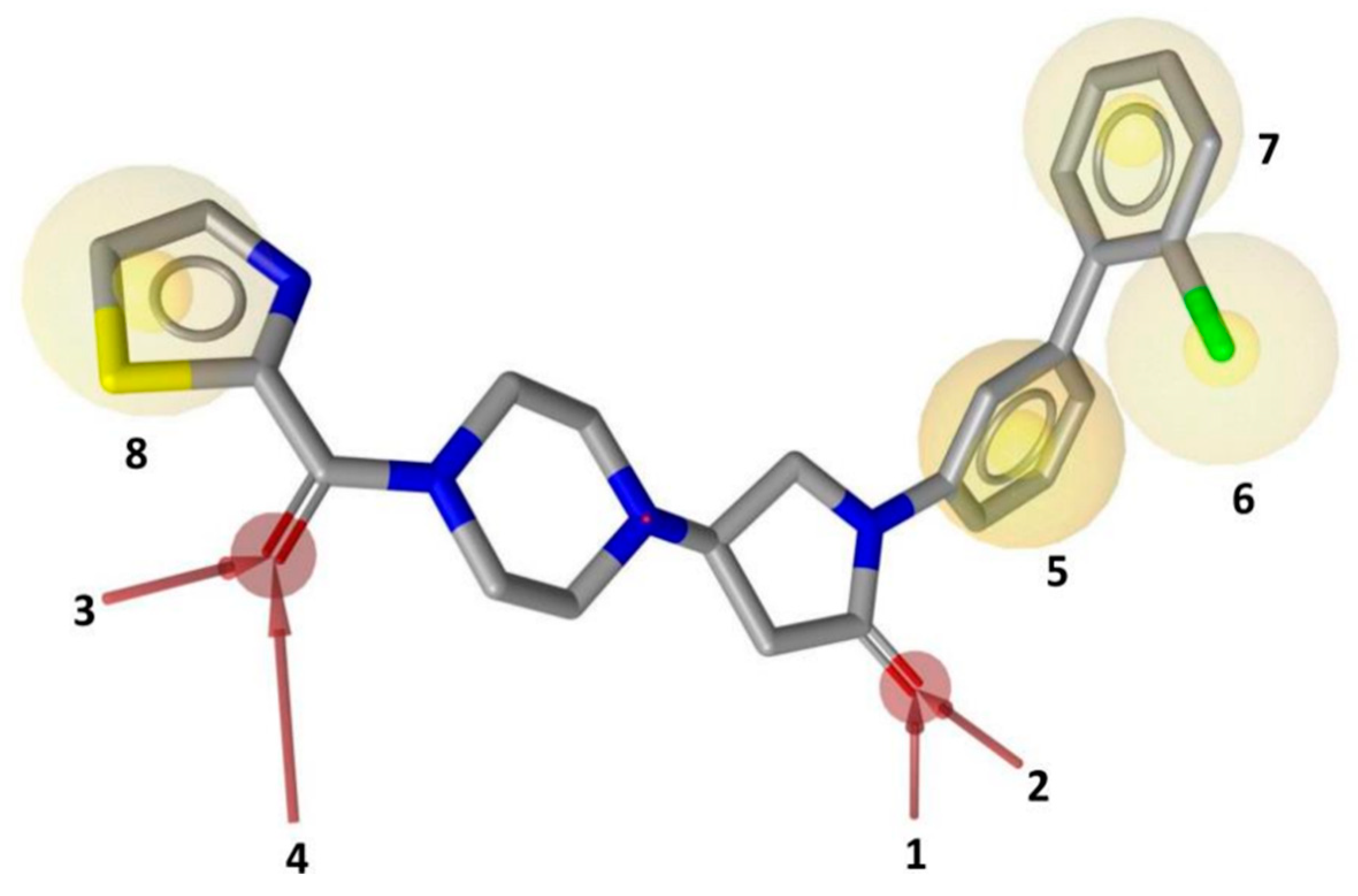
2. Results
3. Conclusions
4. Materials and Methods
4.1. Pharmacophore Model Generation
4.2. Database Generation and Pharmacophore Screening
4.3. Molecular Docking and Pose Filtering
4.4. Molecular Dynamics Simulations
4.5. Similarity Search
4.6. Enzymatic Assays
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Pertwee, R.G. Cannabinoid pharmacology: The first 66 years. Br. J. Pharmacol. 2006, 147, S163–S171. [Google Scholar] [CrossRef] [PubMed]
- Zoerner, A.A.; Gutzki, F.-M.; Batkai, S.; May, M.; Rakers, C.; Engeli, S.; Jordan, J.; Tsikas, D. Quantification of endocannabinoids in biological systems by chromatography and mass spectrometry: A comprehensive review from an analytical and biological perspective. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2011, 1811, 706–723. [Google Scholar] [CrossRef]
- Gonsiorek, W.; Lunn, C.; Fan, X.; Narula, S.; Lundell, D.; Hipkin, R.W. Endocannabinoid 2-arachidonyl glycerol is a full agonist through human type 2 cannabinoid receptor: Antagonism by anandamide. Mol. Pharmacol. 2000, 57, 1045–1050. [Google Scholar] [PubMed]
- Pertwee, R.G.; Howlett, A.C.; Abood, M.E.; Alexander, S.P.H.; Di Marzo, V.; Elphick, M.R.; Greasley, P.J.; Hansen, H.S.; Kunos, G.; Mackie, K.; et al. International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid Receptors and Their Ligands: Beyond CB1 and CB2. Pharmacol. Rev. 2010, 62, 588–631. [Google Scholar] [CrossRef] [PubMed]
- Blankman, J.L.; Simon, G.M.; Cravatt, B.F. A Comprehensive Profile of Brain Enzymes that Hydrolyze the Endocannabinoid 2-Arachidonoylglycerol. Chem. Biol. 2007, 14, 1347–1356. [Google Scholar] [CrossRef] [PubMed]
- Granchi, C.; Bononi, G.; Ferrisi, R.; Gori, E.; Mantini, G.; Glasmacher, S.; Poli, G.; Palazzolo, S.; Caligiuri, I.; Rizzolio, F.; et al. Design, synthesis and biological evaluation of second-generation benzoylpiperidine derivatives as reversible monoacylglycerol lipase (MAGL) inhibitors. Eur. J. Med. Chem. 2021, 209, 112857. [Google Scholar] [CrossRef]
- Nomura, D.K.; Morrison, B.E.; Blankman, J.L.; Long, J.Z.; Kinsey, S.G.; Marcondes, M.C.G.; Ward, A.M.; Hahn, Y.K.; Lichtman, A.H.; Conti, B.; et al. Endocannabinoid hydrolysis generates brain prostaglandins that promote neuroinflammation. Science 2011, 334, 809–813. [Google Scholar] [CrossRef]
- Nomura, D.K.; Long, J.Z.; Niessen, S.; Hoover, H.S.; Ng, S.W.; Cravatt, B.F. Monoacylglycerol Lipase Regulates a Fatty Acid Network that Promotes Cancer Pathogenesis. Cell 2010, 140, 49–61. [Google Scholar] [CrossRef]
- Granchi, C.; Caligiuri, I.; Minutolo, F.; Rizzolio, F.; Tuccinardi, T. A patent review of Monoacylglycerol Lipase (MAGL) inhibitors (2013–2017). Expert Opin. Ther. Pat. 2017, 27, 1341–1351. [Google Scholar] [CrossRef]
- Pacher, P.; Bátkai, S.; Kunos, G. The Endocannabinoid System as an Emerging Target of Pharmacotherapy. Pharmacol. Rev. 2006, 58, 389–462. [Google Scholar] [CrossRef]
- Deng, H.; Li, W. Monoacylglycerol lipase inhibitors: Modulators for lipid metabolism in cancer malignancy, neurological and metabolic disorders. Acta Pharm. Sin. B 2020, 10, 582–602. [Google Scholar] [CrossRef] [PubMed]
- Chanda, P.K.; Gao, Y.; Mark, L.; Btesh, J.; Strassle, B.W.; Lu, P.; Piesla, M.J.; Zhang, M.-Y.; Bingham, B.; Uveges, A.; et al. Monoacylglycerol Lipase Activity Is a Critical Modulator of the Tone and Integrity of the Endocannabinoid System. Mol. Pharmacol. 2010, 78, 996–1003. [Google Scholar] [CrossRef] [PubMed]
- Schlosburg, J.E.; Blankman, J.L.; Long, J.Z.; Nomura, D.K.; Pan, B.; Kinsey, S.G.; Nguyen, P.T.; Ramesh, D.; Booker, L.; Burston, J.J.; et al. Chronic monoacylglycerol lipase blockade causes functional antagonism of the endocannabinoid system. Nat. Neurosci. 2010, 13, 1113–1119. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Torres, G.; Cipriano, M.; Hedén, E.; Björklund, E.; Canales, A.; Zian, D.; Feliú, A.; Mecha, M.; Guaza, C.; Fowler, C.J.; et al. A reversible and selective inhibitor of monoacylglycerol lipase ameliorates multiple sclerosis. Angew. Chemie-Int. Ed. 2014, 53, 13765–13770. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.Z.; Ahenkorah, S.; Vaara, M.; Staszewski, M.; Adams, Y.; Laitinen, T.; Navia-Paldanius, D.; Parkkari, T.; Savinainen, J.R.; Walczyński, K.; et al. Loratadine analogues as MAGL inhibitors. Bioorg. Med. Chem. Lett. 2015, 25, 1436–1442. [Google Scholar] [CrossRef] [PubMed]
- Aida, J.; Fushimi, M.; Kusumoto, T.; Sugiyama, H.; Arimura, N.; Ikeda, S.; Sasaki, M.; Sogabe, S.; Aoyama, K.; Koike, T. Design, Synthesis, and Evaluation of Piperazinyl Pyrrolidin-2-ones as a Novel Series of Reversible Monoacylglycerol Lipase Inhibitors. J. Med. Chem. 2018, 61, 9205–9217. [Google Scholar] [CrossRef]
- Granchi, C.; Lapillo, M.; Glasmacher, S.; Bononi, G.; Licari, C.; Poli, G.; El Boustani, M.; Caligiuri, I.; Rizzolio, F.; Gertsch, J.; et al. Optimization of a Benzoylpiperidine Class Identifies a Highly Potent and Selective Reversible Monoacylglycerol Lipase (MAGL) Inhibitor. J. Med. Chem. 2019, 62, 1932–1958. [Google Scholar] [CrossRef]
- Zhu, B.; Connolly, P.J.; Zhang, Y.-M.; McDonnell, M.E.; Bian, H.; Lin, S.-C.; Liu, L.; Zhang, S.-P.; Chevalier, K.M.; Brandt, M.R.; et al. The discovery of azetidine-piperazine di-amides as potent, selective and reversible monoacylglycerol lipase (MAGL) inhibitors. Bioorg. Med. Chem. Lett. 2020, 30, 127243. [Google Scholar] [CrossRef]
- Granchi, C.; Rizzolio, F.; Palazzolo, S.; Carmignani, S.; Macchia, M.; Saccomanni, G.; Manera, C.; Martinelli, A.; Minutolo, F.; Tuccinardi, T. Structural Optimization of 4-Chlorobenzoylpiperidine Derivatives for the Development of Potent, Reversible, and Selective Monoacylglycerol Lipase (MAGL) Inhibitors. J. Med. Chem. 2016, 59, 10299–10314. [Google Scholar] [CrossRef]
- Wolber, G.; Langer, T. LigandScout: 3-D Pharmacophores Derived from Protein-Bound Ligands and Their Use as Virtual Screening Filters. J. Chem. Inf. Model. 2005, 45, 160–169. [Google Scholar] [CrossRef]
- Tuccinardi, T.; Poli, G.; Romboli, V.; Giordano, A.; Martinelli, A. Extensive consensus docking evaluation for ligand pose prediction and virtual screening studies. J. Chem. Inf. Model. 2014, 54, 2980–2986. [Google Scholar] [CrossRef] [PubMed]
- Poli, G.; Martinelli, A.; Tuccinardi, T. Reliability analysis and optimization of the consensus docking approach for the development of virtual screening studies. J. Enzyme Inhib. Med. Chem. 2016, 31, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Poli, G.; Seidel, T.; Langer, T. Conformational Sampling of Small Molecules With iCon: Performance Assessment in Comparison With OMEGA. Front. Chem. 2018, 6, 229. [Google Scholar] [CrossRef] [PubMed]
- Pini, E.; Poli, G.; Tuccinardi, T.; Chiarelli, L.; Mori, M.; Gelain, A.; Costantino, L.; Villa, S.; Meneghetti, F.; Barlocco, D. New Chromane-Based Derivatives as Inhibitors of Mycobacterium tuberculosis Salicylate Synthase (MbtI): Preliminary Biological Evaluation and Molecular Modeling Studies. Molecules 2018, 23, 1506. [Google Scholar] [CrossRef]
- Chiarelli, L.R.; Mori, M.; Barlocco, D.; Beretta, G.; Gelain, A.; Pini, E.; Porcino, M.; Mori, G.; Stelitano, G.; Costantino, L.; et al. Discovery and development of novel salicylate synthase (MbtI) furanic inhibitors as antitubercular agents. Eur. J. Med. Chem. 2018, 155, 754–763. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E., III; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Tuccinardi, T.; Granchi, C.; Rizzolio, F.; Caligiuri, I.; Battistello, V.; Toffoli, G.; Minutolo, F.; Macchia, M.; Martinelli, A. Identification and characterization of a new reversible MAGL inhibitor. Bioorganic Med. Chem. 2014, 22, 3285–3291. [Google Scholar] [CrossRef]




| Number of Matched Features | Number of Compounds |
|---|---|
| 8 | 5707 |
| 7 | 19,314 |
| 6 | 182,236 |
| 5 | 276,150 |
| Docking Method | RMSD (Å) |
|---|---|
| Autodock 4.2 | 1.5 |
| Dock6 | 0.5 |
| Fred | 3.8 |
| Glamdock | 1.3 |
| Glide SP | 0.6 |
| Glide XP | 1.5 |
| Gold ASP | 1.4 |
| Gold Chemscore | 3.1 |
| Gold Goldscore | 0.6 |
| Gold PLP | 0.7 |
| Plants | 3.2 |
| rDock | 1.6 |
| Vina | 9.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jha, V.; Biagi, M.; Spinelli, V.; Di Stefano, M.; Macchia, M.; Minutolo, F.; Granchi, C.; Poli, G.; Tuccinardi, T. Discovery of Monoacylglycerol Lipase (MAGL) Inhibitors Based on a Pharmacophore-Guided Virtual Screening Study. Molecules 2021, 26, 78. https://doi.org/10.3390/molecules26010078
Jha V, Biagi M, Spinelli V, Di Stefano M, Macchia M, Minutolo F, Granchi C, Poli G, Tuccinardi T. Discovery of Monoacylglycerol Lipase (MAGL) Inhibitors Based on a Pharmacophore-Guided Virtual Screening Study. Molecules. 2021; 26(1):78. https://doi.org/10.3390/molecules26010078
Chicago/Turabian StyleJha, Vibhu, Marzia Biagi, Valeria Spinelli, Miriana Di Stefano, Marco Macchia, Filippo Minutolo, Carlotta Granchi, Giulio Poli, and Tiziano Tuccinardi. 2021. "Discovery of Monoacylglycerol Lipase (MAGL) Inhibitors Based on a Pharmacophore-Guided Virtual Screening Study" Molecules 26, no. 1: 78. https://doi.org/10.3390/molecules26010078
APA StyleJha, V., Biagi, M., Spinelli, V., Di Stefano, M., Macchia, M., Minutolo, F., Granchi, C., Poli, G., & Tuccinardi, T. (2021). Discovery of Monoacylglycerol Lipase (MAGL) Inhibitors Based on a Pharmacophore-Guided Virtual Screening Study. Molecules, 26(1), 78. https://doi.org/10.3390/molecules26010078


















