Abstract
Anion binding and extraction from solutions is currently a dynamic research topic in the field of supramolecular chemistry. A particularly challenging task is the extraction of anions with large hydration energies, such as the carbonate ion. Carbonate-binding complexes are also receiving increased interest due to their relevance to atmospheric CO2 fixation. Nanojars are a class of self-assembled, supramolecular coordination complexes that have been shown to bind highly hydrophilic anions and to extract even the most hydrophilic ones, including carbonate, from water into aliphatic solvents. Here we present an expanded nanojar that is able to bind two carbonate ions, thus doubling the previously reported carbonate-binding capacity of nanojars. The new nanojar is characterized by detailed single-crystal X-ray crystallographic studies in the solid state and electrospray ionization mass spectrometric (including tandem MS/MS) studies in solution.
1. Introduction
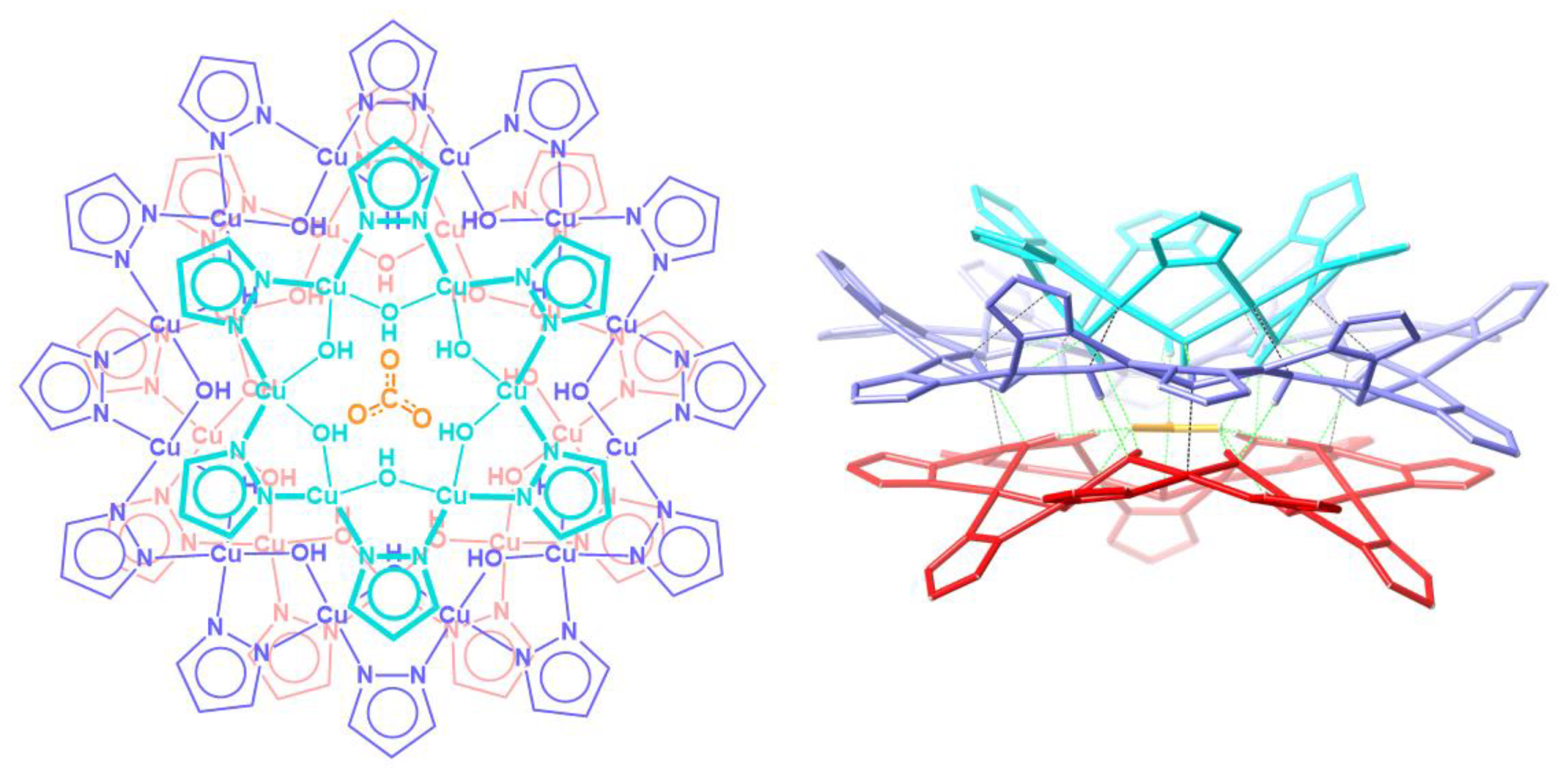
Nanojars are a family of supramolecular coordination complexes that form from a solution of Cu2+, OH− and pyrazolate (pz = C3H3N2−) ions in the presence of a hydrophilic anion, such as carbonate [1], sulfate [2], phosphate [3], arsenate [3] or chloride [4]. The anion templates the formation of {cis-CuII(μ-OH)(μ-pz)}x metallamacrocycles (x = 6–14, except 11). Three (in the case of carbonate, sulfate, phosphate, arsenate) or four (in the case of chloride) of these metallamacrocycles self-assemble around a central anion into nanojars of the formula [anion⊂{CuII(μ-OH)(μ-pz)}n] (n = 27–33), via inter-metallamacrocycle and anion-metallamacrocycle hydrogen bonding, as well as inter-metallamacrocycle Cu∙∙∙O interactions. The incarcerated anion appears to be crucial for the formation of nanojars, as the neutral nanojar host does not exist on its own without an anion guest. Figure 1 illustrates the structure of the nanojar with n = 27.
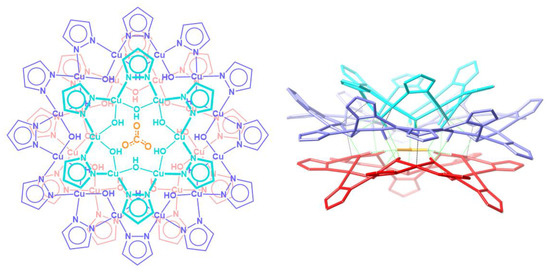
Figure 1.
Schematic representation (left, top-view) of the crystal structure (right, side-view) of the [CO3⊂{CuII(μ-OH)(μ-pz)}6 + 12 + 9]2− nanojar [1].
The recognition and binding of anions has been receiving increased interest in recent years [5,6,7], as the supramolecular binding of anions finds applications in anion sensing, extraction and separation of anions, transmembrane anion transport and anion-driven architectonics and organocatalysis [8]. We have recently shown that nanojars bind the incarcerated oxoanions (carbonate, sulfate, phosphate, arsenate) with unprecedented strength by wrapping a multitude of hydrogen bonds around the anion and totally isolating it from its surrounding medium (as in the sulfate [9] and phosphate [10] binding proteins). Indeed, an aqueous Ba2+ solution is unable to precipitate the corresponding barium salt (e.g., BaSO4, Ksp = 1.08 × 10−10 at 25 °C in H2O) when stirred with a solution of the nanojars. We have also demonstrated that nanojars are able to transfer these anions, including one of the most hydrophilic ones, carbonate, from water into aliphatic solvents [11]. Thus, nanojars can be used as extraction agents for the removal of such anions from contaminated aqueous media by liquid–liquid extraction [12].
Herein we report the serendipitous discovery that upon addition of 1,10-phenathroline into the nanojar-forming reaction mixture, expanded nanojars form that bind two carbonate ions instead of one, thus doubling the carbonate-binding capacity of nanojars. As described below, the binding of the second carbonate ion by four copper-centers (μ4-CO3) provides for an interesting new example of an inverse coordination complex, wherein the bridging ligand is the coordination center surrounded by metal ions [13,14,15,16,17,18,19,20].
2. Results and Discussion
Nanojars have so far been characterized almost exclusively with tetrabutylammonium as counterion, such as in (Bu4N+)2[CO32−⊂{CuII(μ-OH)(μ-pz)}6+12+9] (1), with the single exception of (K+ ⊂ 18-crown-6)2[SO42−⊂{CuII(μ-OH)(μ-pz)}8+14+9] [2]. In an attempt to prepare nanojars with [Cu(phen)3]2+ formed in situ as the countercation, a mixture of CuSO4·5H2O, pyrazole, NaOH and 1,10-phenanthroline (phen) was stirred in tetrahydrofuran (THF). After filtration and evaporation, the resulting product was crystallized from a nitrobenzene solution by heptane vapor diffusion. Instead of the expected [Cu(phen)3][SO4⊂{CuII(μ-OH)(μ-pz)}n], X-ray crystallography showed the formation of neutral, expanded nanojars, [CO3⊂{Cu29(μ-OH)27(μ-pz)27(phen)2(μ4-CO3)(H2O)}] (2). Atmospheric CO2 was apparently absorbed during the reaction, leading to the binding of two CO32− ions by the resulting nanojar. After rationalizing the obtained structure, the reaction was also repeated using Cu(NO3)2·2.5H2O and Na2CO3·H2O instead of CuSO4·5H2O.
2.1. Crystallographic Description
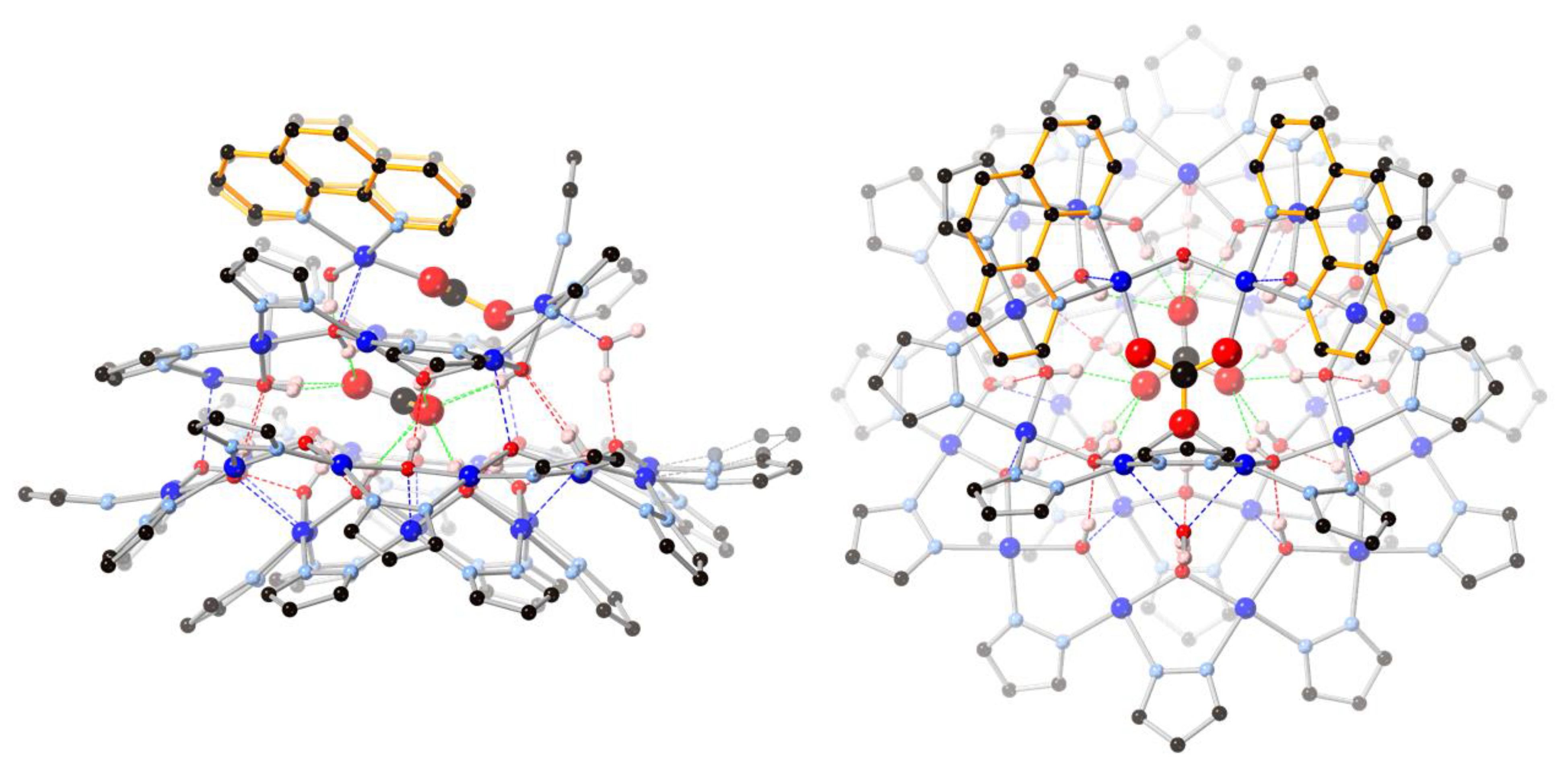
Located on a general position, nanojar 2 (triclinic, Pī) has pseudo-mirror symmetry (Figure 2). Its structure is closely related to that of 1, in which three neutral [cis-CuII(μ-OH)(μ-pz)]n rings, with a larger one (n = 12) sandwiched by two smaller ones (n = 6 and 9), define the nanojar, with its cavity occupied by an incarcerated carbonate ion (Figure 1). The same Cu6 + Cu12 + Cu9 ring combination is found in both 1 and 2, with the exception that one OH− group of the Cu9-ring in 2 is replaced by an O-atom of a second CO32− ion. The central, larger ring is approximately flat, with the pyrazolate units symmetrically alternating slightly above and below the ring mean-plane and not forming hydrogen bonds to the carbonate ion. The smaller side-rings are bowl-shaped, with their pyrazolate moieties pointing away from the central ring and their OH groups pointing toward the center of the nanojar and forming multiple hydrogen bonds with the incarcerated CO32− ion. Although there is no direct bonding between the two smaller rings, they are both involved in multiple H-bonds and weak axial Cu–O interactions with the larger central ring. In the [cis-CuII(μ-OH)(μ-pz)]n rings, Cu–O and Cu–N bond-lengths are within normal ranges, 1.893(3)–2.007(3) and 1.943(6)–2.06(2) Å, respectively (Table 1). While in 1 the 2—charge of the incarcerated carbonate ion is balanced by two Bu4N+ counterions, in 2 it is the additional bonded [Cu2(phen)2CO3]2+ moiety that renders the assembly neutral.
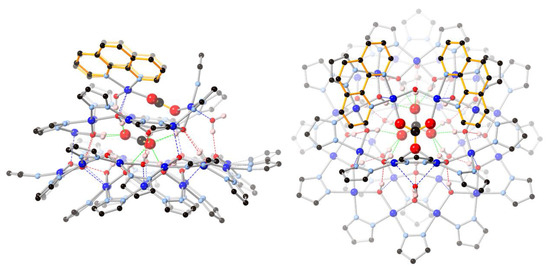
Figure 2.
Ball-and-stick representation of the crystal structure of 2(C6H5NO2)6.74(C7H16)0.76: (left) side-view; (right) top-view. Color code: dark blue—Cu; light blue—N; red—O; black—C; pink—H. The 1,10-phenanthroline moieties are highlighted in orange. The nitrobenzene/heptane solvent molecules and C–H hydrogen atoms are omitted for clarity, and only the major components of the disordered carbonate and pyrazolate moieties are shown.

Table 1.
Selected bond lengths for 2.
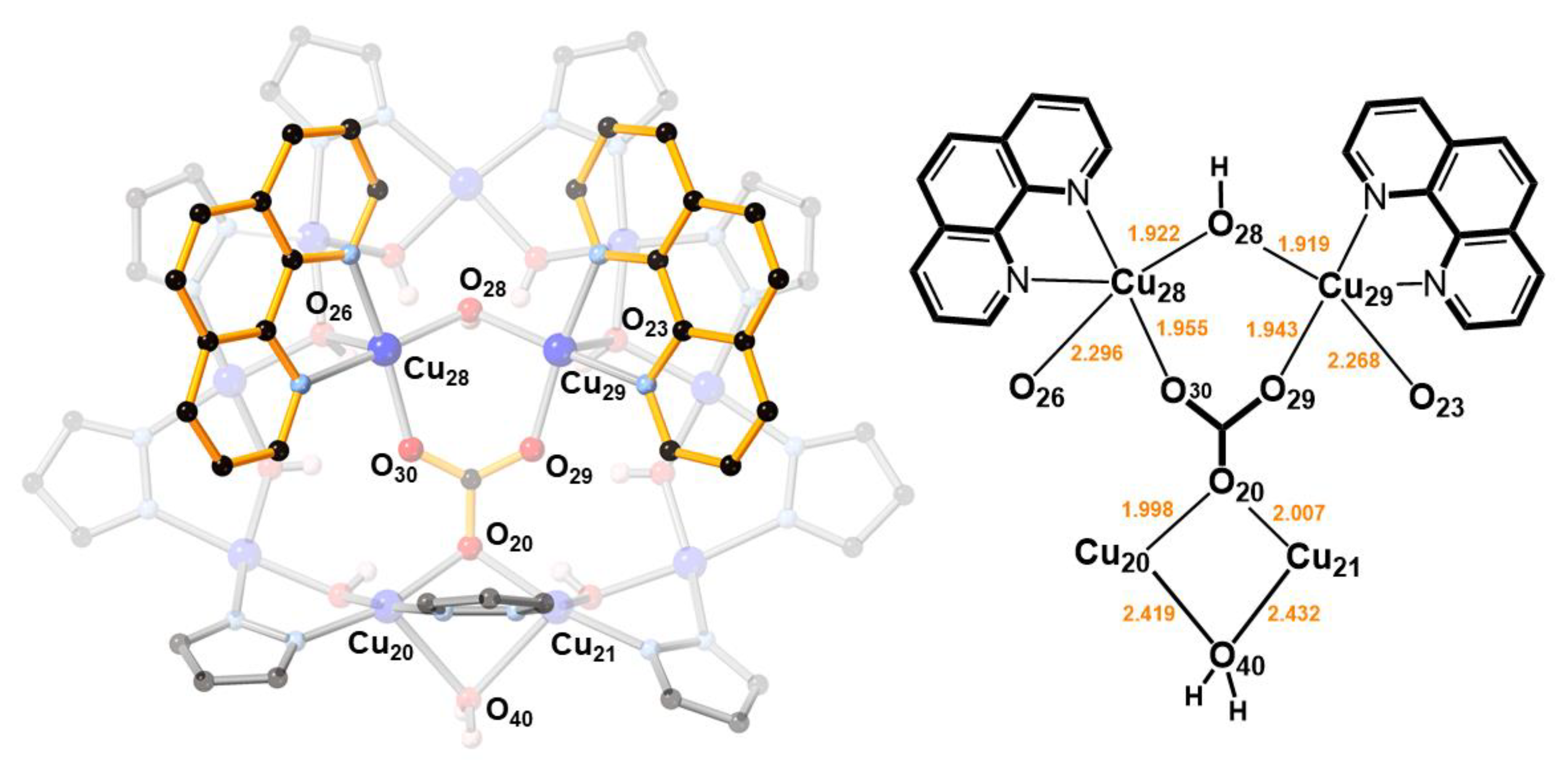
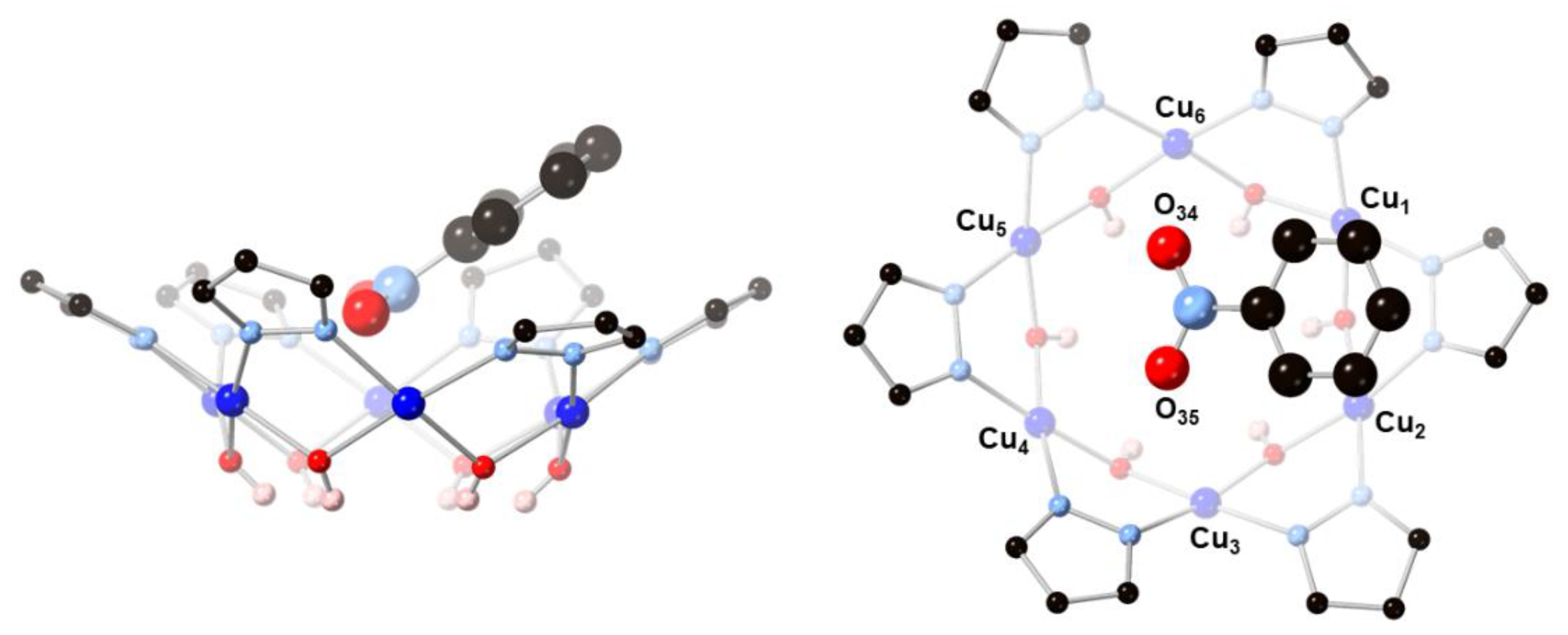
In 2, two O-atoms of the additional carbonate ion are bound to two CuII(1,10-phen) units, which are bridged by a OH− group (O28) and form weak Cu–O bonds (2.296(3) and 2.268(3) Å) with the Cu9-ring (Figure 3). The OH− group (O28) is H-bonded to the central carbonate ion. As a consequence of binding the second carbonate ion, a pyrazolate group of the Cu9-ring is pulled away from the Cu12-ring, opening up a cavity that becomes occupied by a water molecule. This H2O molecule bridges two Cu-atoms of the Cu9-ring (Cu···O: 2.419(3) and 2.432(3) Å) and donates a H-bond to a OH-group of the Cu12-ring (O40···O8: 2.684(5) Å).
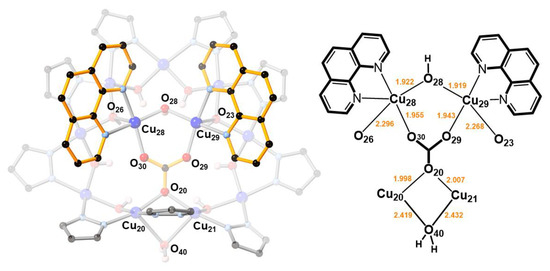
Figure 3.
A [Cu2(phen)2CO3]2+ moiety binds to the [Cu(μ-OH)(μ-pz)]9 ring in 2 by replacing a OH− group (O28) with an O-atom of its CO32− ion (O20), and by two axial Cu–O bonds (Cu28–O26 and Cu29–O23).
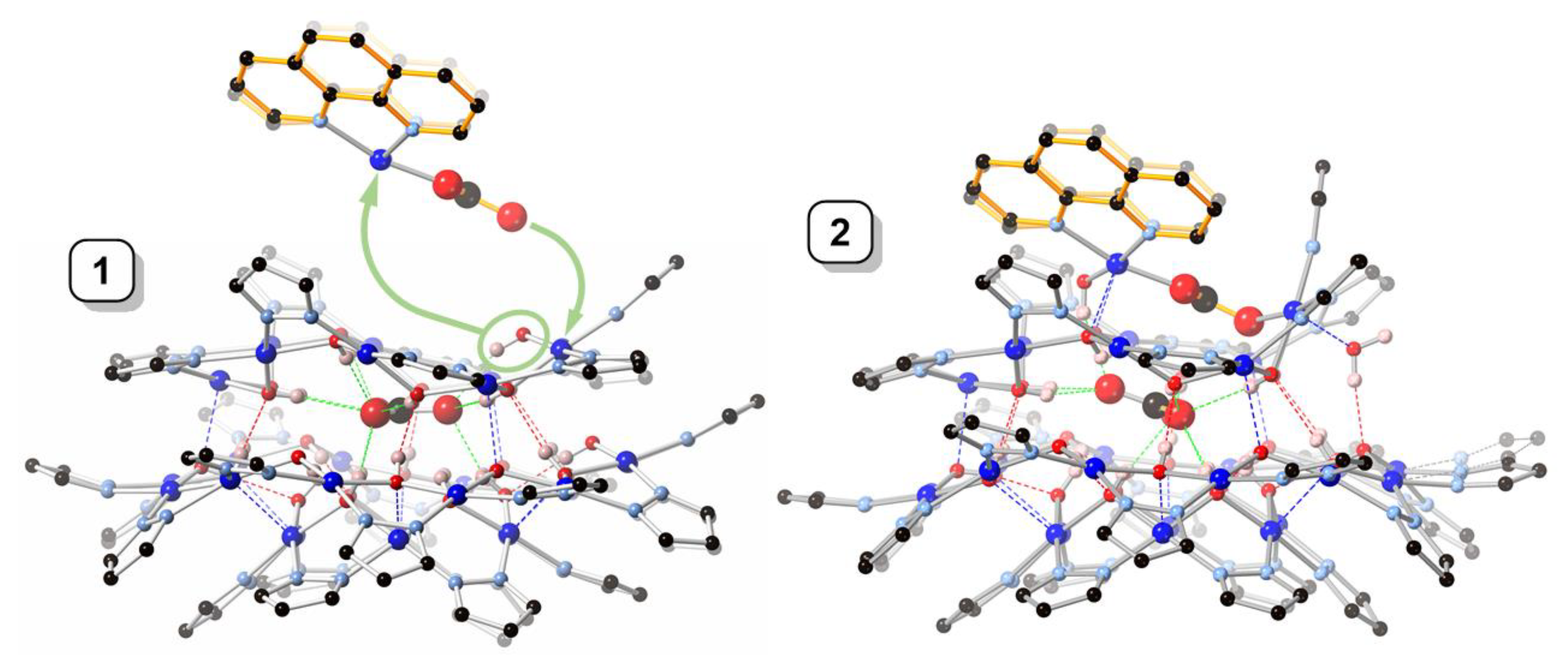
While in 1 the central carbonate ion is approximately parallel to the [Cu(μ-OH)(μ-pz)]n rings, in 2 it is found tilted: the angle between the CO32− and Cu12-ring mean-planes is 2.2(1)° in 1 and 22.2(2)° in 2 (Figure 4). As a consequence of the tilting, some of the H-bonding distances to CO32− in 2 (Table 2) are shorter (down to 2.657(5) Å) and others are longer (up to 3.088(5) Å) than in 1 (2.746(5)–2.915(5) Å). Nonetheless, the average of the twelve H-bonds to carbonate (four to each O-atom) is virtually identical in 1 (2.842(5) Å) and 2 (2.838(5) Å). The second CO32− ion in 2 is coordinate-covalently bound to the Cu9 ring and the two additional Cu-atoms, almost parallel to the central CO32− ion (angle between mean-planes: 6.6(2)°), with a C···C separation of only 3.071(6) Å. Another very closely-spaced, head-to-head pair of CO32− ions (C···C: 3.664(1), O···O: 1.946(1) Å) has been reported in which both CO32− ions are bound to multiple metal centers [21]. The tetranuclear Cu4(μ4-CO3) moiety has also been reported with a few other ligand systems [22,23,24,25,26,27,28,29].
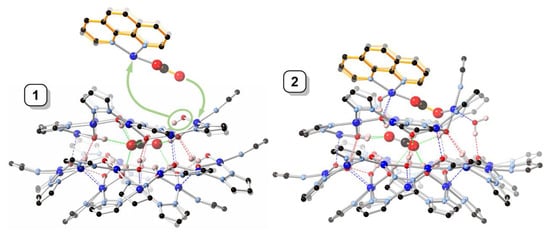
Figure 4.
Comparison of the crystal structures of 1 and 2, illustrating the formation of 2 from 1 by incorporation of a [Cu2(phen)2CO3]2+ moiety. Color code: dark blue—Cu; light blue—N; red—O; black—C; pink—H. The 1,10-phenanthroline moieties are highlighted in orange.

Table 2.
Hydrogen bonding data for 2. O1–O6: Cu6-ring; O7–O18: Cu12-ring; O19–O27: Cu9-ring; O28: Cu2(OH)(phen)2 unit; O31–O33 and O931–O933: central carbonate (disordered, 91/9).
As in 1, the OH-groups of the Cu12-ring in 2 donate twelve alternating H-bonds, six to the Cu6-ring (O···O: 2.716(4)–2.786(4) Å, average: 2.742(5) Å) and six to the Cu9-ring (O···O: 2.721(4)–2.914(5) Å, average: 2.780(5) Å), with an overall average of 2.762(4) Å. The corresponding overall average of the twelve O···O distances in 1 is virtually identical (2.761(5) Å).
The Cu6- and Cu9-rings in 2 each form six Cu···O contacts <3.00(8) Å with O-atoms of the Cu12-ring (Cu6-ring, Cu···O: 2.410(3)–2.609(3) Å, average: 2.476(3) Å; Cu9-ring, Cu···O: 2.318(3)–3.079(3) Å; average: 2.698(3) Å). Two Cu-atoms of the Cu9-ring (bridged by an O-atom of the second carbonate ion) bind the bridging H2O molecule. All other Cu-atoms, including those of the Cu12-ring, are at distances larger than 3.196(3) Å from the closest nonbonding O-atoms. Overall, there are twelve Cu···O distances <3.00(8) Å between Cun-rings, with an average of 2.587(3) Å. The corresponding value for 1 is 2.564(4) Å.
In addition to the two carbonate ions, nanojar 2 also binds a nitrobenzene solvent molecule in the outer cavity of the Cu6-ring (Figure 5) by a close π-π stacking interaction between the phenyl group and a pyrazolate moiety (centroid···centroid: 3.593(3) Å, angle between mean-planes: 6.7(2)°) and by weak, bifurcated interactions between the O-atoms of the nitro group and four Cu-atoms (Cu···O: 2.647(4) and 3.033(4) Å, and 2.827(4) and 2.844(4) Å, respectively).
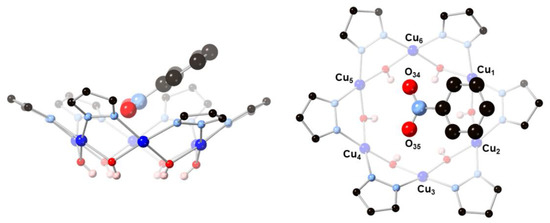
Figure 5.
Illustration of the binding of nitrobenzene by the [Cu(μ-OH)(μ-pz)]6 ring of 2: (left) side-view; (right) top-view.
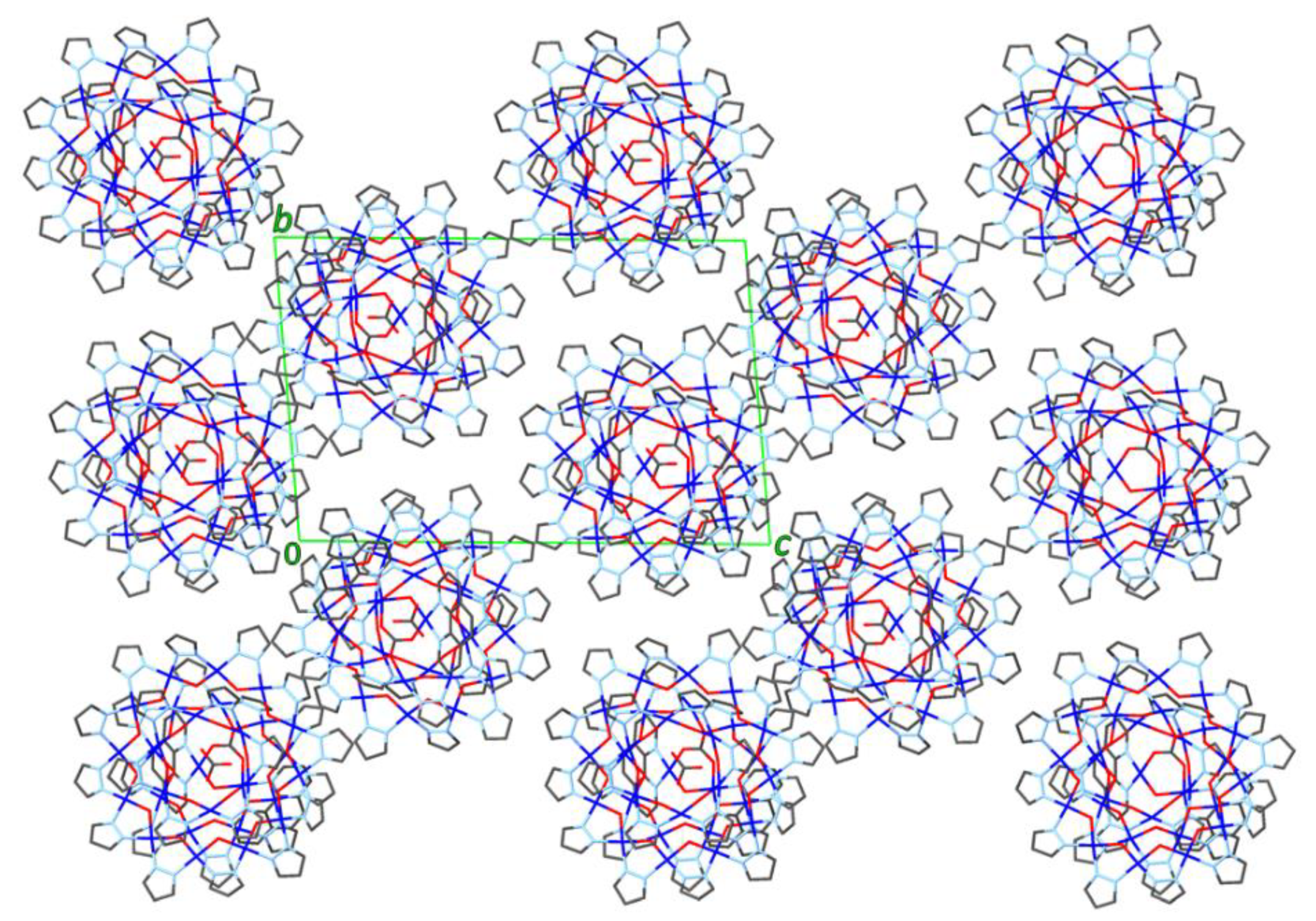
As shown in Figure 6, the close-packing of nanojars leaves relatively large void spaces in the crystal lattice, which are filled by multiple solvent molecules (see also Figure 7). In addition to the nitrobenzene molecule bound in the outer cavity of the Cu6-ring of the nanojar, there are five more nitrobenzene molecules filling up the void spaces, as well as a seventh nitrobenzene molecule disordered with a heptane molecule. The presence of aromatic moieties in the included solvent molecules appears to be crucial for the formation of nanojar crystals, as they form multiple aromatic interactions with the nanojar molecules and with each other. Nevertheless, the crystal lattice is not robust: the crystals quickly become opaque and disintegrate if removed from the mother liquor at ambient conditions, requiring low-temperature conditions for X-ray diffraction measurement.
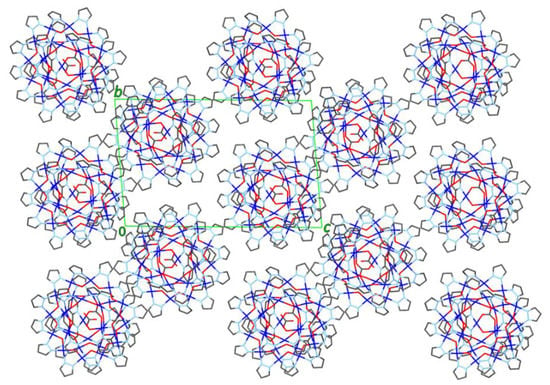
Figure 6.
View of the packing diagram of 2 down the a-axis (solvent molecules and H-atoms are not shown).
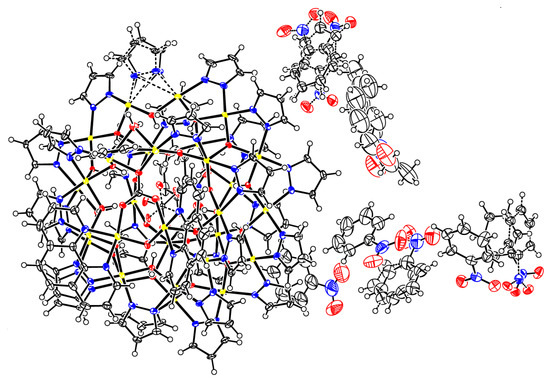
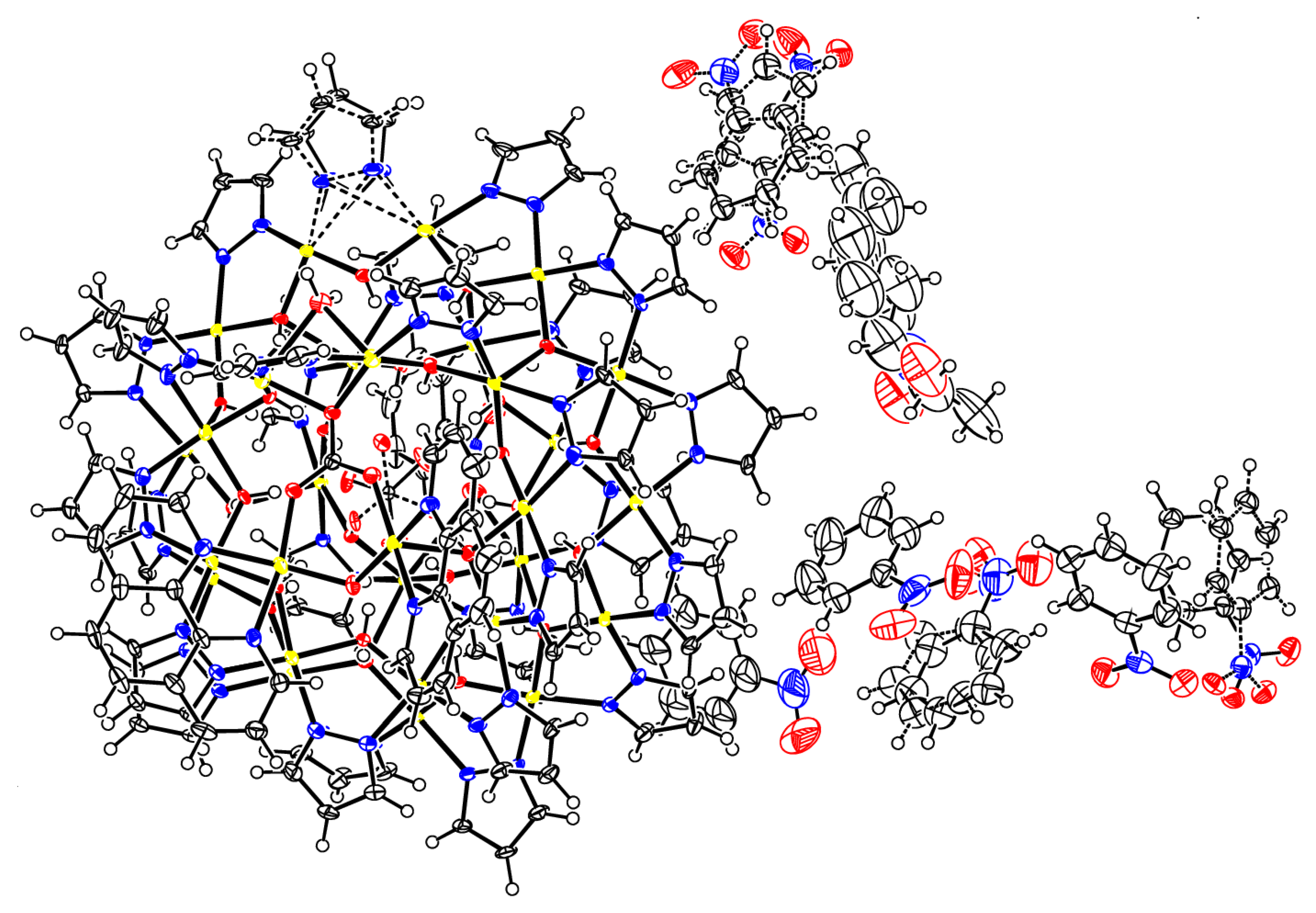
Figure 7.
Thermal ellipsoid plot of the crystal structure of 2(C6H5NO2)6.74(C7H16)0.76.
2.2. Mass Spectrometric Studies
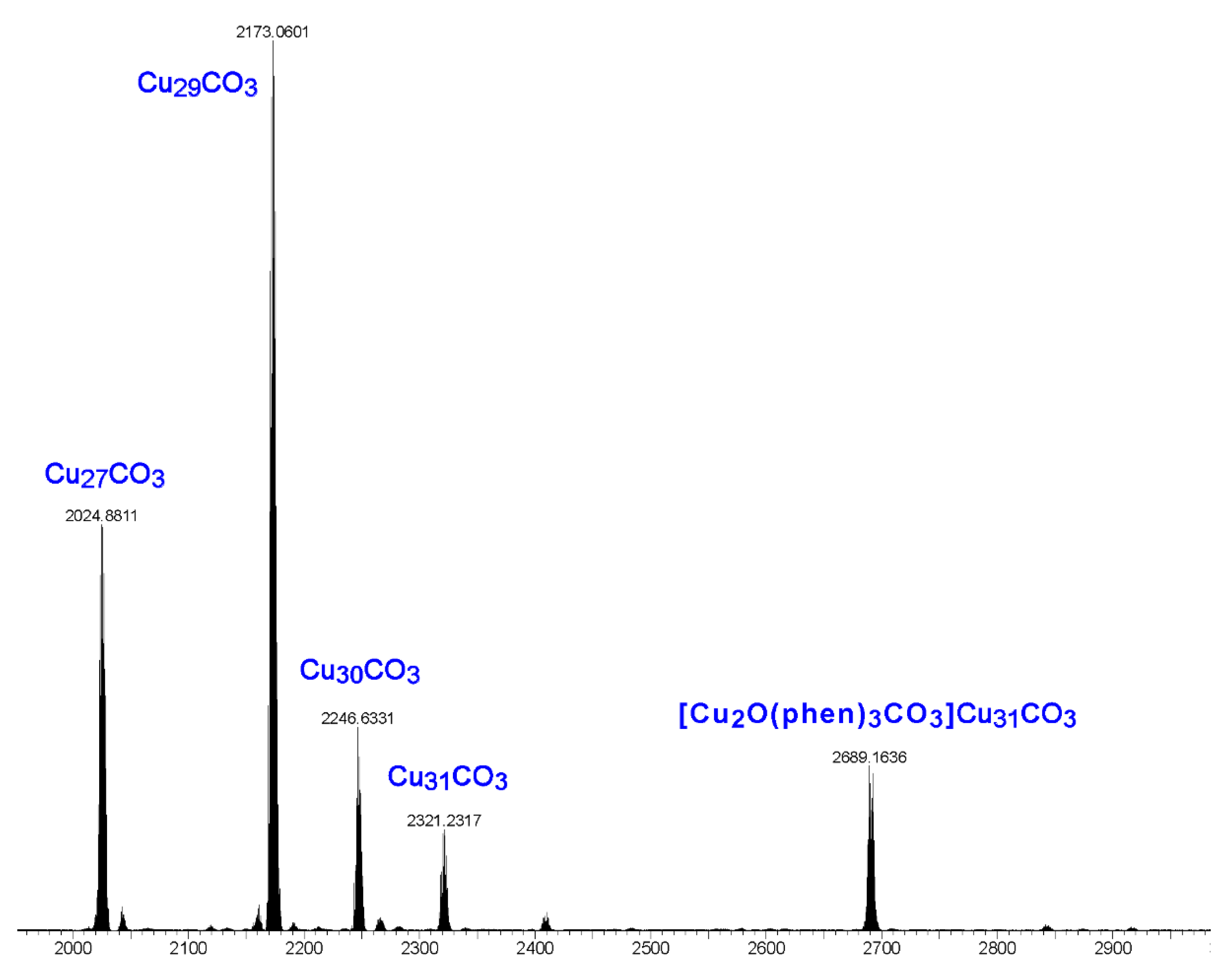
Electrospray ionization mass spectrometry (ESI-MS) studies show that the product obtained from the reaction of copper nitrate, pyrazole, sodium hydroxide, sodium carbonate and 1,10-phenanthroline is a mixture of nanojars (Figure 8). The spectrum shows the following peaks in the negative mode (no nanojar peaks are observable in the positive mode): [CO3⊂{Cu(μ-OH)(μ-pz)}n]2− (CunCO3; n = 27, m/z = 2023; n = 29, m/z = 2171; n = 30, m/z = 2245; n = 31, m/z = 2318) and [{Cu2O(phen)3CO3}CO3⊂{Cu(μ-OH)(μ-pz)}31]2− (m/z = 2690). The first four peaks correspond to nanojars without the additional [Cu2(phen)2CO3]2+ moiety. Obviously, neutral [Cu2(phen)2(CO3)2{Cu(OH)(pz)}n] nanojar peaks cannot be expected in the mass spectrum. It is apparent that during ionization in the mass spectrometer, the neutral nanojars lose the [Cu2(phen)2CO3]2+ moiety and become [CO3⊂{Cu(μ-OH)(μ-pz)}n]2−. Although the parent, neutral nanojars cannot be observed directly by ESI-MS, several independent observations indirectly support their assumed structure. First, the crystal structure of [Cu2(phen)2(CO3)2{Cu(OH)(pz)}n] with n = 27 has been unambiguously established. In this Cu6+Cu12+Cu9 nanojar, the Cu2(phen)2CO3 moiety is bound to the Cu9-ring. Other nanojar sizes, such as with n = 29 (Cu7+Cu13+Cu9), n = 30 (Cu8+Cu13+Cu9) and n = 31 (Cu8+Cu14+Cu9) also have Cu9-rings to which the Cu2(phen)2CO3 moiety can bind. Second, a peak corresponding to [CuI(phen)2]+ (m/z = 424) is observed in the positive mode ESI-MS spectrum of the as-synthesized neutral nanojar mixture, which originates from the [Cu2(phen)2CO3]2+ moiety detached from the nanojar upon ionization. Third, the neutral nanojar mixture has significantly decreased solubility in organic solvents, compared to the 2− charged analogs in (Bu4N+)2[CO3⊂{CuII(μ-OH)(μ-pz)}n]2−. Thus, the neutral nanojar mixture only dissolves well in solvents such as DMF, nitrobenzene and THF, but not in toluene, chlorobenzene, butyl acetate, methanol, acetone and dioxane, which are good solvents for the 2− charged nanojars. Finally, the peak at m/z = 2690 corresponding to [{Cu2O(phen)3CO3}CO3⊂{Cu(μ-OH)(μ-pz)}31]2− clearly demonstrates the existence of expanded nanojars in solution.
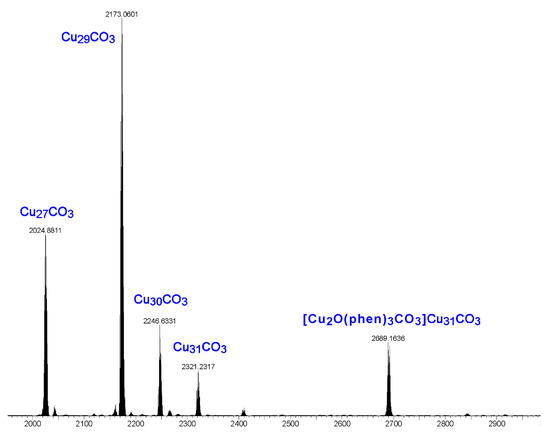
Figure 8.
ESI-MS(–) spectrum in DMF of the product obtained from the reaction of copper nitrate, pyrazole, sodium hydroxide, sodium carbonate and 1,10-phenanthroline.
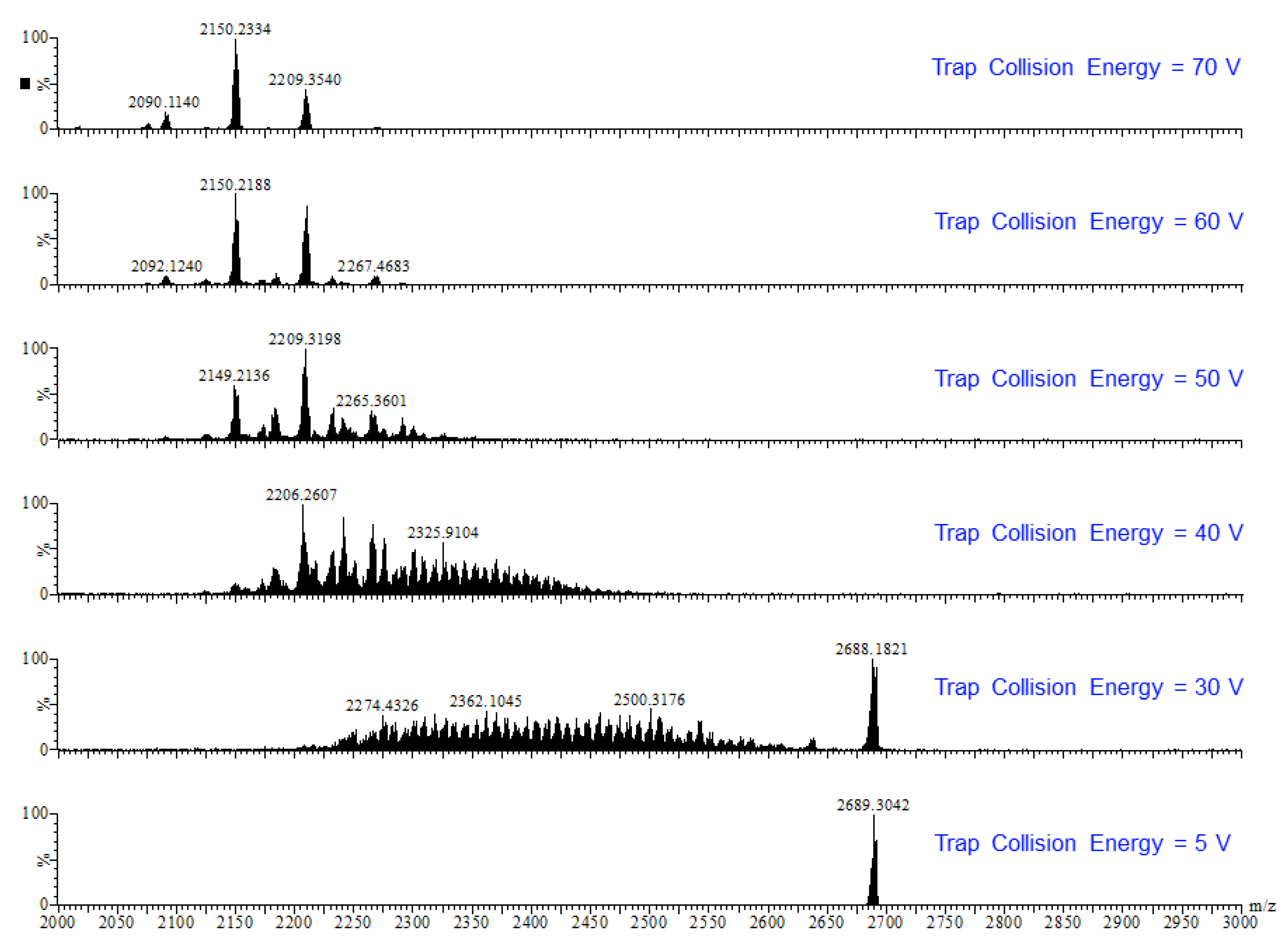
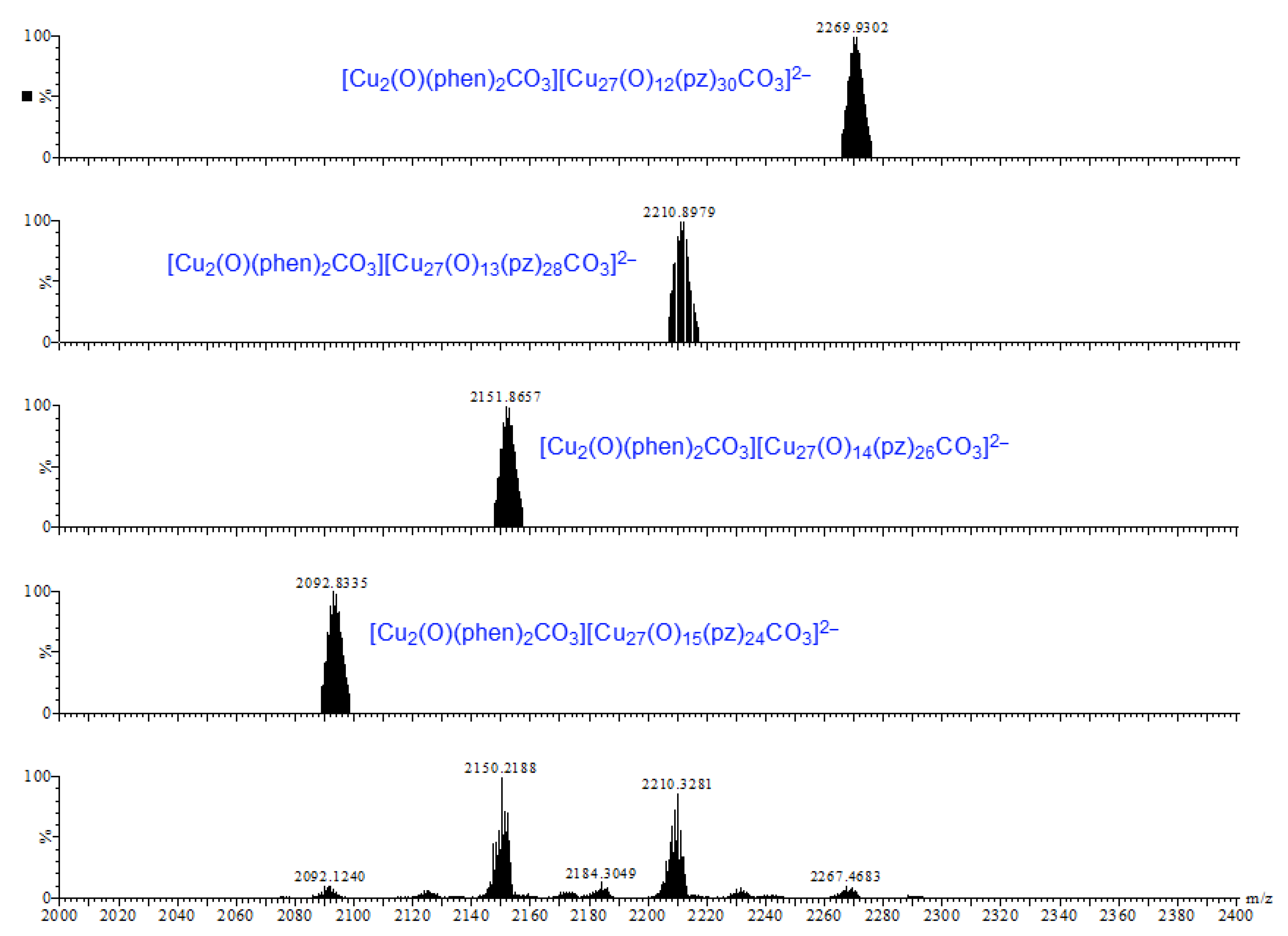
Earlier tandem mass spectrometry (MS-MS) studies showed that the various [CO3⊂{Cu(μ-OH)(μ-pz)}n]2− nanojar species shrink as the trap collision energy is increased, by losing neutral Cu5(OH)10(Hpz)10−y(H2O)(n+y−20)/2 fragments (y = 4–12; y has the same parity as n [1]. Thus, four to five shrunken daughter-nanojar species of the formula [Cun−5O(n − y)/2(pz)n + y − 10CO3]2− were observed for each parent nanojar. Similarly, the peak at m/z = 2690 corresponding to [{Cu2O(phen)3CO3}CO3⊂{Cu(μ-OH)(μ-pz)}31]2− gradually disappears upon increasing the trap collision energy, by losing a neutral Cu4(OH)y(Hpz)8 − y(H2O)(31 − y)/2(phen) fragment and giving rise to peaks at m/z 2092, 2151, 2269 and 2210 (Figure 9), which correspond to [Cu27O(31 − y)/2(pz)31 + y − 8CO3]2− (y = 1, 3, 5, 7) species (Figure 10). So far, these shrunken daughter-nanojars have only been observed by mass spectrometry and have not been isolated; therefore, their detailed structure is yet unknown. As with other nanojars, a peak at m/z = 198, corresponding to [CuI(pz)2]−, is also observed in the tandem mass spectrum.
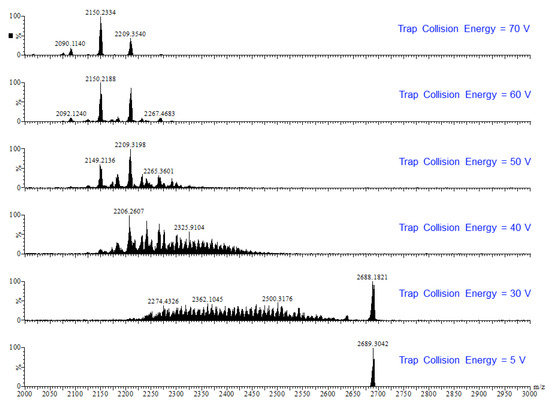
Figure 9.
ESI-MS-MS(–) spectra of the isolated peak with m/z = 2690.
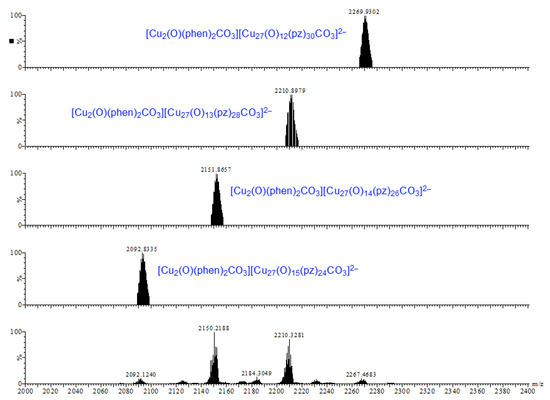
Figure 10.
Comparison of the calculated (upper four) and observed (bottom) daughter-peaks of the isolated peak with m/z = 2690.
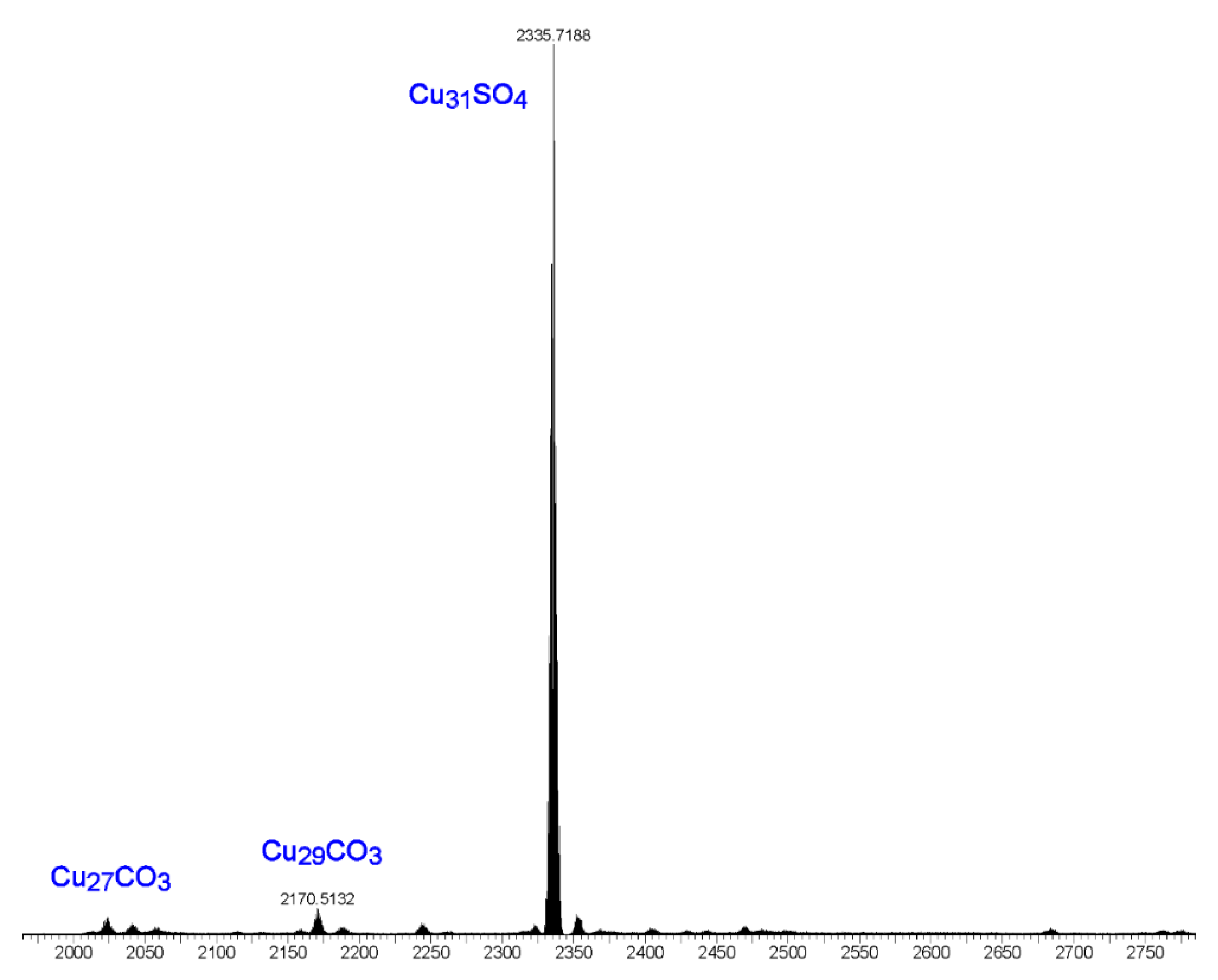
The following observations further support the assumption that the [Cu2(phen)2CO3]2+ moiety binds only to the Cu9-ring in nanojars. The ESI-MS(–) spectrum of the product obtained from the reaction of copper sulfate, pyrazole, sodium hydroxide and 1,10-phenanthroline is shown in Figure 11. The major peak in this spectrum corresponds to [SO4⊂{Cu(μ-OH)(μ-pz)}31]2− (Cu31SO4), assumingly derived from [{Cu2(phen)2CO3}SO4⊂{Cu(μ-OH)(μ-pz)}31]. Smaller peaks are observed at m/z 2023 (Cu27CO3) and m/z 2171 (Cu29CO3). These latter species formed as side-products upon absorption of small amounts of atmospheric CO2 during the reaction and explain the serendipitous formation of a few crystals of 2. Noteworthy is the absence of significant peaks at m/z 2041 (Cu27SO4), m/z 2115 (Cu28SO4), m/z 2189 (Cu29SO4) and m/z 2262 (Cu30SO4). As shown earlier, Cu27SO4 and Cu29SO4 species form in very small amounts under similar reaction conditions from copper sulfate, pyrazole, sodium hydroxide and tetrabutylammonium hydroxide in THF, whereas Cu30SO4 was not observed at all (the major species observed were Cu28SO4 and Cu31SO4) [1]. The absence of a peak at m/z 2115, corresponding to Cu28SO4, suggests that [{Cu2(phen)2CO3}SO4⊂{Cu(μ-OH)(μ-pz)}28] nanojars do not form, probably due to the fact that the Cu28SO4 nanojar (Cu6+Cu12+Cu10) lacks a Cu9-ring and cannot accommodate the Cu2(phen)2CO3 moiety.
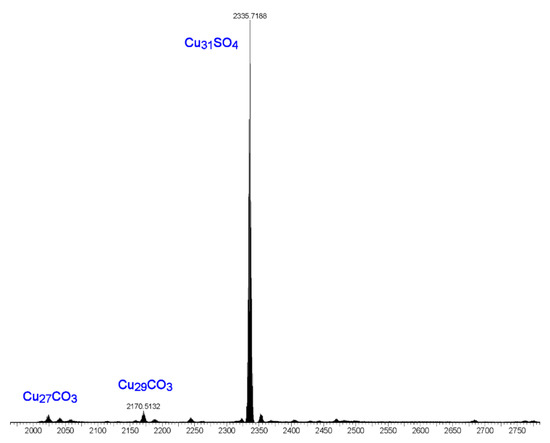
Figure 11.
ESI-MS(–) spectrum in DMF of the product obtained from the reaction of copper sulfate, pyrazole, sodium hydroxide and 1,10-phenanthroline.
3. Materials and Methods
All commercially available chemicals were used as received. Reactions were carried out in closed vessels, but not under strictly air-free conditions.
Synthesis of [{Cu2(phen)2CO3}CO3⊂{Cu(μ-OH)(μ-pz)}n] (n = 27, 29, 30, 31). Cu(NO3)2·2.5H2O (0.4652 g, 2.00 mmol), pyrazole (0.132 g, 1.94 mmol), NaOH (0.155 g, 3.88 mmol), 1,10-phenanthroline (0.037 g, 0.21 mmol) and Na2CO3·H2O (0.2481, 2.00 mmol) were stirred for six days in 15 mL of THF. The reaction mixture was filtered, and the solvent was left to evaporate. The solid product obtained was washed with water, methanol and acetone and was dried under high vacuum to yield 0.283 g of a dark blue powder.
Synthesis of [{Cu2(phen)2CO3}SO4⊂{Cu(μ-OH)(μ-pz)}31]. This compound was prepared as described above, using CuSO4·5H2O (1.0000 g, 4.00 mmol), pyrazole (0.2640 g, 3.88 mmol), NaOH (0.3100 g, 7.75 mmol) and 1,10-phenanthroline (0.0740 g, 0.41 mmol) in 30 mL of THF. Yield: 0.5348 g dark blue powder.
3.1. Mass Spectrometry
Mass spectrometric analysis of the nanojars was performed with a Waters Synapt G1 HDMS instrument using electrospray ionization (ESI). 10−4–10−5 M solutions were prepared in N,N-dimethylformamide (DMF). Samples were infused by a syringe pump at 5 μL/min, and nitrogen was supplied as nebulizing gas at 500 L/h. The electrospray capillary voltage was set to –2.5 or +3.0 kV, respectively, with a desolvation temperature of 150 °C. The sampling and extraction cones were maintained at 40 V and 4.0 V, respectively, at 80 °C. The MS/MS conditions were the same, except the transfer collision energy was 5 V and the trap collision energies were 5, 30, 40, 50, 60 and 70 V.
3.2. X-ray Crystallography
A few single-crystals of 2 were grown from a nitrobenzene solution by heptane vapor diffusion. Once removed from the mother liquor, the crystals are very sensitive to solvent loss at ambient conditions and were mounted quickly under a cryostream (100 K) to prevent decomposition. X-ray diffraction data were collected at 100 K from a single-crystal mounted atop a glass fiber under Paratone-N oil with a Bruker SMART APEX II diffractometer using graphite-monochromated Mo-Kα (λ = 0.71073 Å) radiation. The structure was solved by employing SHELXTL direct methods and refined by full-matrix least squares on F2 using the APEX2 v2014.9-0 software package [30]. C–H hydrogen atoms were placed in idealized positions and refined using the riding model. Hydroxyl and water H atom positions were located from difference density maps and were refined with O–H distance restraints of 0.82(2) Å. A pyrazolate ligand was refined as disordered. The two disordered moieties were restrained to have similar geometries. Uij components of ADPs for disordered atoms closer to each other than 2.0 Å were restrained to be similar. Subject to these conditions, the occupancy ratio refined to 0.805(13)/0.195(4). The oxygen atoms of the carbonate ion were refined as disordered. The two disordered moieties were restrained to have similar geometries. Uij components of ADPs for disordered atoms closer to each other than 2.0 Å were restrained to be similar. Subject to these conditions, the occupancy ratio refined to 0.913(4)/0.087(4). Three nitrobenzene solvate molecules were disordered with two alternative orientations (one by two-fold symmetry, two in general positions), one with three orientations, and one was disordered with a heptane molecule. The disordered nitrobenzene moieties were restrained to have similar geometries (SAME commands). C926 of one nitrobenzene moiety was restrained to be coplanar with its neighboring atoms. Bond distances within the heptane molecule were restrained to be similar to each other (SADI command). Uij components of ADPs for disordered atoms closer to each other than 2.0 Å were restrained to be similar. Subject to these conditions, the occupancy rates refined to 0.910(4)/0.090(4) for the two moieties of the nitrobenzene of N60, to 0.489(14)/0.511(14) for the two moieties of the nitrobenzene of N62, to 0.502(3)/0.313(3)/0.185(3) for the three moieties of the nitrobenzene of N64 and to 0.765(6)/0.235(6) for the disorder of heptane and nitrobenzene (in favor of heptane). A thermal ellipsoid plot of the crystal structure is shown in Figure 7.
Summary of the crystallographic data. Chemical formula, C152.76H171.91Cu29N64.74O47.47; formula weight, 5517.12; crystal system, triclinic; space group, P ī (No. 2); a = 14.7855(3) Å; b = 20.6120(4) Å; c = 31.7417(6) Å; α = 95.026(1)°; β = 92.124(1)°; γ = 93.936(1); V = 9604.6(3) Å3; Z = 2; Dcalc = 1.908 g/cm3; μ = 3.225 mm−1; no. of reflns collected, 299360; no. of unique reflns, 39404; no. of obsd reflns (I > 2σ(I)), 29554; R(int), 0.0723; data/parameters/restrains, 39404/3255/2672; goodness-of-fit (on F2): 1.032; R(F) (I > 2σ(I)), 0.0384; Rw(F) (I > 2σ(I)), 0.0793; R(F) (all data), 0.0641; Rw(F) (all data), 0.0891; residual electron density, max/min (e/Å3), 2.122/–1.143. Crystallographic data for 2(C6H5NO2)6.74(C7H16)0.76 were deposited with the Cambridge Crystallographic Data Center (CCDC 2078120). Copies of the data can be obtained free of charge at http://www.ccdc.cam.ac.uk/products/csd/request (accesed on 20 May 2021).
4. Conclusions
In summary, we present a new, expanded nanojar of the formula [CO3⊂{Cu29(μ-OH)27(μ-pz)27(phen)2(μ4-CO3)(H2O)}], which is able to bind two carbonate ions, compared to only one carbonate ion in previously reported nanojars [CO3⊂{Cu(μ-OH)(μ-pz)}n]2− (n = 27, 29, 30, 31). Single-crystal X-ray crystallographic studies in the solid state show that the new nanojar is an extension of the [CO3⊂{Cu(μ-OH)(μ-pz)}6+12+9]2− motif, with a [Cu2(phen)2CO3]2+ moiety bound to the Cu9-ring of the nanojar. Upon binding of this additional moiety, one OH− group of the Cu9-ring is displaced by an O-atom of the second carbonate ion and becomes a bridging ligand for the Cu2-moiety. Thus, the expanded nanojar can also be described as [{Cu2(μ-OH)(phen)2(μ4-CO3)}CO3⊂{Cu27(μ-OH)26(μ-pz)27}]. Solution studies by electrospray ionization mass spectrometry indicate that homologous nanojar species based on [CO3⊂{Cu(μ-OH)(μ-pz)}n]2− (n = 29, 30, 31) also form in the reaction of copper nitrate, pyrazole, sodium hydroxide, sodium carbonate and 1,10-phenanthroline. Although the expanded nanojars described above cannot be observed directly by ESI-MS due to their overall neutral charge, ionization by loss of the [Cu2(phen)2CO3]2+ moiety does lead to [CO3⊂{Cu(μ-OH)(μ-pz)}n]2− (n = 27, 29, 30, 31) daughter-species. Additionally, a peak in the mass spectrum corresponding to [{Cu2O(phen)3CO3}CO3⊂{Cu(μ-OH)(μ-pz)}31]2− does provide direct evidence and demonstrates the existence of expanded nanojars in solution.
Author Contributions
Conceptualization, G.M.; methodology, G.M. and W.A.A.I.; software, G.M. and W.A.A.I.; validation, G.M. and W.A.A.I.; formal analysis, G.M.; investigation, W.A.A.I. and G.M.; resources, G.M.; data curation, G.M.; writing—original draft preparation, G.M.; writing—review and editing, G.M. and W.A.A.I.; visualization, G.M. and W.A.A.I.; supervision, G.M.; project administration, G.M.; funding acquisition, G.M. Both authors have read and agreed to the published version of the manuscript.
Funding
This material is based upon work supported by the National Science Foundation under Grant No. CHE-1808554.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Matthias Zeller for the final refinement of the disorder in the crystal structure.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are not available from the authors.
References
- Ahmed, B.M.; Szymczyna, B.R.; Jianrattanasawat, S.; Surmann, S.A.; Mezei, G. Survival of the Fittest Nanojar: Stepwise Breakdown of Polydisperse Cu27–Cu31 Nanojar Mixtures into Monodisperse Cu27(CO3) and Cu31(SO4) Nanojars. Chem. Eur. J. 2016, 22, 5499–5503. [Google Scholar] [CrossRef]
- Fernando, I.R.; Surmann, S.A.; Urech, A.A.; Poulsen, A.M.; Mezei, G. Selective total encapsulation of the sulfate anion by neutral nano-jars. Chem. Commun. 2012, 48, 6860–6862. [Google Scholar] [CrossRef] [PubMed]
- Mezei, G. Incarceration of one or two phosphate or arsenate species within nanojars, capped nanojars and nanohelicages: Helical chirality from two closely-spaced, head-to-head PO43− or AsO43− ions. Chem. Commun. 2015, 51, 10341–10344. [Google Scholar] [CrossRef] [PubMed]
- Mezei, G.; Baran, P.; Raptis, R.G. Anion Encapsulation by Neutral Supramolecular Assemblies of Cyclic CuII Complexes: A Series of Five Polymerization Isomers, [{cis-CuII(μ-OH)(μ-pz)}n], n = 6, 8, 9, 12, and 14. Angew. Chem. Int. Ed. 2004, 43, 574–577. [Google Scholar] [CrossRef] [PubMed]
- Hen, L.; Berry, S.N.; Wu, X.; Howe, E.N.W.; Gale, P.A. Advances in Anion Receptor Chemistry. Chem 2020, 6, 61–141. [Google Scholar]
- Liu, Y.; Sengupta, A.; Raghavachari, K.; Flood, A.H. Anion Binding in Solution: Beyond the Electrostatic Regime. Chem 2017, 3, 411–427. [Google Scholar] [CrossRef]
- Molina, P.; Zapata, F.; Caballero, A. Anion Recognition Strategies Based on Combined Noncovalent Interactions. Chem. Rev. 2017, 117, 9907–9972. [Google Scholar] [CrossRef]
- Busschaert, N.; Caltagirone, C.; Van Rossom, W.; Gale, P.A. Applications of Supramolecular Anion Recognition. Chem. Rev. 2015, 115, 8038–8155. [Google Scholar] [CrossRef]
- Jacobson, B.L.; Quiocho, F.A. Sulfate-Binding Protein Dislikes Protonated Oxyacids: A Molecular Explanation. J. Mol. Biol. 1988, 204, 783–787. [Google Scholar] [CrossRef]
- Gonzalez, D.; Richez, M.; Bergonzi, C.; Chabriere, E.; Elias, M. Crystal structure of the phosphate-binding protein (PBP-1) of an ABC-type phosphate transporter from Clostridium perfringens. Sci. Rep. 2014, 4, 6636. [Google Scholar] [CrossRef]
- Ahmed, B.M.; Calco, B.; Mezei, G. Tuning the structure and solubility of nanojars by peripheral ligand substitution, leading to unprecedented liquid–liquid extraction of the carbonate ion from water into aliphatic solvents. Dalton Trans. 2016, 45, 8327–8339. [Google Scholar] [CrossRef] [PubMed]
- Mezei, G. Selective Extraction of Anions from Solutions. U.S. Patent 10,087,197 B2, 2 October 2018. U.S. Patent 9,901,901 B2, 27 February 2018; European Patent 2852558 B1, 9 September 2020. [Google Scholar]
- Haiduc, I. Inverse coordination—An emerging new chemical concept. Oxygen and other calchogens as coordination centers. Coord. Chem. Rev. 2017, 338, 1–26. [Google Scholar] [CrossRef]
- Haiduc, I. Inverse coordination—An emerging new chemical concept. II. Halogens as coordination centers. Coord. Chem. Rev. 2017, 348, 71–91. [Google Scholar] [CrossRef]
- Haiduc, I. Nitrogen centered inverse coordination complexes. A survey of molecular topologies. J. Coord. Chem. 2018, 71, 3139–3179. [Google Scholar] [CrossRef]
- Haiduc, I. Review. Inverse coordination. Inorganic open and cyclic nitrogen heteroatom molecules as coordination centers. A survey of molecular topologies. J. Coord. Chem. 2019, 72, 35–52. [Google Scholar] [CrossRef]
- Haiduc, I. Review. Inverse coordination. Organic nitrogen heterocycles as coordination centers. A survey of molecular topologies and systematization. Part 1. Five-membered and smaller rings. J. Coord. Chem. 2019, 72, 2127–2159. [Google Scholar] [CrossRef]
- Haiduc, I. Review. Inverse coordination. Organic nitrogen heterocycles as coordination centers. A survey of molecular topologies and systematization. Part 2. Six-membered rings. J. Coord. Chem. 2019, 72, 2805–2903. [Google Scholar] [CrossRef]
- Haiduc, I. Inverse coordination metal complexes with oxalate and sulfur, selenium and nitrogen analogues as coordinationcenters. Topology and systematization. J. Coord. Chem. 2020, 73, 1619–1700. [Google Scholar] [CrossRef]
- Haiduc, I.; Tiekink, E.R.T. Inverse Coordination. A Novel Chemical Concept; Sunway University Press: Sunway City, Selangor, Malaysia, 2020. [Google Scholar]
- Velasco, V.; Aguilà, D.; Barrios, L.A.; Borilovic, I.; Roubeau, O.; Ribas-Ariño, J.; Fumanal, M.; Teat, S.J.; Aromí, G. New coordination features; a bridging pyridine and the forced shortest non-covalent distance between two CO32− species. Chem. Sci. 2015, 6, 123–131. [Google Scholar] [CrossRef]
- Einstein, F.W.B.; Willis, A.C. Crystal and molecular structure of μ-carbonato-di-μ-chloro-tetrakis(bis(3-aminopropyl)amine)tetracopper(II) chloride hydrate. Inorg. Chem. 1981, 20, 609–614. [Google Scholar] [CrossRef]
- Escuer, A.; Peñalba, E.; Vicente, R.; Solans, X.; Font-Bardía, M. Two new tetranuclear µ4-carbonato copper(II) complexes. Syntheses, crystal structure and magnetic behaviour of [(µ4-CO3)(µ-Br)2{Cu4(bapa)4}]Br4 and [(µ4-CO3)(µ-Cl)2{Cu4(bapma)4}]Cl4·12H2O [bapa = bis(aminopropyl)amine and bapma = bis(aminopropyl)methylamine]. J. Chem. Soc. Dalton Trans. 1997, 2315–2320. [Google Scholar] [CrossRef]
- Rodríguez, M.; Llobet, A.; Corbella, M.; Müller, P.; Usón, M.A.; Martelle, A.E.; Reibenspiese, J. Solvent controlled nuclearity in Cu(II) complexes linked by the CO32− ligand: Synthesis, structure and magnetic properties. J. Chem. Soc. Dalton Trans. 2002, 2900–2906. [Google Scholar] [CrossRef]
- Fondo, M.; García-Deibe, A.M.; Corbella, M.; Ruiz, E.; Tercero, J.; Sanmartín, J.; Bermejo, M.R. Unexpected Ferromagnetic Interaction in a New Tetranuclear Copper(II) Complex: Synthesis, Crystal Structure, Magnetic Properties, and Theoretical Studies. Inorg. Chem. 2005, 44, 5011–5020. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Chen, L.; Chen, R.; Peng, Z.-H.; Song, Y.; Pan, Z.-Q.; Huang, Q.-M.; Hua, X.-L.; Bai, Z.-W. Hexamine copper(II) coordination polymers: Synthesis, structure and magnetic properties. CrystEngComm 2009, 11, 671–679. [Google Scholar] [CrossRef]
- Ho, Y.-H.; Chang, M.-C.; Yu, K.-H.; Liu, Y.-H.; Wang, Y.; Cheng, Y.-C.; Chen, J.-T. CO2 fixation by dicopper(II) complexes in hypodentate framework of N8O2. Dalton Trans. 2014, 43, 6287–6290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Nishihara, S.; Nakano, Y.; Maryunina, K.Y.; Inoue, K. A Cuprate Spin Ladder Linked by a Pyridyl Ligand. Chem. Lett. 2014, 43, 1713–1715. [Google Scholar] [CrossRef]
- Uraev, A.I.; Popov, L.D.; Levchenkov, S.I.; Shcherbakov, I.N.; Suponitsky, K.Y.; Garnovskii, D.A.; Lukov, V.V.; Kogan, V.A. Crystal structure and magnetic properties of a tetranuclear carbonate-bridged CuII complex with a Schiff base compartmental ligand with the N2OS2 donor set. Mendeleev Commun. 2015, 25, 62–64. [Google Scholar] [CrossRef]
- APEX2 v2014.9-0; Bruker AXS Inc.: Madison, WI, USA, 2014.
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).