Structural Insights of Three 2,4-Disubstituted Dihydropyrimidine-5-carbonitriles as Potential Dihydrofolate Reductase Inhibitors
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemical Synthesis
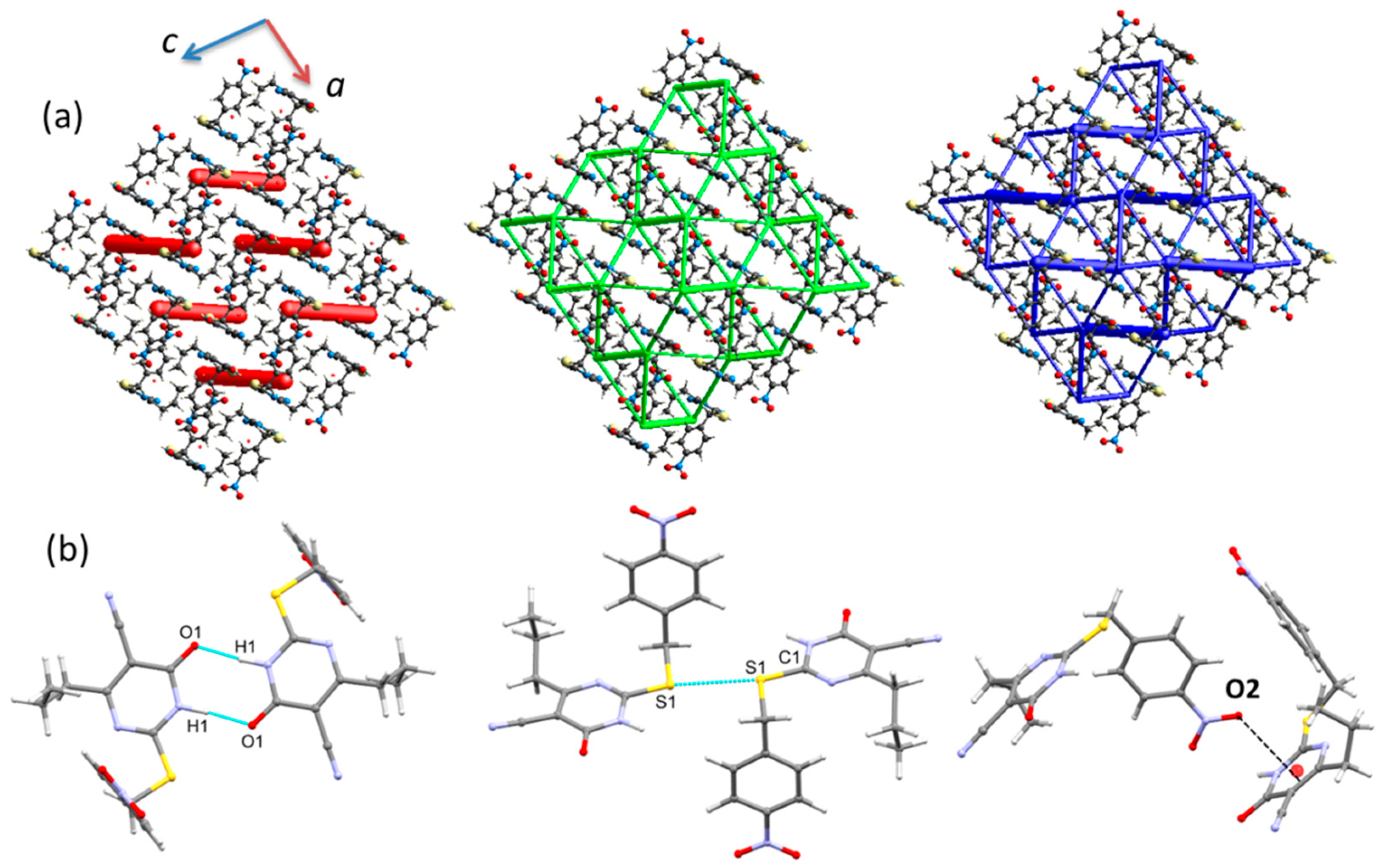
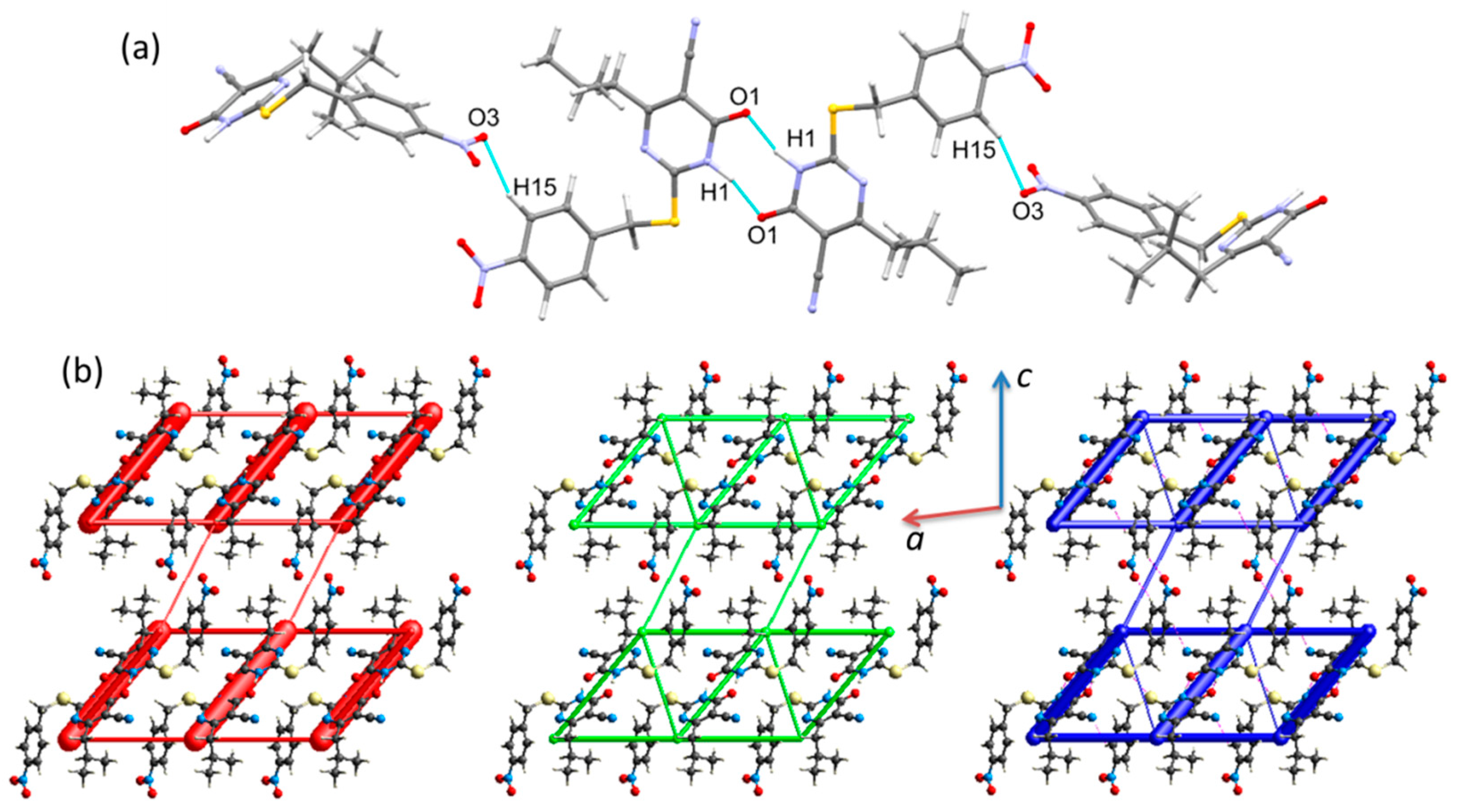
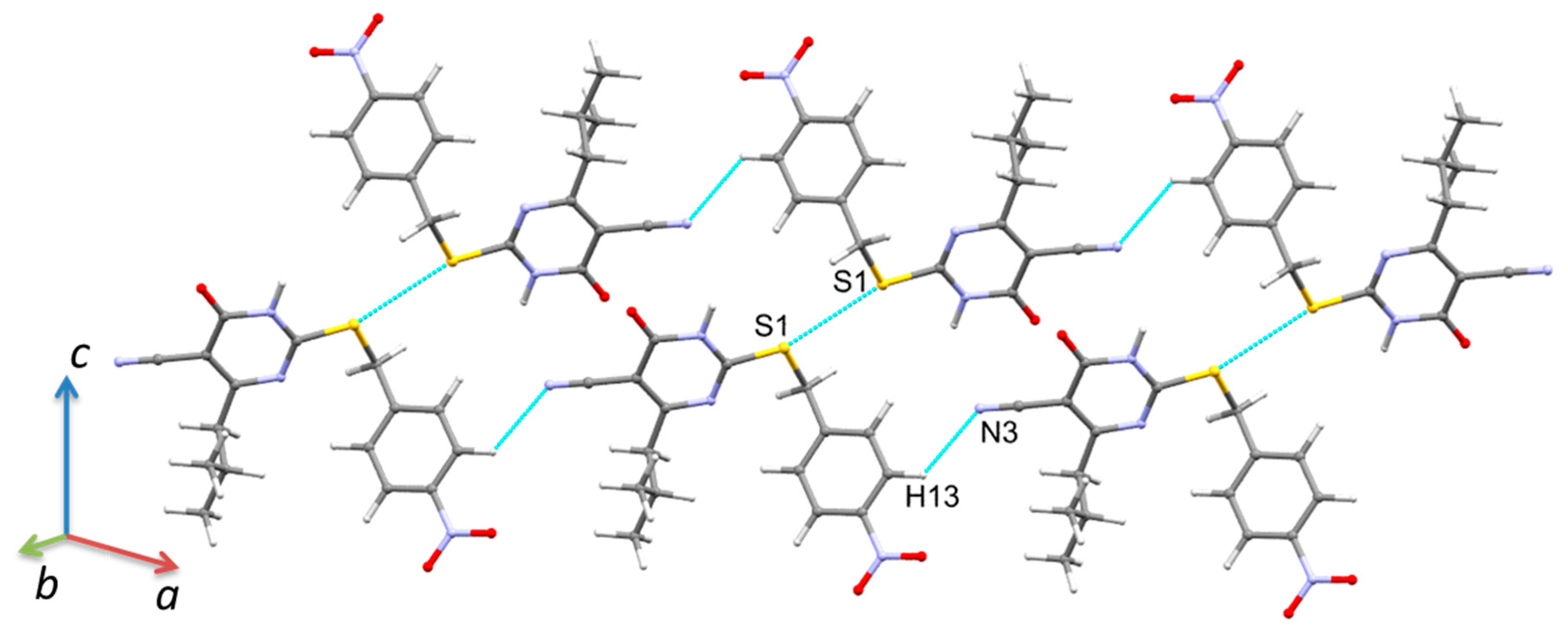
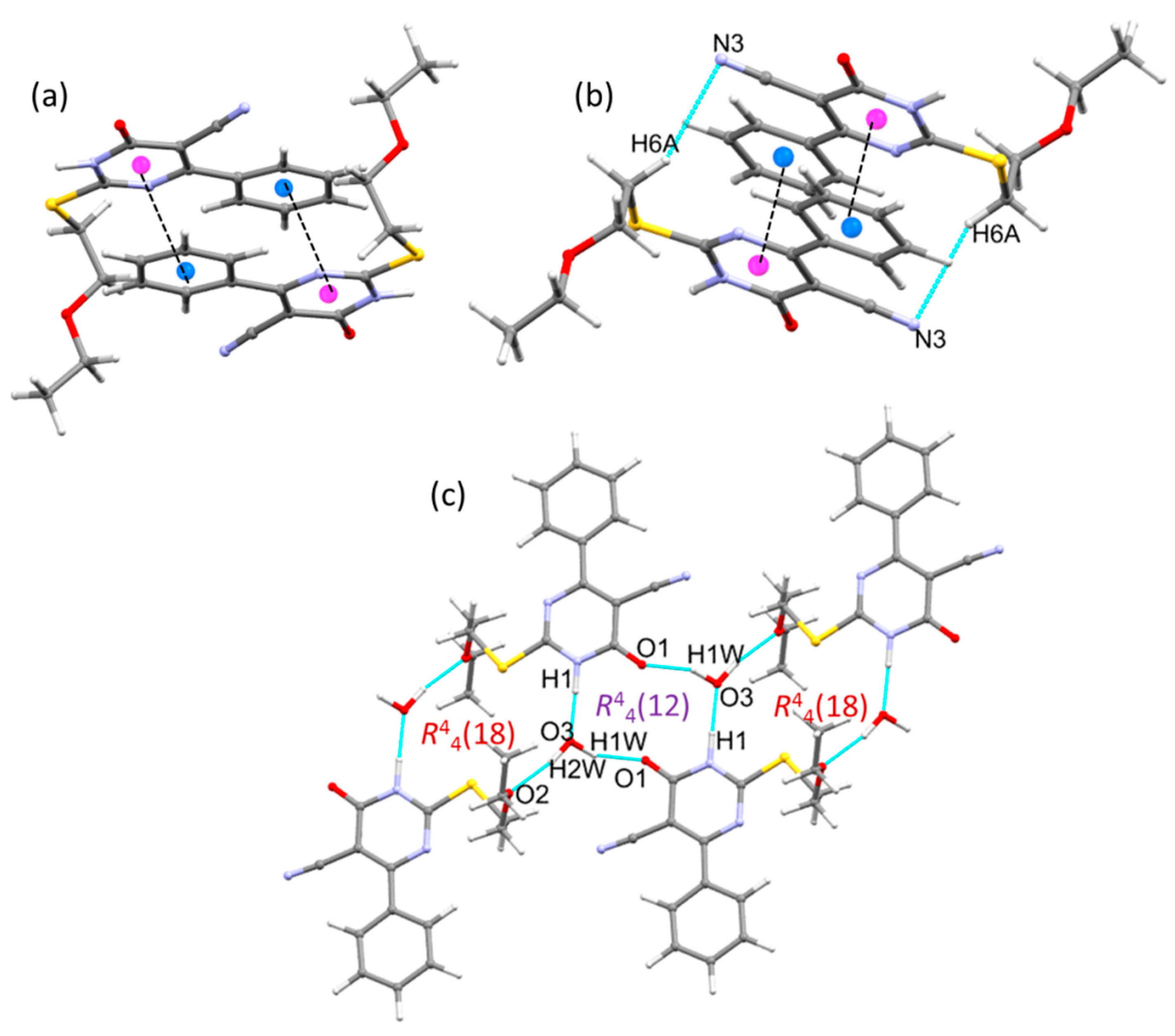
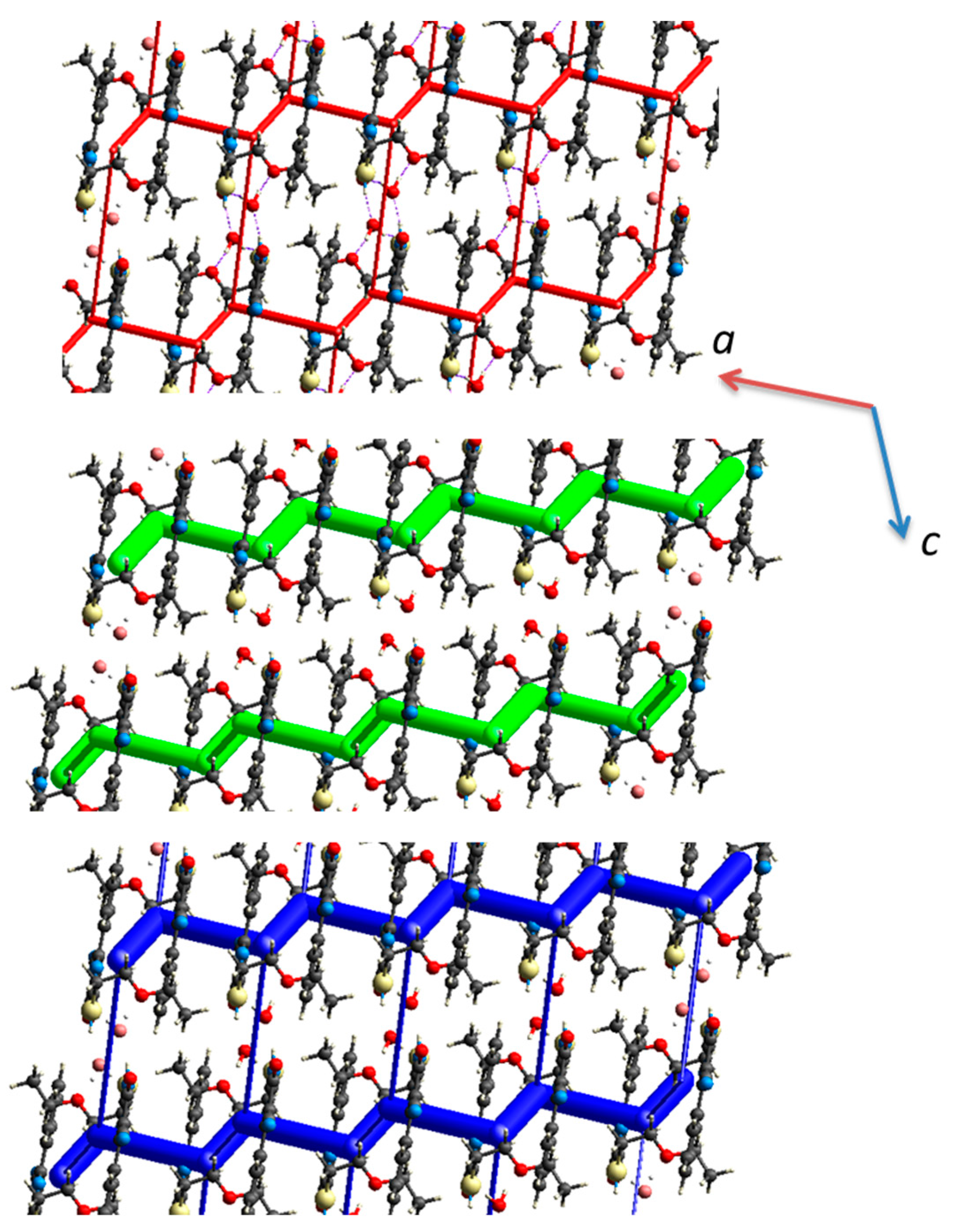
2.2. Crystal Structures
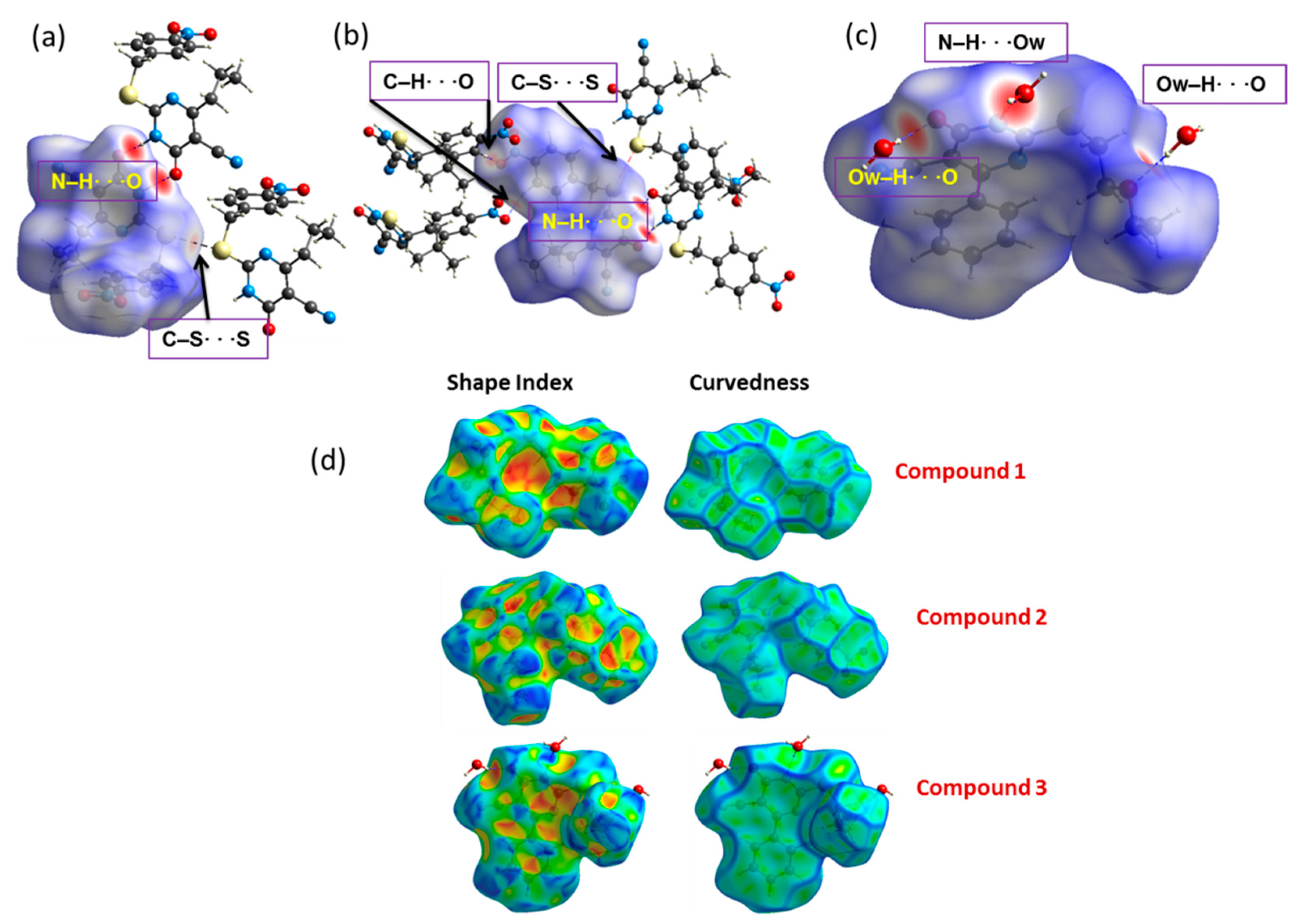
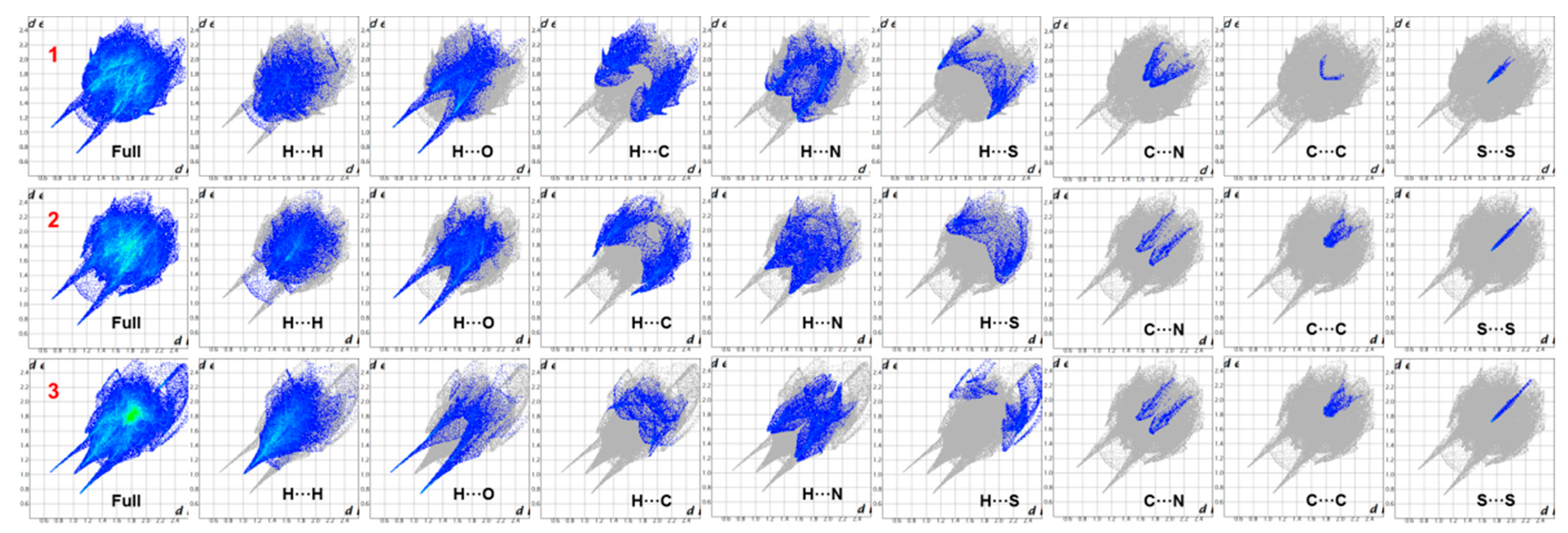
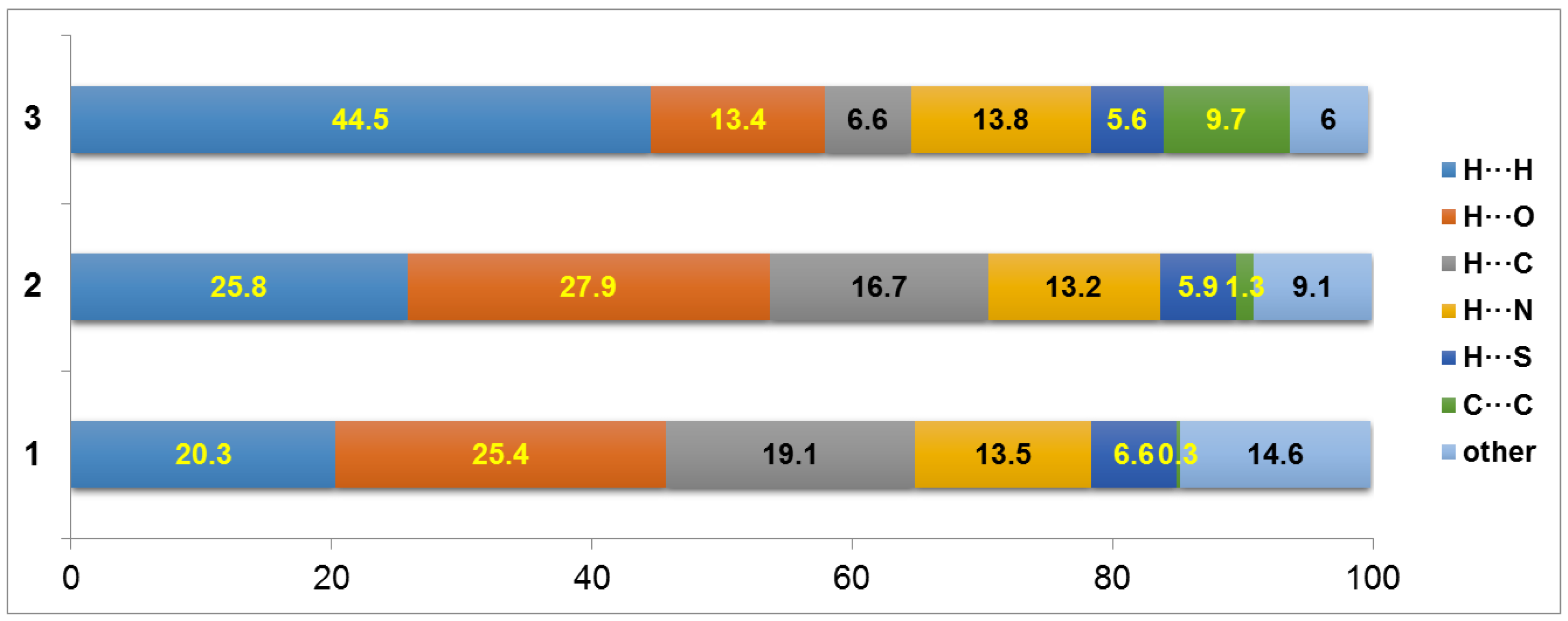
2.3. Hirshfeld Surface and 2D Fingerprint Plot Analysis
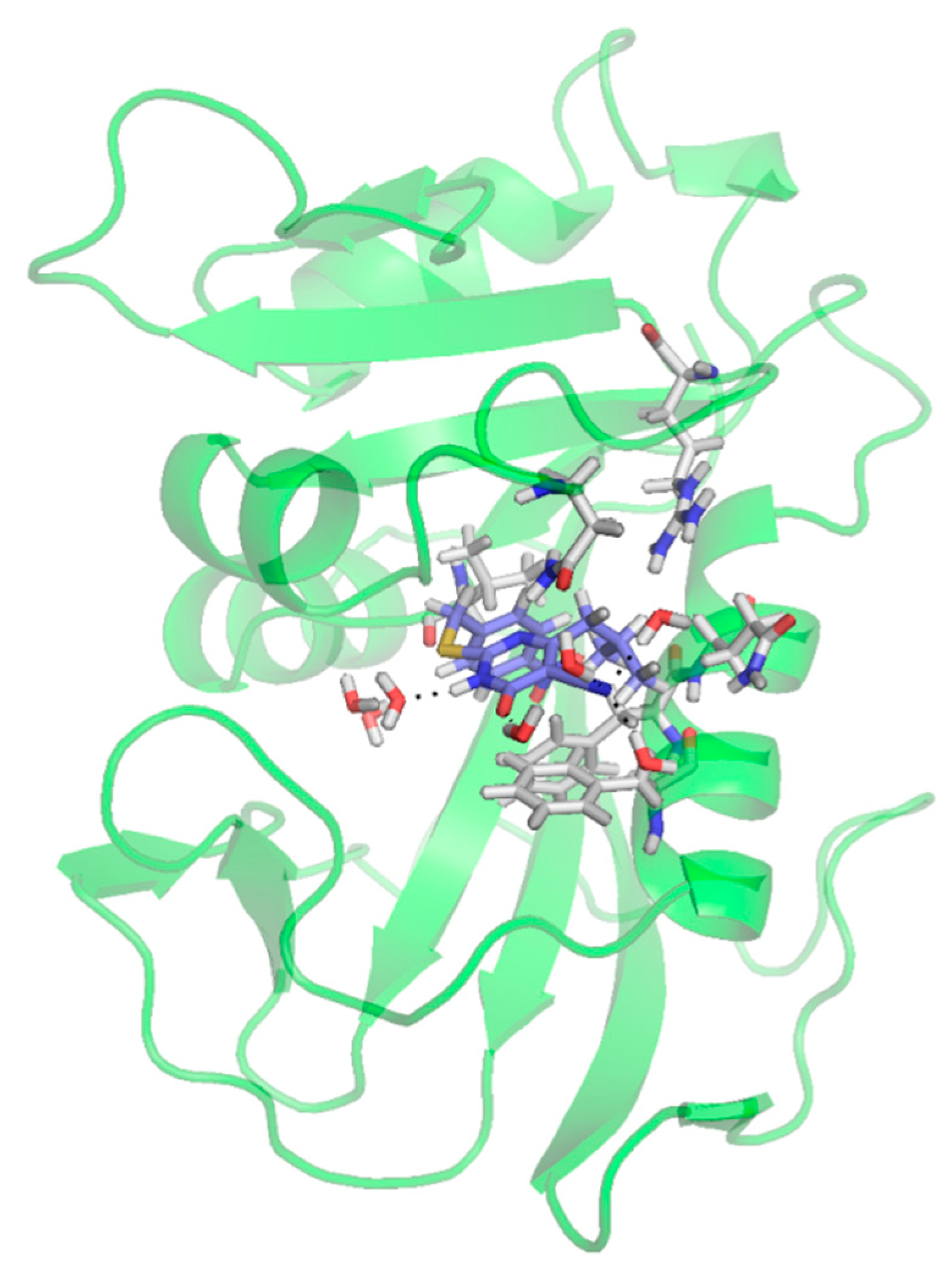
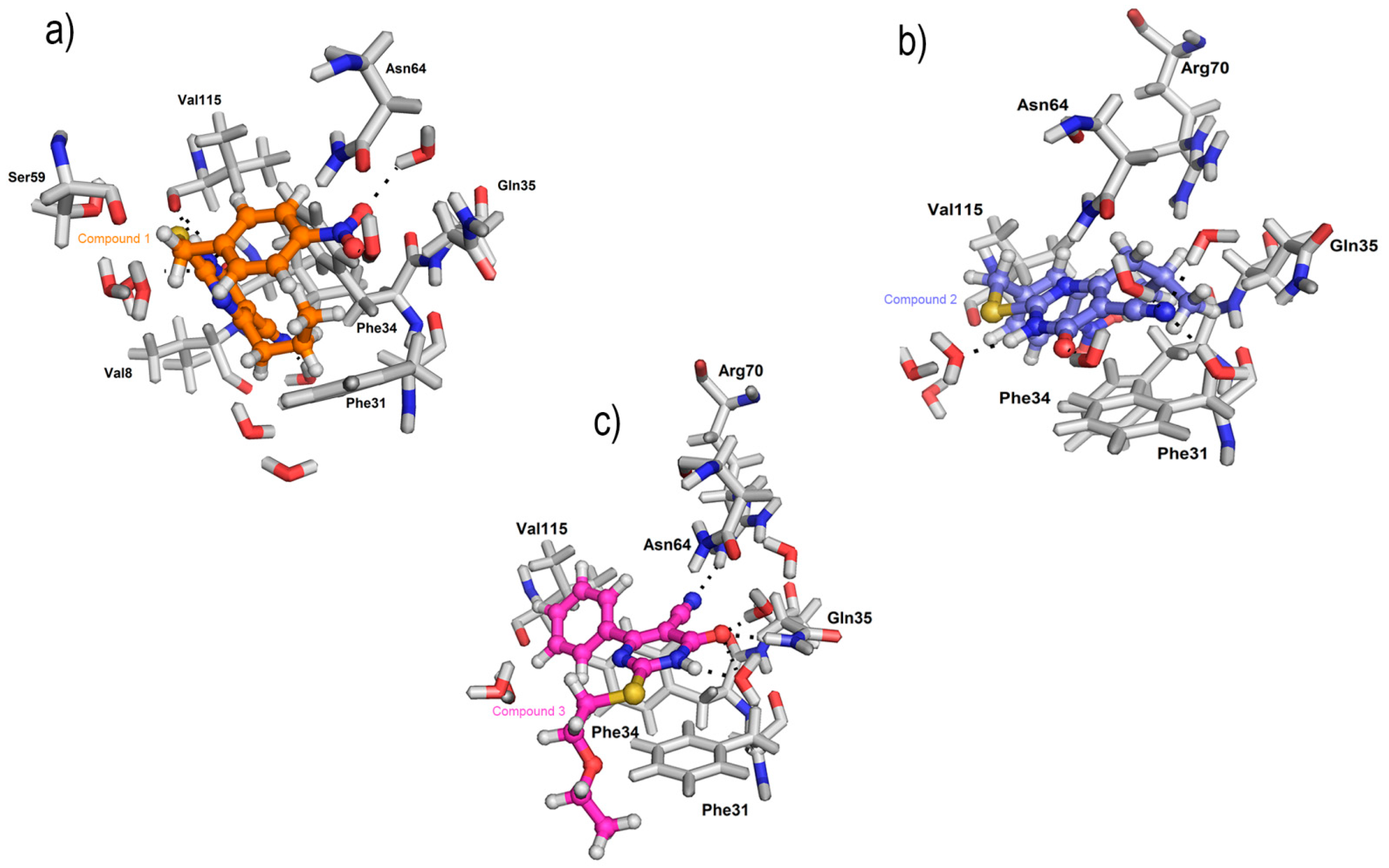
2.4. Molecular Docking Studies as Human Dihydrofolate Reductase (hDHFR) Inhibitors
3. Materials and Methods
3.1. Synthesis and Crystallization
3.2. Single Crystal X-ray Diffraction Determination
3.3. Computational Details
3.3.1. Hirshfeld Surface Analysis
3.3.2. Molecular Docking Studies
Protein Preparation and Grid Generation
Ligand Preparation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Kumar, S.; Narasimhan, B. Therapeutic potential of heterocyclic pyrimidine scaffolds. Chem. Cent. J. 2018, 12, 38. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.S.; Lenzi, M.; Lim, T.H.; Hotchkiss, K.A.; Wilson, P.; Schwartz, E.L. Novel 6-substituted uracil analogs as inhibitors of the angiogenic actions of thymidine phosphorylase. Biochem. Pharmacol. 2001, 62, 1257–1263. [Google Scholar] [CrossRef]
- Matsushita, S.; Nitanda, T.; Furukawa, T.; Sumizawa, T.; Tani, A.; Nishimoto, K.; Akiba, S.; Miyadera, K.; Fukushima, M.; Yamada, Y.; et al. The effect of a thymidine phosphorylase inhibitor on angiogenesis and apoptosis in tumors. Cancer Res. 1999, 59, 1911–1916. [Google Scholar] [PubMed]
- Boisdron-Celle, M.; Remaud, G.; Traore, S.; Poirier, A.L.; Gamelin, L.; Morel, A.; Gamelin, E. 5-Fluorouracil-related severe toxicity: A comparison of different methods for the pretherapeutic detection of dihydropyrimidine dehydrogenase deficiency. Cancer Lett. 2007, 249, 271–282. [Google Scholar] [CrossRef]
- Cole, C.; Reigan, P.; Gbaj, A.; Edwards, P.N.; Douglas, K.T.; Stratford, I.J.; Freeman, S.; Jaffar, M. Potent tumor-selective nitroimidazolylmethyluracil prodrug derivatives: Inhibitors of the angiogenic enzyme thymidine phosphorylase. J. Med. Chem. 2003, 46, 207–209. [Google Scholar] [CrossRef]
- Pedikian, A.Y.; Stroihlein, J.; Karlin, D.; Bodey, G.P. A comparative study of oral tegafur and intravenous 5-fluorouracil in patients with metastatic colorectal cancers. Am. J. Clin. Oncol. 1983, 6, 181–186. [Google Scholar] [CrossRef]
- De Corte, B.L. From 4,5,6,7-tetrahydro-5-methylimidazo[4,5,1-jk](1,4)-benzodiazepin-2(1H)-one (TIBO) to etravirine (TMC125): Fifteen years of research on non-nucleoside inhibitors of HIV-1 reverse transcriptase. J. Med. Chem. 2005, 48, 1689–1696. [Google Scholar] [CrossRef]
- Andries, K.; Azijn, H.; Thielemans, T.; Ludovici, D.; Kukla, M.; Heeres, J.; Janssen, P.; De Corte, B.; Vingerhoets, J.; Pauwels, R.; et al. TMC125, a novel next-generation nonnucleoside reverse transcriptase inhibitor active against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 2004, 48, 4680–4686. [Google Scholar] [CrossRef] [Green Version]
- Ji, L.; Chen, F.-E.; De Clercq, E.; Balzarini, J.; Pannecouque, C. Synthesis and anti-HIV-1 activity evaluation of 5-alkyl-2-alkylthio-6-(arylcarbonyl or alpha-cyanoarylmethyl)-3,4-dihydropyrimidin-4(3H)-ones as novel non-nucleoside HIV-1 reverse transcriptase inhibitors. J. Med. Chem. 2007, 50, 1778–1786. [Google Scholar] [CrossRef]
- Mai, A.; Artico, M.; Rotili, D.; Tarantino, D.; Clotet-Codina, I.; Armand-Ugón, M.; Ragno, R.; Simeoni, S.; Sbardella, G.; Nawrozkij, M.B.; et al. Synthesis and biological properties of novel 2-aminopyrimidin-4(3H)-ones highly potent against HIV-1 mutant strains. J. Med. Chem. 2007, 50, 5412–5424. [Google Scholar] [CrossRef]
- Mai, A.; Artico, M.; Ragno, R.; Sbardella, G.; Massa, S.; Musiu, C.; Mura, M.; Marturana, F.; Cadeddu, A.; Maga, G.; et al. 5-Alkyl-2-alkylamino-6-(2,6-difluorophenylalkyl)-3,4-dihydropyrimidin-4(3H)-ones, a new series of potent, broad-spectrum non-nucleoside reverse transcriptase inhibitors belonging to the DABO family. Bioorg. Med. Chem. 2005, 13, 2065–2077. [Google Scholar] [CrossRef]
- Yang, S.; Chen, F.E.; De Clercq, E. Dihydro-alkoxyl-benzyl-oxopyrimidine derivatives (DABOs) as non-nucleoside reverse transcriptase inhibitors: An update review (2001–2011). Curr. Med. Chem. 2012, 19, 152–162. [Google Scholar] [CrossRef]
- Gauni, K.K.; Kohlhage, H. In vitro and in vivo virostatic properties of alkylated pyrimidines against DNA and RNA viruses. Chemotherapy 1969, 14, 158–169. [Google Scholar] [CrossRef]
- Semaine, W.; Johar, M.; Tyrrell, D.L.; Kumar, R.; Agrawal, B. Inhibition of hepatitis B virus (HBV) replication by pyrimidines bearing an acyclic moiety: Effect on wild-type and mutant HBV. J. Med. Chem. 2006, 49, 2049–2054. [Google Scholar] [CrossRef]
- Ramajayam, R.; Tan, K.P.; Liu, H.G.; Liang, P.H. Synthesis, docking studies, and evaluation of pyrimidines as inhibitors of SARS-CoV 3CL protease. Bioorg. Med. Chem. Lett. 2010, 20, 3569–3572. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer, B.I.; Dicker, A.P.; Bertino, J.R. Dihydrofolate reductase as a therapeutic target. FASEB J. 1990, 4, 2441–2452. [Google Scholar] [CrossRef] [PubMed]
- Kompis, I.M.; Islam, K.; Then, R.L. DNA and RNA synthesis: Antifolates. Chem. Rev. 2005, 105, 593–620. [Google Scholar] [CrossRef]
- Amyes, S.G. Comparative antibacterial spectrum of trimethoprim and brodimoprim. J. Chemother. 1993, 5, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Sincak, C.A. Iclaprim, a novel diaminopyrimidine for the treatment of resistant Gram-positive infections. Ann. Pharmacother. 2009, 43, 1107–1114. [Google Scholar] [CrossRef]
- Locher, H.H.; Schlunegger, H.; Hartman, P.G.; Anghern, P.; Then, R.L. Antibacterial activities of epiroprim, a new dihydrofolate reductase inhibitor, alone and in combination with dapsone. Antimicrob. Agents Chemother. 1996, 40, 1376–1381. [Google Scholar] [CrossRef] [Green Version]
- Walzer, P.D.; Kim, C.K.; Foy, J.M.; Linke, M.J.; Cushion, M.T. Inhibitors of folic acid synthesis in the treatment of experimental Pneumocystis carinii pneumonia. Antimicrob. Agents Chemother. 1988, 32, 96–103. [Google Scholar] [CrossRef] [Green Version]
- Cowman, A.F.; Morry, M.J.; Biggs, B.A.; Cross, G.A.; Foote, S.J. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 1988, 85, 9109–9113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunyarataphan, S.; Leartsakulpanich, U.; Taweechai, S.; Tarnchompoo, B.; Kamchonwongpaisan, S.; Yuthavong, Y. Evaluation of the activities of pyrimethamine analogs against Plasmodium vivax and Plasmodium falciparum dihydrofolate reductase-thymidylate synthase using in vitro enzyme inhibition and bacterial complementation assays. Antimicrob. Agents Chemother. 2006, 50, 3631–3637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suryawanshi, S.N.; Bhat, B.A.; Pandey, S.; Chandra, N.; Gupta, S. Chemotherapy of leishmaniasis. Part VII: Synthesis and bioevaluation of substituted terpenyl pyrimidines. Eur. J. Med. Chem. 2007, 42, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Mc Carthy, O.K.; Schipani, A.; Buendía, A.M.; Ruiz-Perez, L.M.; Kaiser, M.; Brun, R.; Pacanowska, D.G.; Gilbert, I.H. Design, synthesis and evaluation of novel uracil amino acid conjugates for the inhibition of Trypanosoma cruzi dUTPase. Bioorg. Med. Chem. Lett. 2006, 16, 3809–3812. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, M.B.; Salunkhe, S.M.; Patil, D.R.; Anbhule, P.V. A novel and efficient one step synthesis of 2-amino-5-cyano-6-hydroxy-4-aryl pyrimidines and their anti-bacterial activity. Eur. J. Med. Chem. 2009, 44, 2651–2654. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, N.; Srivastava, P.; Raghuwanshi, S.K.; Upadhyay, D.N.; Sinha, S.; Shukla, P.K.; Ji Ram, V. Chloropyrimidines as a new class of antimicrobial agents. Bioorg. Med. Chem. 2002, 10, 869–874. [Google Scholar] [CrossRef]
- Agarwal, N.; Raghuwanshi, S.K.; Upadhyay, D.N.; Shukla, P.K.; Ram, V.J. Suitably functionalised pyrimidines as potential antimycotic agents. Bioorg. Med. Chem. Lett. 2000, 10, 703–706. [Google Scholar] [CrossRef]
- Taher, A.T.; Abou-Seri, S.M. Synthesis and bioactivity evaluation of new 6-aryl-5-cyano thiouracils as potential antimicrobial and anticancer agents. Molecules 2012, 17, 9868–9886. [Google Scholar] [CrossRef]
- Al-Abdullah, E.S.; Al-Obaid, A.-R.M.; Al-Deeb, O.A.; Habib, E.E.; El-Emam, A.A. Synthesis of novel 6-phenyl-2,4-disubstituted pyrimidine-5-carbonitriles as potential antimicrobial agents. Eur. J. Med. Chem. 2011, 46, 4642–4647. [Google Scholar] [CrossRef]
- Al-Abdullah, E.S.; Al-Turkistani, A.A.; Al-Deeb, O.A.; El-Brollosy, N.R.; Habib, E.E.; El-Emam, A.A. Pyrimidine-5-carbonitriles II: Synthesis and antimicrobial activity of novel 6-alkyl-2,4-disubstituted pyrimidine-5-carbonitriles. Drug Res. 2014, 64, 31–39. [Google Scholar] [CrossRef]
- Al-Deeb, O.A.; Al-Turkistani, A.A.; Al-Abdullah, E.A.; El-Brollosy, N.R.; Habib, E.E.; El-Emam, A.A. Pyrimidine-5-carbonitriles—Part III: Synthesis and antimicrobial activity of novel 6-(2-substituted propyl)-2,4-disubstituted pyrimidine-5-carbonitriles. Heterocycl. Commun. 2013, 19, 411–419. [Google Scholar] [CrossRef]
- Kambe, S.; Saito, K.; Kishi, H. A one-step synthesis of 4-oxo-2-thioxopyrimidine derivatives by ternary condensation of ethyl cyanoacetate, aldehydes, and thiourea. Synthesis 1979, 287–289. [Google Scholar] [CrossRef]
- El-Emam, A.A.; Demirtaş, G.; Dege, N.; Al-Deeb, O.A.; El-Brollosy, N.R. 2-[(2-Meth-oxyethyl)sulfanyl]-4-(2-methyl-propyl)- 6-oxo-1,6-dihydropyrimidine-5-carbonitrile. Acta Crystallogr. 2012, E68, o1379. [Google Scholar] [CrossRef]
- Al-Tamimi, A.-M.S.; Ghabbour, H.A.; El-Emam, A.A. Crystal structure of 6-oxo-4-propyl-2-(propylthio)-1,6-dihydropyri- midine-5-carbonitrile, C11H15N3OS. Z. Kristallogr. NCS 2016, 231, 583–585. [Google Scholar] [CrossRef] [Green Version]
- Sert, Y.; El-Emam, A.A.; Al-Deeb, O.A.; Al-Turkistani, A.A.; Ucun, F.; Cırak, C. The biomolecule, 2-[(2-methoxyl)sulfanyl]-4- (2-methylpropyl)-6-oxo-1,6-dihydropyrimidine-5-carbonitrile: FT-IR, Laser Raman spectra and DFT. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2014, 126, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Sert, Y.; Al-Turkistani, A.A.; Al-Deeb, O.A.; El-Emam, A.A.; Ucun, F.; Çırak, Ç. Experimental FT-IR, Laser-Raman and DFT spectroscopic analysis of a potential chemotherapeutic agent 6-(2-methylpropyl)-4-oxo-2-sulfanylidene-1,2,3,4-tetrahydropy-rimidine-5-carbonitrile. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2014, 120, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.J.; Thomas, S.P.; Shi, M.W.; Jayatilaka, D.; Spackman, M.A. Energy frameworks: Insights into interaction anisotropy and the mechanical properties of molecular crystals. Chem. Commun. 2015, 51, 3735–3738. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Jayatilaka, D.; Spackman, M.A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Commun. 2007, 3814–3816. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, J.J.; Fabbiani, F.P.A.; Spackman, M.A. Comparison of polymorphic molecular crystal structures through Hirshfeld surface snalysis. Cryst. Growth Des. 2007, 7, 755–769. [Google Scholar] [CrossRef]
- Spackman, M.A.; McKinnon, J.J. Fingerprinting intermolecular interactions in molecular crystals. CrystEngComm 2002, 4, 378–392. [Google Scholar] [CrossRef]
- LigPrep; v2.5; Schrödinger Inc.: Portland, OR, USA, 2011.
- Volpato, J.P.; Yachnin, B.J.; Blanchet, J.; Guerrero, V.; Poulin, L.; Fossati, E.; Berghuis, A.M.; Pelletier, J.N. Multiple conformers in active site of human dihydrofolate reductase F31R/Q35E double mutant suggest structural basis for methotrexate resistance. J. Biol. Chem. 2009, 284, 20079–20089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, R.C.; Reid, J.S. The analytical calculation of absorption in multifaceted crystals. Acta Cryst. A 1995, 51, 887–897. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. A complete structure solution, refinement and analysis program. J. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Cryst. C 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer17; University of Western Australia: Perth, Australia, 2017. [Google Scholar]
- Oefner, C.; D’Arcy, A.; Winkler, F.K. Crystal structure of human dihydrofolate reductase complexed with folate. Eur. J. Biochem. 1988, 174, 377–385. [Google Scholar] [CrossRef]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aid. Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef]

| Compound 1 | Compound 2 | Compound 3 | |
|---|---|---|---|
| Empirical formula | C15H14N4O3S | C16H16N4O3S | C15H17N3O3S |
| Formula weight | 330.36 | 344.39 | 319.38 |
| Temperature (K) | 293 (2) | ||
| Crystal system | Monoclinic | Monoclinic | Triclinic |
| Space group | P21/n | C2/c | P-1 |
| a/Å b/Å c/Å | 12.5792 (7) 9.5493 (5) 13.3158 (8) | 18.3676 (7) 5.7996 (2) 31.8592 (12) | 7.1456 (4) 10.8642 (6) 11.2566 (7) |
| α/° β/° γ/° | 90 106.432 (7) 90 | 90 95.343 (4) 90 | 104.672 (5) 108.159 (5) 95.421 (5) |
| Volume/Å3 | 1534.20 (16) | 3379.0 (2) | 788.84 (8) |
| Z | 4 | 8 | 2 |
| Calculated density (g/cm3) | 1.430 | 1.354 | 1.345 |
| Absorption coefficient (mm−1) | 2.068 | 1.899 | 1.968 |
| F(000) | 688 | 1440 | 336 |
| Crystal size (mm3) | 0.16 × 0.12 × 0.08 | 0.16 × 0.05 × 0.05 | 0.26 × 0.14 × 0.10 |
| Radiation | Cu Kα (λ = 1.54184) | ||
| 2Θ range for data collection | 8.5 to 151.4 | 5.6 to 151.4 | 8.6 to 151.1 |
| Index ranges | −15 ≤ h ≤ 15, −11 ≤ k ≤ 11, −16 ≤ l ≤ 16 | −22 ≤ h ≤ 22, −7 ≤ k ≤ 6, −39 ≤ l ≤ 40 | −8 ≤ h ≤ 8, −13 ≤ k ≤ 13, −13 ≤ l ≤ 11 |
| Reflections collected | 15422 | 16836 | 12241 |
| Independent reflections | 3163 [Rint = 0.0971, Rsigma = 0.0654] | 3418 [Rint = 0.1098, Rsigma = 0.0941] | 3220 [Rint = 0.0442, Rsigma = 0.0377] |
| Data/restraints/parameter | 3163/0/213 | 3418/2/211 | 3220/2/212 |
| Goodness-of-fit on F2 | 1.030 | 0.998 | 1.035 |
| Final R indices [I>2 σ(I)] | R1 = 0.0569, wR2 = 0.1209 | R1 = 0.0766, wR2 = 0.1777 | R1 = 0.0432, wR2 = 0.1040 |
| Final R indices (all data) | R1 = 0.1087, wR2 = 0.1479 | R1 = 0.1565, wR2 = 0.2358 | R1 = 0.0689, wR2 = 0.1210 |
| Largest diff. peak and hole (e.Å−3) | 0.20/−0.25 | 0.52/−0.36 | 0.16/−0.18 |
| CCDC number | 2063317 | 2063318 | 2063320 |
| Compound | Docking Score | Glide Energy | No. of Interactions | No. of H-Bonding Residues | Interacting Residues |
|---|---|---|---|---|---|
| 1 | −8.53 kcal/mol | −93.57 | 1 | 0 | Phe34 (pi.cation) Interaction with Gln35, Asn64, and Ser59 via water |
| 2 | −8.34 kcal/mol | −84.82 | 1 | 1 | H-bond: Val115, Phe34 (pi…pi), Phe31 (pi…cation) Interaction with Gln35, Asn64, and Val8 via water |
| 3 | −7.34 kcal/mol | −75.03 | 2 | 2 | H-bond: Arg 70, and Gln35; water-mediated interactions with Asn4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Wahaibi, L.H.; Shaik, A.; Elmorsy, M.A.; Abdelbaky, M.S.M.; Garcia-Granda, S.; Thamotharan, S.; Thiruvenkatam, V.; El-Emam, A.A. Structural Insights of Three 2,4-Disubstituted Dihydropyrimidine-5-carbonitriles as Potential Dihydrofolate Reductase Inhibitors. Molecules 2021, 26, 3286. https://doi.org/10.3390/molecules26113286
Al-Wahaibi LH, Shaik A, Elmorsy MA, Abdelbaky MSM, Garcia-Granda S, Thamotharan S, Thiruvenkatam V, El-Emam AA. Structural Insights of Three 2,4-Disubstituted Dihydropyrimidine-5-carbonitriles as Potential Dihydrofolate Reductase Inhibitors. Molecules. 2021; 26(11):3286. https://doi.org/10.3390/molecules26113286
Chicago/Turabian StyleAl-Wahaibi, Lamya H., Althaf Shaik, Mohammed A. Elmorsy, Mohammed S. M. Abdelbaky, Santiago Garcia-Granda, Subbiah Thamotharan, Vijay Thiruvenkatam, and Ali A. El-Emam. 2021. "Structural Insights of Three 2,4-Disubstituted Dihydropyrimidine-5-carbonitriles as Potential Dihydrofolate Reductase Inhibitors" Molecules 26, no. 11: 3286. https://doi.org/10.3390/molecules26113286
APA StyleAl-Wahaibi, L. H., Shaik, A., Elmorsy, M. A., Abdelbaky, M. S. M., Garcia-Granda, S., Thamotharan, S., Thiruvenkatam, V., & El-Emam, A. A. (2021). Structural Insights of Three 2,4-Disubstituted Dihydropyrimidine-5-carbonitriles as Potential Dihydrofolate Reductase Inhibitors. Molecules, 26(11), 3286. https://doi.org/10.3390/molecules26113286








