The Advantage of Automatic Peer-Reviewing of 13C-NMR Reference Data Using the CSEARCH-Protocol †
Abstract
:1. Introduction
2. Results and Discussion
2.1. The CSEARCH-Robot-Referee—General Overview
- Formal check of the structure (valency, charge, and stereocenter);
- Check formal correctness of the supplied peak list (symmetry and exchangeable assigned signals);
- Perform statistical analysis based on underlying data used during the spectrum prediction process to allow for the evaluation of the quality of the result;
- Assign signals if unassigned signals are given;
- Calculate and visualize the coincidence between the structure proposal and the given experimental data;
- Perform a search for identical structures contained in the underlying knowledge base;
- Perform a search for an identical spectral pattern associated with different structures;
- Detect positions with a large deviation between the experimental and predicted values;
- Start structure generator, which modifies the topology of the given structure exclusively at the positions with a large deviation;
- Perform dereplication based on the given peak list using the database of 520 million predicted spectra.
2.2. Examples of Wrong/Useless 13C-NMR Data
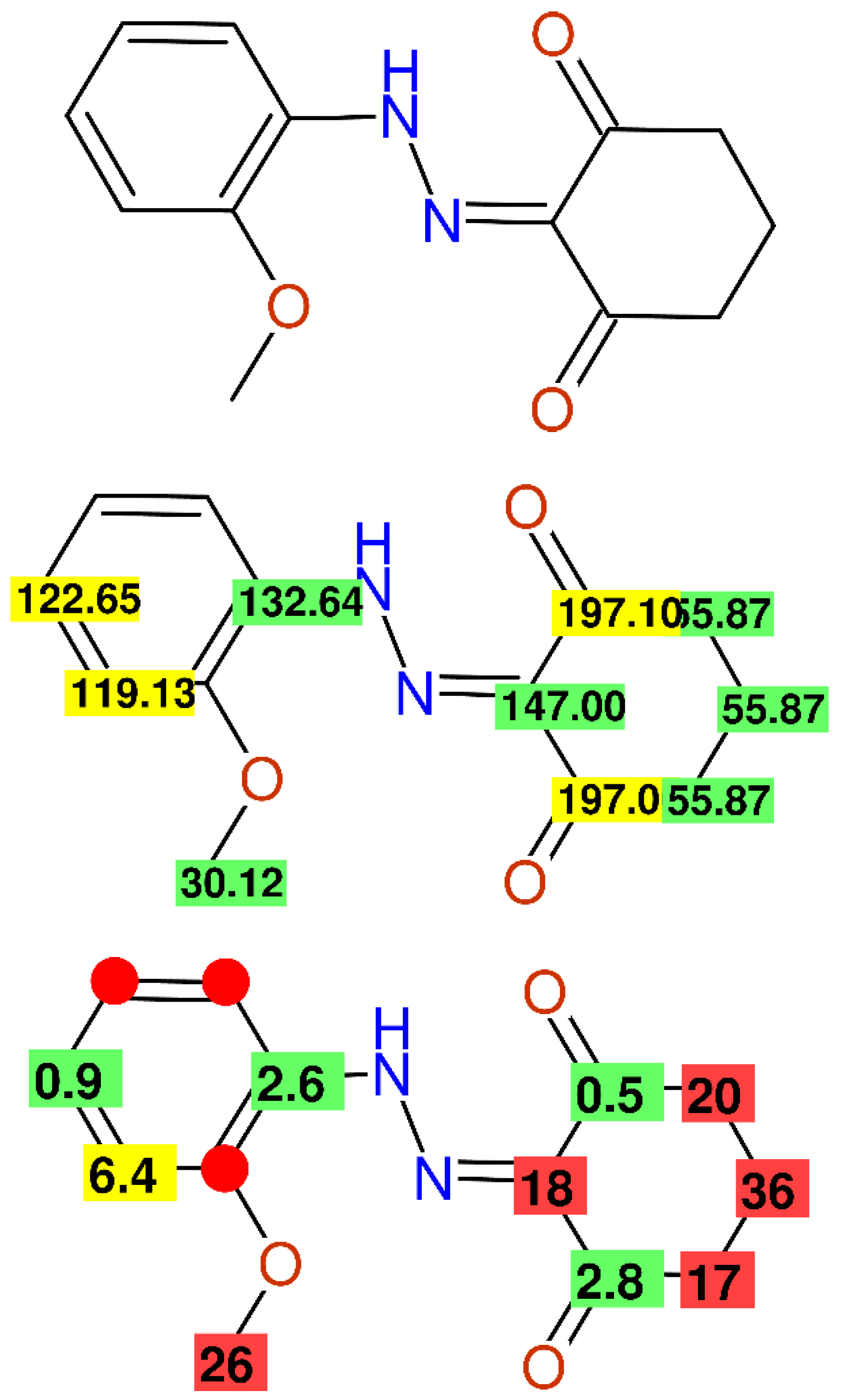
2.2.1. Using the Same Data Twice
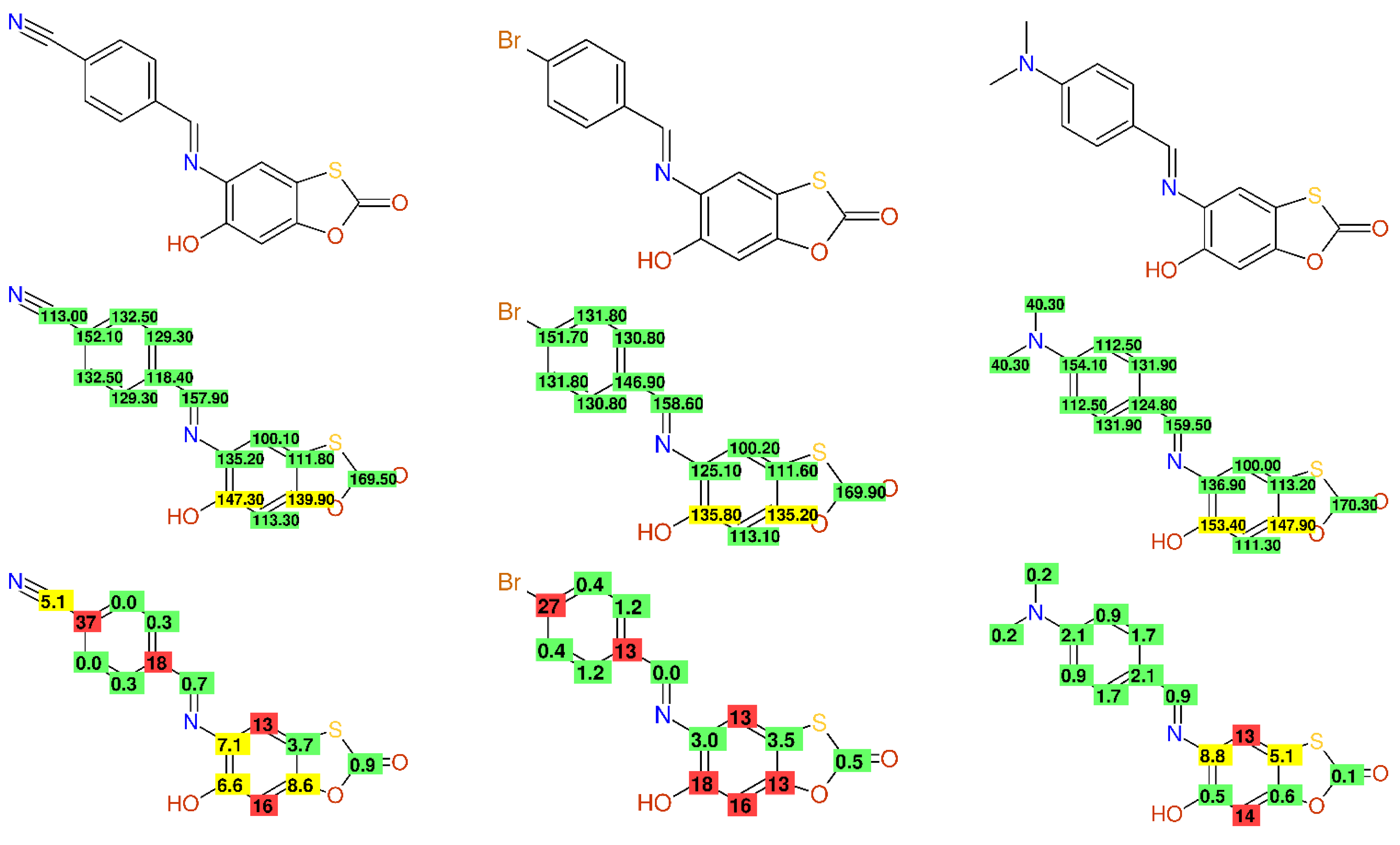
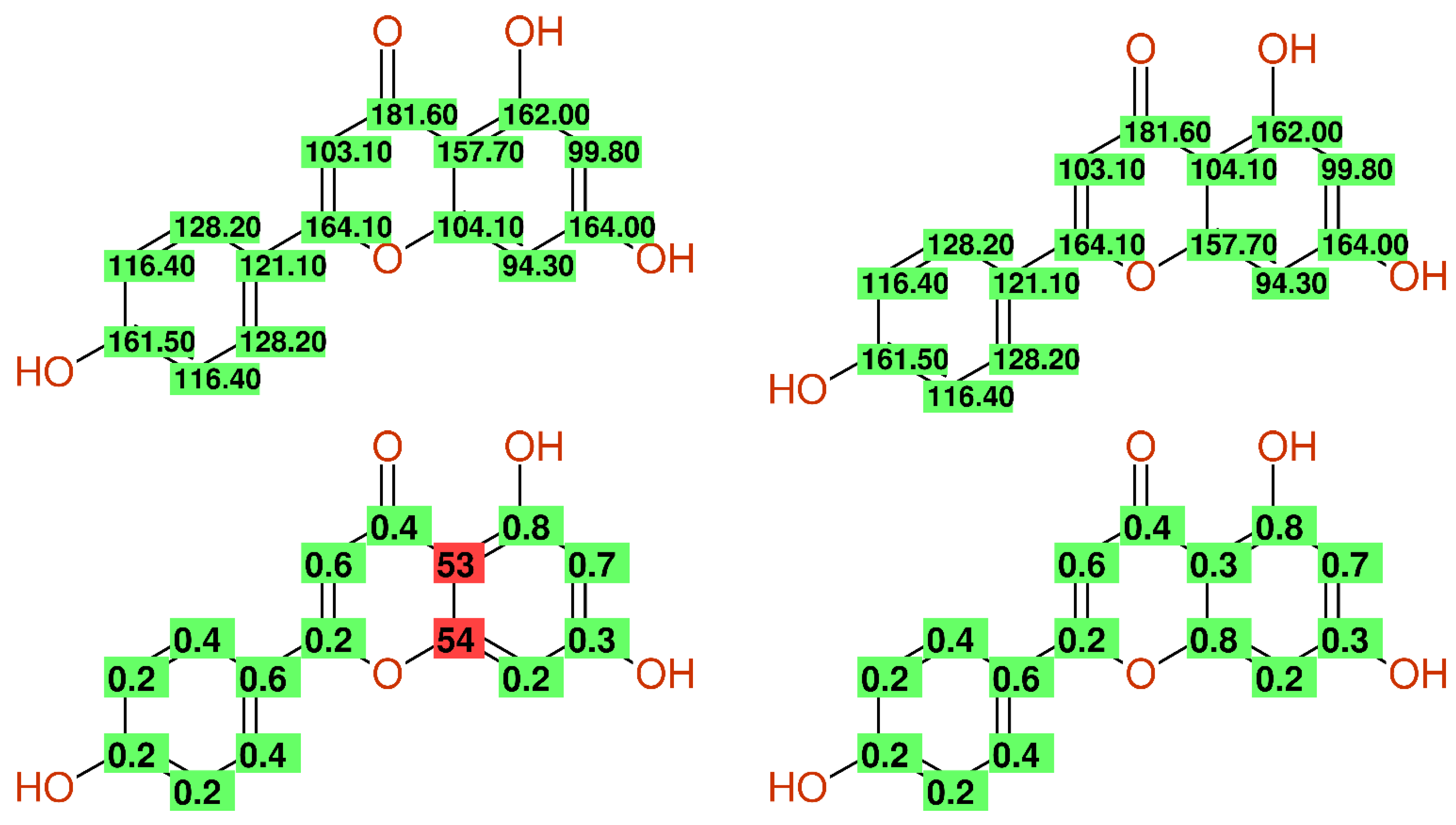
2.2.2. Wrong Values—Strange Substituent Effects
2.2.3. Typos and Transmission Errors
2.2.4. Trivial Assignment Errors
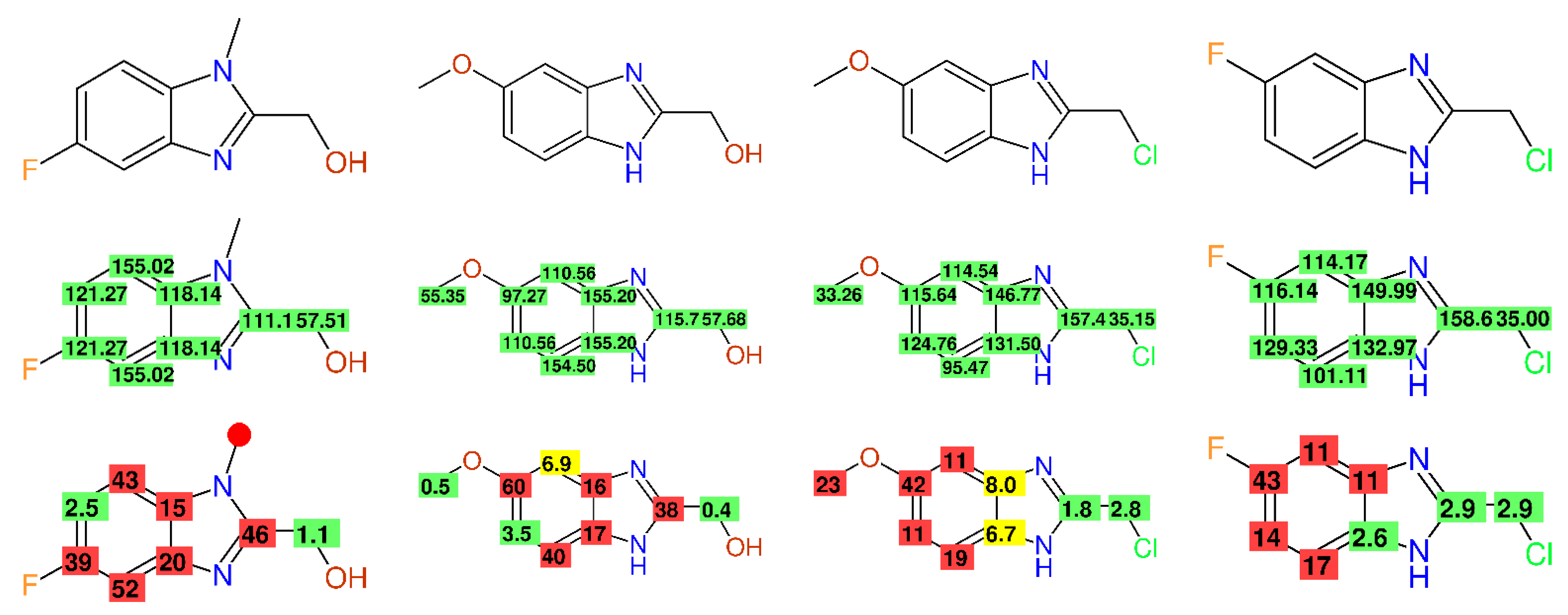
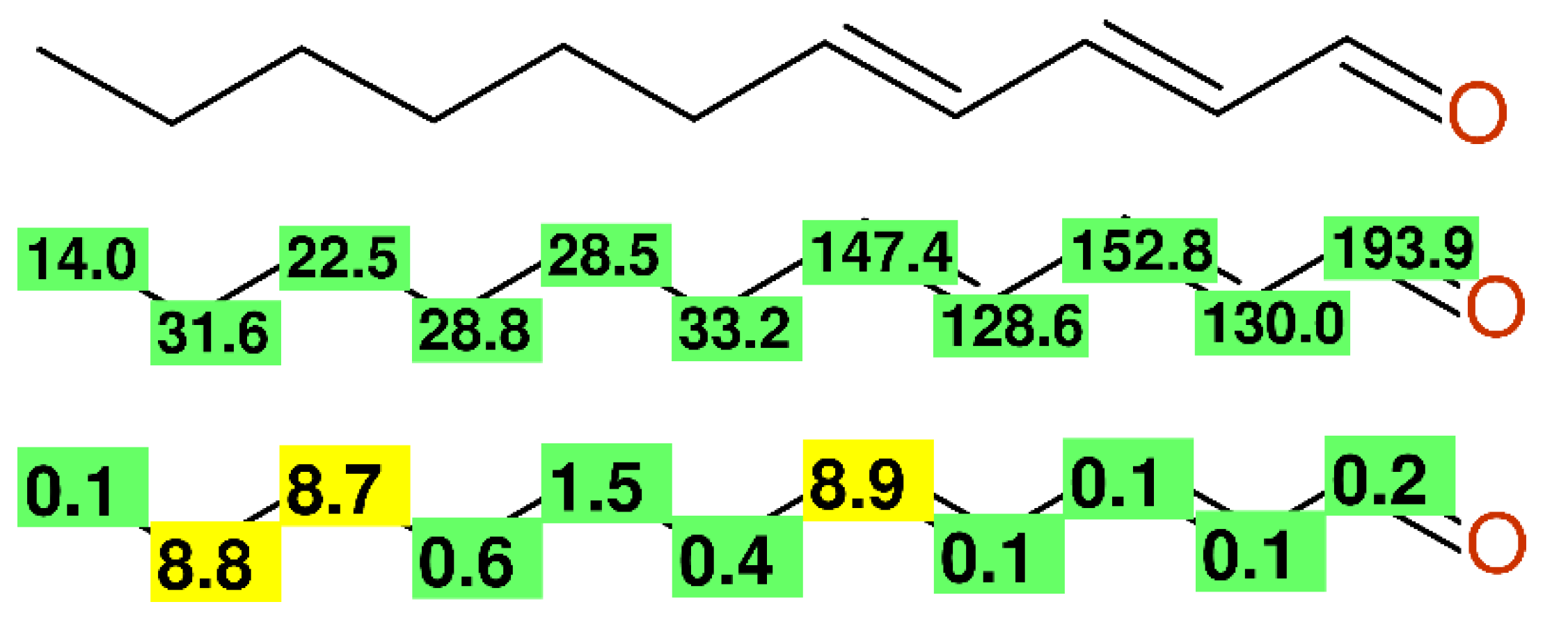
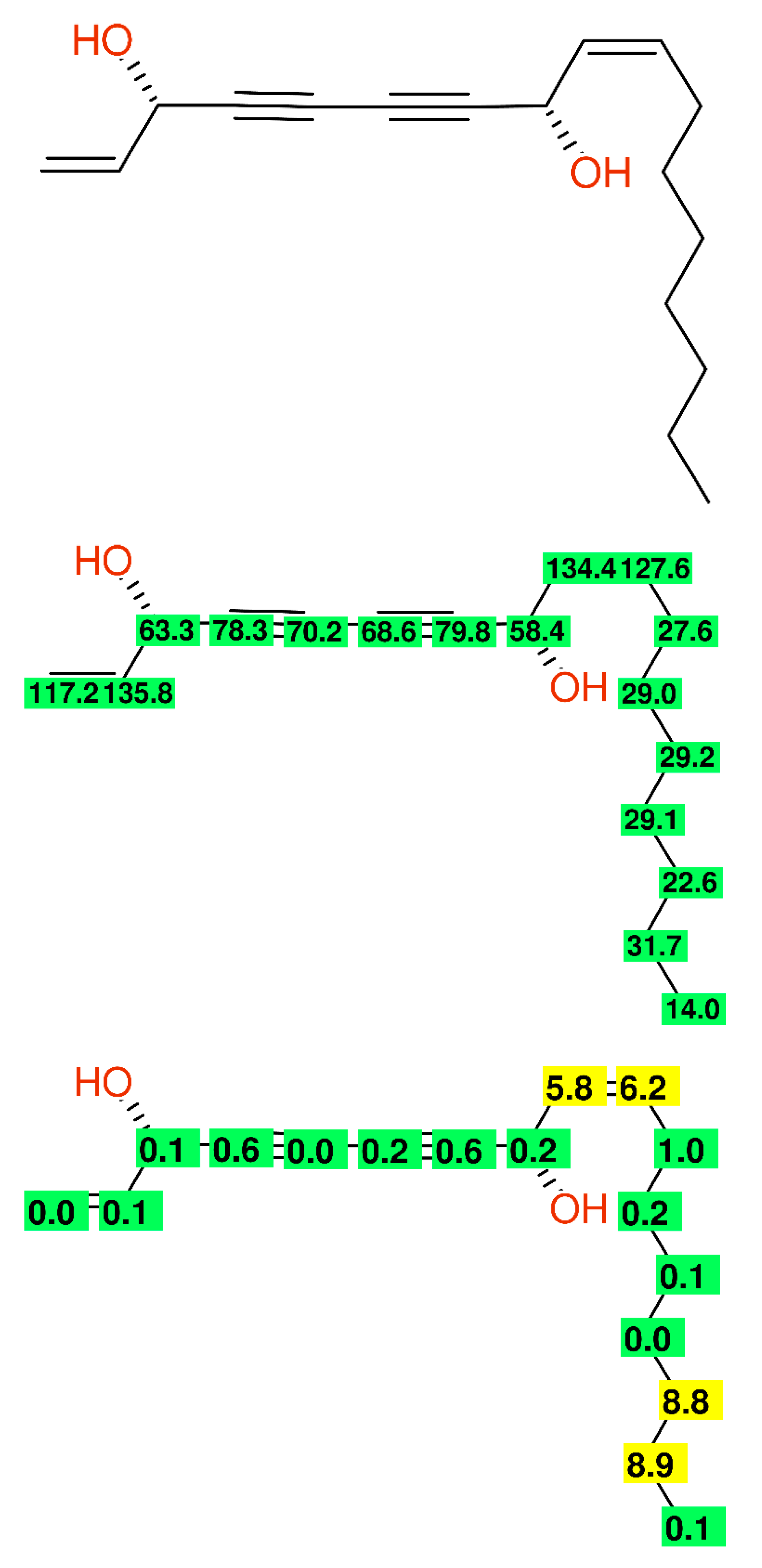
2.2.5. Alkyl-Chains
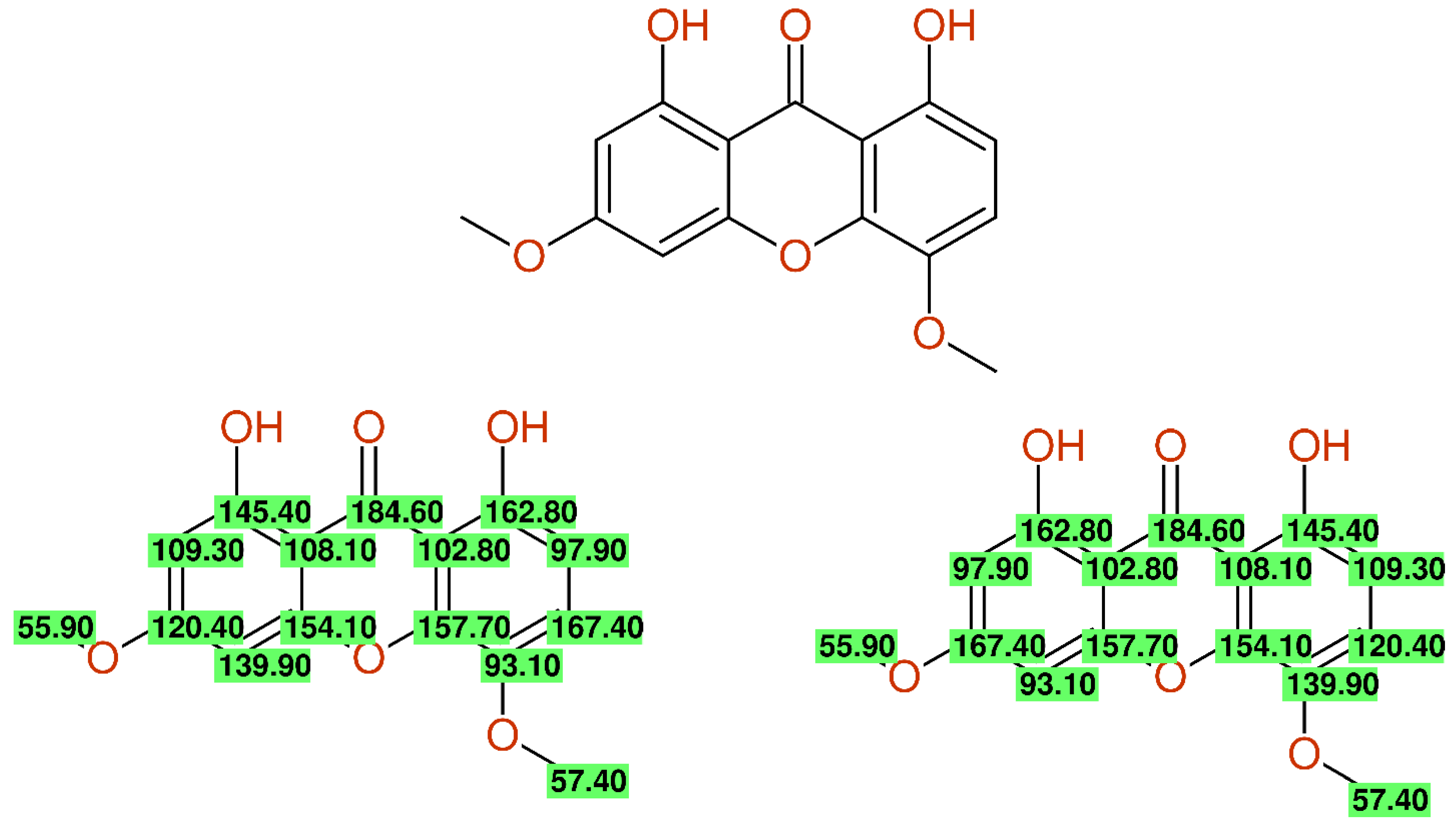
2.2.6. Wrong Structure Drawing
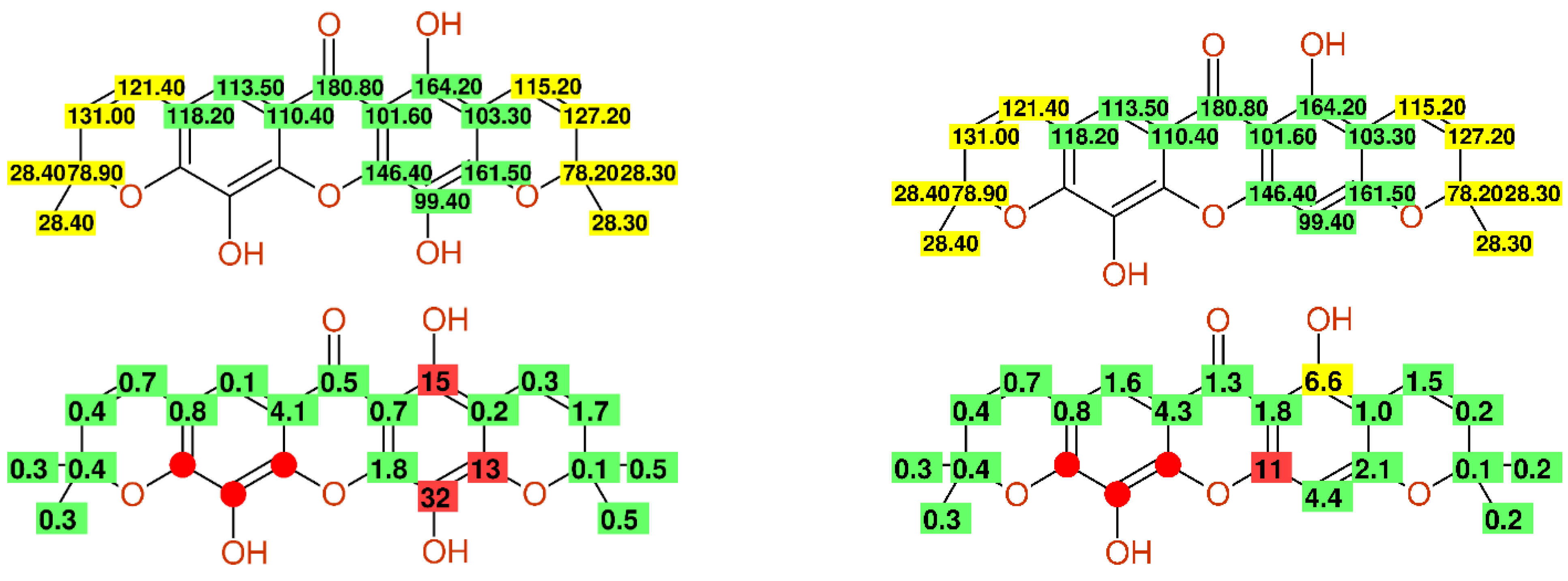
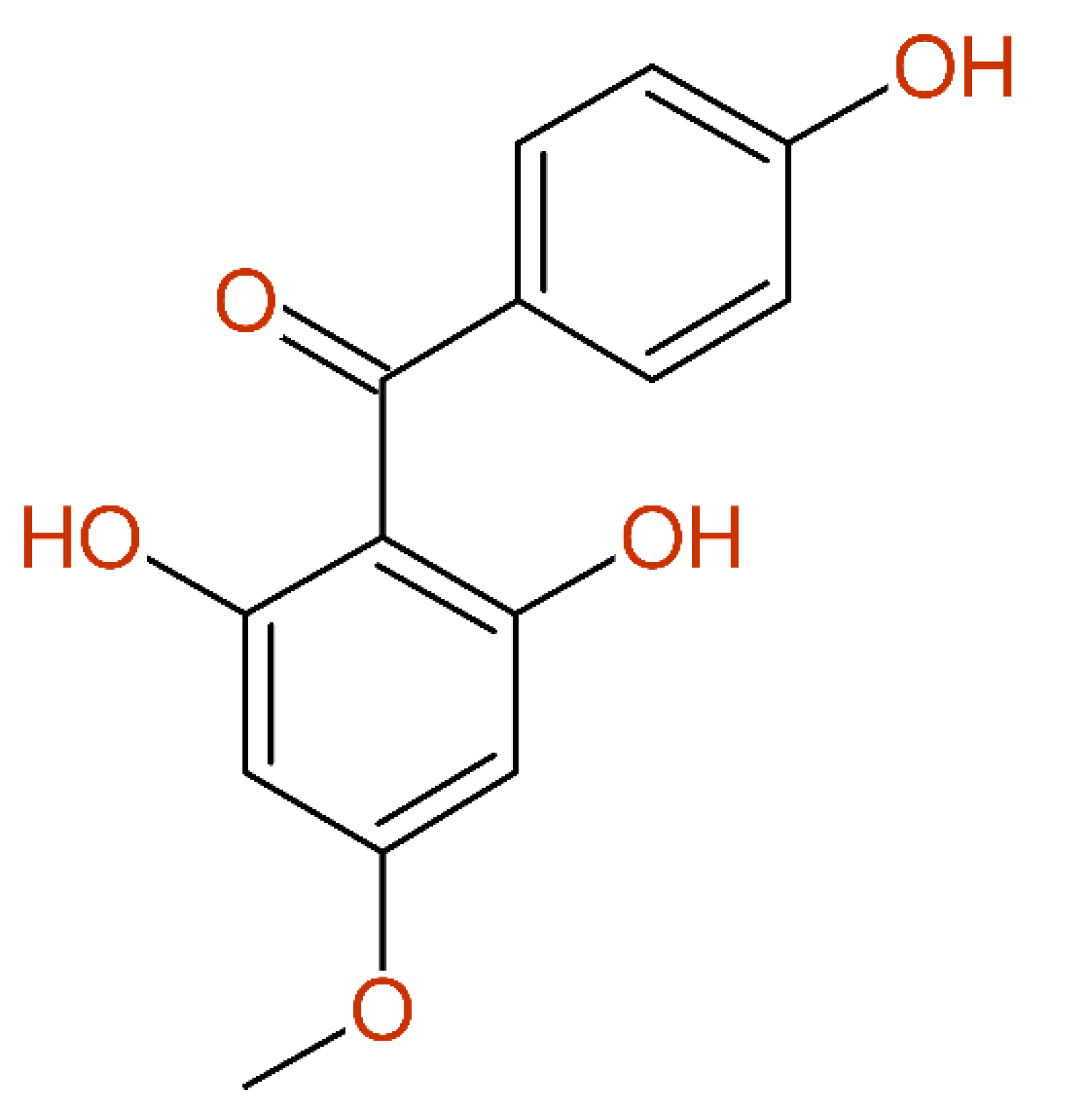
2.2.7. Multiple Inconsistencies
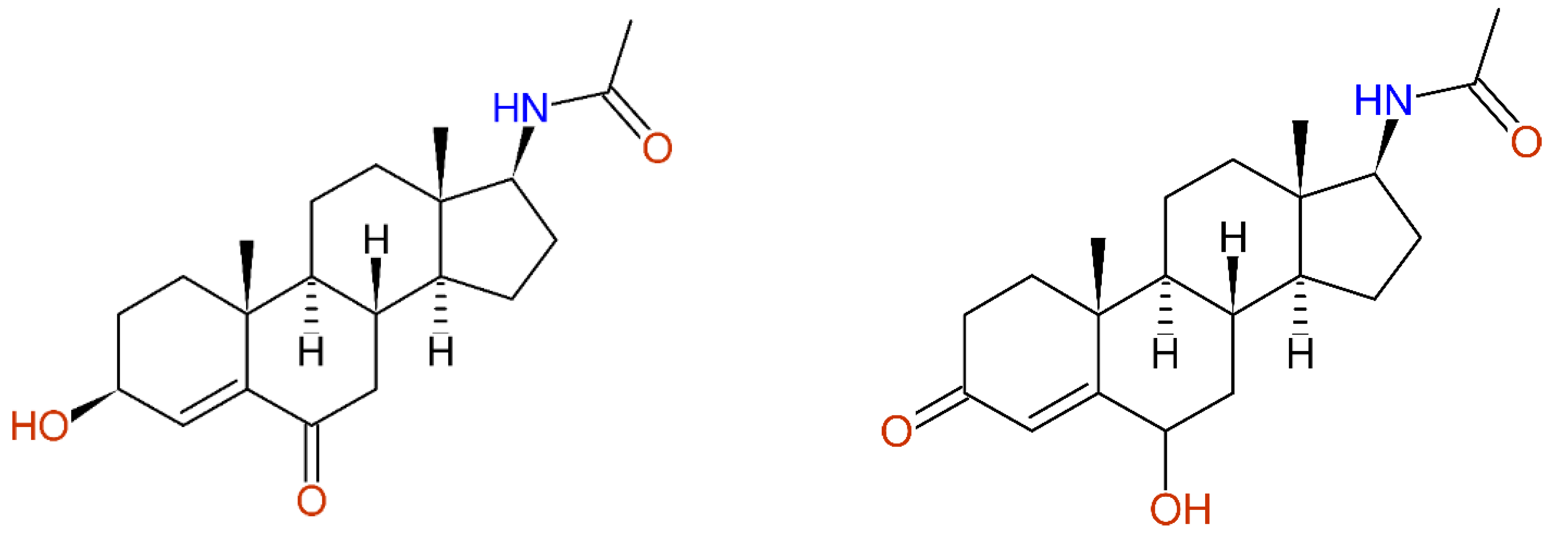
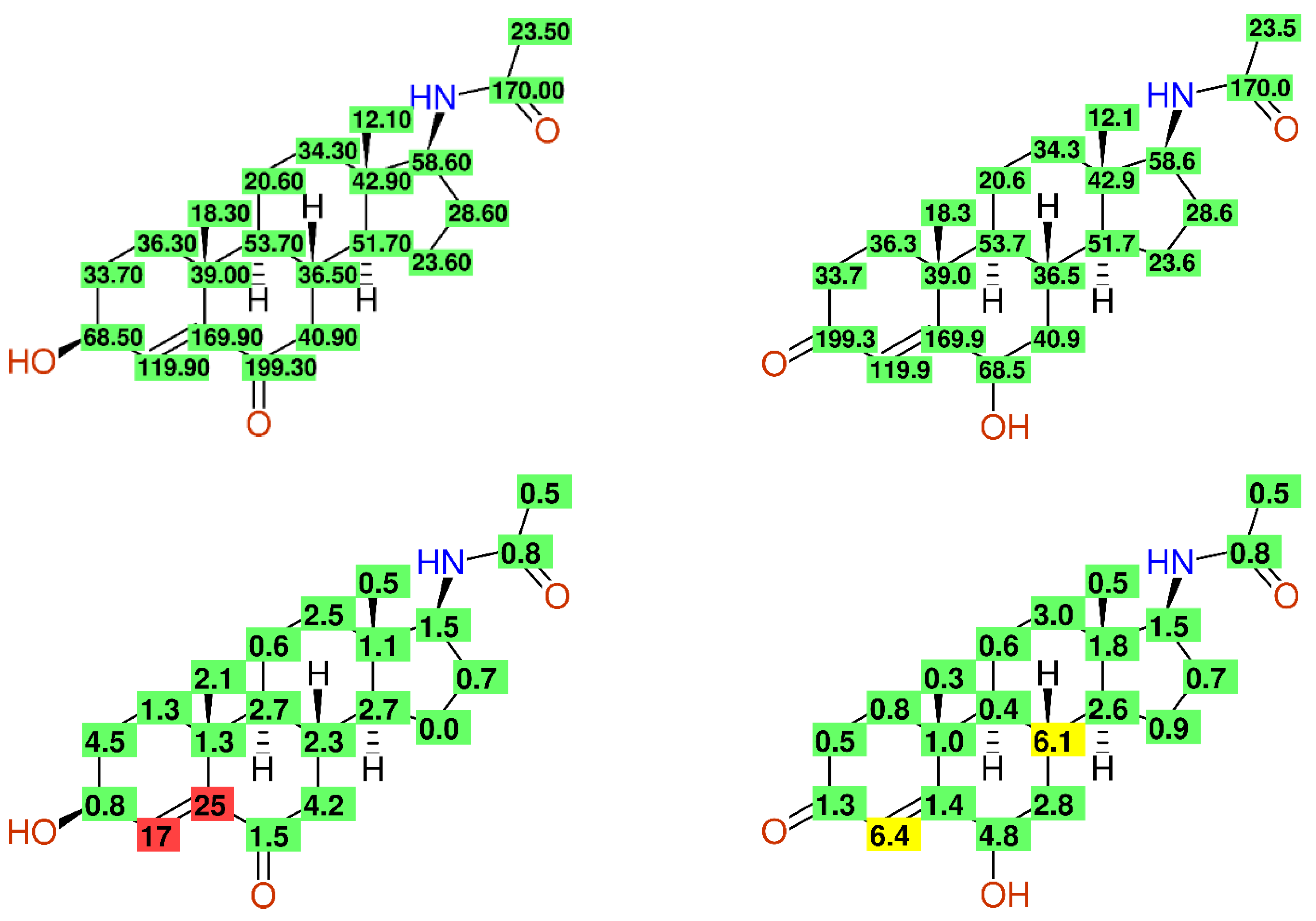
2.2.8. Fully Automatic Structure Revisions
3. Conclusions
- Structures must be deposited as computer-readable files (e.g., MOLfile)—every structure drawing must be derived thereof in order to avoid drawing errors.
- Every structure must be accompanied by a unique identifier (e.g., INCHIKEYS), avoiding transmission errors and allowing “identical structure search” via text-based search-engines.
- As many steps as possible in the process of publishing results must be done in a software-supported way. It must be mandatory for authors to provide all of the necessary experimental data (free induction decays for NMR) so that every conclusion can be reproduced, and it must be mandatory for the publishers to allow for the uploading of such data. Furthermore, these data must be made searchable and downloadable for later use.
- Improvements in the actual “peer-reviewing” workflow, including massive computer-supported technologies, every set of spectral data has to be automatically checked during upload, and the associated protocol as described here must be an integral part of the manuscript available to the reviewer(s) and, upon publication, to the readers.
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Nicolaou, K.C.; Snyder, S.A. Chasing molecules that were never there: Misassigned natural products and the role of chemical synthesis in modern structure elucidation. Angew. Chem. Int. Ed. 2005, 44, 1012–1044. [Google Scholar] [CrossRef]
- McAlpine, J.B.; Chen, S.N.; Kutateladze, A.; MacMillan, J.B.; Appendino, G.; Barison, A.; Beniddir, M.A.; Biavatti, M.W.; Bluml, S.; Boufridi, A.; et al. The value of universally available raw NMR data for transparency, reproducibility, and integrity in natural product research. Nat. Prod. Rep. 2019, 36, 35–107. [Google Scholar] [CrossRef] [Green Version]
- Robien, W. A Critical Evaluation of the Quality of Published 13C NMR Data in Natural Product Chemistry. In Progress in the Chemistry of Organic Natural Products; Kinghorn, A.D., Falk, H., Gibbons, S., Kobayashi, J., Eds.; Springer International Publishing: Cham, Switzerland, 2017; Volume 105, pp. 137–215. [Google Scholar] [CrossRef]
- Bremser, W. HOSE—A novel substructure code. Anal. Chim. Acta 1978, 103, 355–365. [Google Scholar] [CrossRef]
- Purtuc, V. Abschätzung von 13C-NMR-Spektren Mittels Neuronaler Netze. Master’s Thesis, University of Vienna, Vienna, Austria, 1997. [Google Scholar]
- Jonas, E.; Kuhn, S. Rapid prediction of NMR spectral properties with quantified uncertainty. J. Cheminform 2019, 11, 50. [Google Scholar] [CrossRef] [Green Version]
- Schütz, V.; Purtuc, V.; Felsinger, S.; Robien, W. CSEARCH-STEREO: A new generation of NMR database system allowing three-dimensional spectrum prediction. Fresenius J. Anal. Chem. 1997, 359, 33–41. [Google Scholar] [CrossRef]
- Technical Note: ACD/Labs NMR Predictors. Advanced Chemistry Development, Inc.: Toronto, ON, Canada, 21 November 2019. Available online: https://www.acdlabs.com/download/technotes/2019/Technical-Note-Drawing-with-NMR-Predictors.pdf (accessed on 5 May 2021).
- Kuhn, S.; Johnson, S.R. Stereo-aware extension of HOSE-codes. ACS Omega 2019, 4, 7323–7329. [Google Scholar] [CrossRef] [PubMed]
- NMR Predictor Software. Available online: https://www.acdlabs.com/products/adh/nmr/nmr_pred/ (accessed on 5 May 2021).
- Satoh, H.; Koshino, H.; Uno, T.; Koichi, S.; Iwata, S.; Nakata, T. Effective consideration of ring structures in CAST/CNMR for highly accurate 13C- NMR chemical shift prediction. Tetrahedron 2005, 61, 7431–7437. [Google Scholar] [CrossRef]
- Kalchhauser, H.; Robien, W. CSEARCH: A computer program for identification of organic compounds and fully automated assignment of carbon-13 nuclear magnetic resonance spectra. J. Chem. Inform. Comput. Sci. 1985, 25, 103–108. [Google Scholar] [CrossRef]
- KnowItAll NMR Spectral Database Collection. Available online: https://sciencesolutions.wiley.com/solutions/technique/nmr/knowitall-nmr-collection/ (accessed on 5 May 2021).
- NMRPredict Overview. Available online: http://www.modgraph.co.uk/product_nmr.htm (accessed on 5 May 2021).
- Steinbeck, C.; Krause, S.; Kuhn, S. NMRShiftDB—Constructing a free chemical information system with open-source components. J. Chem. Inf. Comput. Sci. 2003, 43, 1733–1739. [Google Scholar] [CrossRef] [Green Version]
- SpectraBaseTM. Available online: https://spectrabase.com/ (accessed on 5 May 2021).
- IR, UV/VIS and NMR Spectra Predictions. Available online: http://insideinformatics.cambridgesoft.com/videosanddemos/143/ir-uvvis-and-nmr-spectra-predictions (accessed on 5 May 2021).
- Accurate Prediction of 1H and 13C NMR Spectra from a Chemical Structure. Available online: https://mestrelab.com/software/mnova/nmr-predict/ (accessed on 5 May 2021).
- NMR Software. Available online: https://www.bruker.com/products/mr/nmr/software.html (accessed on 5 May 2021).
- Robien, W. Do high-quality 13C-NMR spectral data really come from journals with high impact factors? TrAC Trends Anal. Chem. 2009, 28, 914–922. [Google Scholar] [CrossRef]
- Pupier, M.; Nuzillard, J.M.; Wist, J.; Schlörer, N.E.; Kuhn, S.; Erdelyi, M.; Steinbeck, C.; Williams, A.J.; Butts, C.; Claridge, T.D.W.; et al. NMReDATA, a standard to report the NMR-assignment and parameters of organic compounds. Magn. Reson. Chem. 2018, 56, 703–715. [Google Scholar] [CrossRef] [Green Version]
- NMReDATA Initiative. Available online: http://nmredata.org/ (accessed on 5 May 2021).
- Urbas, A.; Schönberger, T.; Corbett, C.; Lippa, K.; Rudolphi, F.; Robien, W. NPS-Data hub: A web-based community driven analytical data repository for new psychoactive substances. Forensic. Chem. 2018, 9, 76–81. [Google Scholar] [CrossRef]
- Schlörer, N.; Kuhn, S. Moleküle und Strukturbeweise teilen: NMR-Spektroskopie-Datenbank für organische Moleküle. Chem. Unserer. Zeit. 2020, 54, 6–13. [Google Scholar] [CrossRef]
- Burns, D.C.; Reynolds, W.F. Minimizing the risk of deducing wrong natural product structures from NMR data. Magn. Reson. Chem. 2021, 59, 500–533. [Google Scholar] [CrossRef]
- CSEARCH Robot Referee. Available online: https://c13nmr.at/molecules/robot.php (accessed on 5 May 2021).
- Robien, W. Computer-assisted peer reviewing of spectral data: The CSEARCH-protocol. Mh. Chem. 2019, 150, 927–932. [Google Scholar] [CrossRef] [Green Version]
- Haider, N.; Robien, W. Automatisierte Qualitätskontrolle von 13C-NMR-Daten. Nachr. Chem. 2016, 64, 196–198. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef] [Green Version]
- Index of /Pubchem/Compound/CURRENT-Full/SDF. Available online: https://ftp.ncbi.nih.gov/pubchem/Compound/CURRENT-Full/SDF/ (accessed on 5 May 2021).
- ChemSpider: Search and Share Chemistry. Available online: https://www.chemspider.com (accessed on 5 May 2021).
- eMolecules: Empowering Drug Discovery. Available online: https://www.emolecules.com (accessed on 5 May 2021).
- Huang, Y.-H.; Zeng, W.-M.; Li, G.-Y.; Liu, G.-Q.; Zhao, D.-D.; Wang, J.; Zhang, Y.-L. Characterization of a New Sesquiterpene and Antifungal Activities of Chemical Constituents from Dryopteris fragrans (L.) Schott. Molecules 2014, 19, 507–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baba, K.; Kawanishi, H.; Taniguchi, M.; Kozawa, M. Chromones from Cnidium monnieri. Phytochemistry 1992, 31, 1367–1370. [Google Scholar] [CrossRef]
- Chazin, E.D.L.; Sanches, P.D.S.; Lindgren, E.B.; Vellasco Júnior, W.T.; Pinto, L.C.; Burbano, R.M.R.; Yoneda, J.D.; Leal, K.Z.; Gomes, C.R.B.; Wardell, J.L.; et al. Synthesis and Biological Evaluation of Novel 6-Hydroxy-benzo[d][1,3]oxathiol-2-one Schiff Bases as Potential Anticancer Agents. Molecules 2015, 20, 1968–1983. [Google Scholar] [CrossRef] [Green Version]
- Turan, N.; Körkoca, H.; Adigüzel, R.; Çolak, N.; Buldurun, K. Synthesis, Structural Characterization and Biological Activity of Novel Cyclohexane-1,3-dione Ligands and Their Metal Complexes. Molecules 2015, 20, 9309–9325. [Google Scholar] [CrossRef] [Green Version]
- Alasmary, F.A.S.; Snelling, A.M.; Zain, M.E.; Alafeefy, A.M.; Awaad, A.S.; Karodia, N. Synthesis and Evaluation of Selected Benzimidazole Derivatives as Potential Antimicrobial Agents. Molecules 2015, 20, 15206–15223. [Google Scholar] [CrossRef] [PubMed]
- Nagappan, T.; Segaran, T.C.; Wahid, M.E.A.; Ramasamy, P.; Vairappan, C.S. Efficacy of Carbazole Alkaloids, Essential Oil and Extract of Murraya koenigii in Enhancing Subcutaneous Wound Healing in Rats. Molecules 2012, 17, 14449–14463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estork, D.M.; Gusmão, D.F.; Paciencia, M.L.B.; Díaz, I.E.C.; Varella, A.D.; Younes, R.N.; Reis, L.F.L.; Montero, E.F.S.; Bernardi, M.M.; Suffredini, I.B. First Chemical Evaluation and Toxicity of Casinga-cheirosa to Balb-c Male Mice. Molecules 2014, 19, 3973–3987. [Google Scholar] [CrossRef] [PubMed]
- González-Olvera, R.; Espinoza-Vázquez, A.; Negrón-Silva, G.E.; Palomar-Pardavé, M.E.; Romero-Romo, M.A.; Santillan, R. Multicomponent Click Synthesis of New 1,2,3-Triazole Derivatives of Pyrimidine Nucleobases: Promising Acidic Corrosion Inhibitors for Steel. Molecules 2013, 18, 15064–15079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; He, X.-M.; Zhao, M.-M.; Li, L.; Li, C.-B.; Dong, Y. Antioxidant and Nitrite-Scavenging Capacities of Phenolic Compounds from Sugarcane (Saccharum officinarum L.) Tops. Molecules 2014, 19, 13147–13160. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Martínez, F.J.; Razo-Hernández, R.S.; Peraza-Campos, A.L.; Villanueva-García, M.; Sumaya-Martínez, M.T.; Cano, D.J.; Gómez-Sandoval, Z. Synthesis and in Vitro Antioxidant Activity Evaluation of 3-Carboxycoumarin Derivatives and QSAR Study of Their DPPH• Radical Scavenging Activity. Molecules 2012, 17, 14882–14898. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, B.; Jiang, Y.; Liu, Z.; Liu, Y.; Wang, X.; Kuang, H. Studies on Cytotoxic Activity against HepG-2 Cells of Naphthoquinones from Green Walnut Husks of Juglans mandshurica Maxim. Molecules 2015, 20, 15572–15588. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.F.; Yang, K.; Zhang, H.M.; Cao, J.; Fang, R.; Liu, Z.L.; Du, S.S.; Wang, Y.Y.; Deng, Z.W.; Zhou, L. Components and Insecticidal Activity against the Maize Weevils of Zanthoxylum schinifolium Fruits and Leaves. Molecules 2011, 16, 3077–3088. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Mei, W.; Gong, M.; Zuo, W.; Bai, H.; Dai, H. Antibacterial Activity of the Flavonoids from Dalbergia odorifera on Ralstonia solanacearum. Molecules 2011, 16, 9775–9782. [Google Scholar] [CrossRef] [Green Version]
- Han, L.; Ji, L.; Boakye-Yiadom, M.; Li, W.; Song, X.; Gao, X. Preparative Isolation and Purification of Four Compounds from Cistanches deserticola Y.C. Ma by High-Speed Counter-Current Chromatography. Molecules 2012, 17, 8276–8284. [Google Scholar] [CrossRef] [PubMed]
- Kwiecień, A.; Ciunik, Z. Stable Hemiaminals: 2-Aminopyrimidine Derivatives. Molecules 2015, 20, 14365–14376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.Q.; Liu, Q.Z.; Liu, Z.L.; Du, S.S.; Deng, Z.W. Chemical Composition and Nematicidal Activity of Essential Oil of Agastache rugosa against Meloidogyne incognita. Molecules 2013, 18, 4170–4180. [Google Scholar] [CrossRef]
- Popoola, O.K.; Marnewick, J.L.; Rautenbach, F.; Iwuoha, E.I.; Hussein, A.A. Acylphloroglucinol Derivatives from the South African Helichrysum niveum and Their Biological Activities. Molecules 2015, 20, 17309–17324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filho, H.D.S.R.; Pacheco, L.C.; Andrade, E.D.S.; Correa, M.J.C.; Araújo, R.N.M.; Guilhon, G.M.S.P.; Silva, J.K.R.d.; Santos, L.S. Xanthones from the Roots of Moutabea guianensis Aubl. Molecules 2015, 20, 127–134. [Google Scholar] [CrossRef] [Green Version]
- Petroski, R.J.; Vermillion, K.; Cossé, A.A. Two-Carbon Homologation of Aldehydes and Ketones to α,β-Unsaturated Aldehydes. Molecules 2011, 16, 5062–5078. [Google Scholar] [CrossRef]
- Meng, L.-Z.; Huang, W.-H.; Wang, C.-Z.; Yuan, C.-S.; Li, S.-P. Anticancer Activities of Polyynes from the Root Bark of Oplopanax horridus and Their Acetylated Derivatives. Molecules 2014, 19, 6142–6162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marti, G.; Eparvier, V.; Litaudon, M.; Grellier, P.; Guéritte, F. A New Xanthone from the Bark Extract of Rheedia acuminata and Antiplasmodial Activity of Its Major Compounds. Molecules 2010, 15, 7106–7114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Syahri, J.; Yuanita, E.; Nurohmah, B.A.; Wathon, M.H.; Syafri, R.; Armunanto, R.; Purwono, B. Xanthone as Antimalarial: QSAR Analysis, Synthesis, Molecular Docking and In-vitro Antimalarial Evaluation. Orient J. Chem. 2017, 33, 29–40. [Google Scholar] [CrossRef] [Green Version]
- Andriani, Y.; Tengku-Muhammad, T.S.; Mohamad, H.; Saidin, J.; Syamsumir, D.F.; Chew, G.-S.; Abdul Wahid, M.E. Phaleria macrocarpa Boerl. (Thymelaeaceae) Leaves Increase SR-BI Expression and Reduce Cholesterol Levels in Rats Fed a High Cholesterol Diet. Molecules 2015, 20, 4410–4429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Cui, J.; Jia, L.; Gan, C.; Song, H.; Zeng, C.; Zhou, A. Synthesis and Evaluation of Some 17-Acetamidoandrostane and N,N-Dimethyl-7-deoxycholic Amide Derivatives as Cytotoxic Agents: Structure/Activity Studies. Molecules 2013, 18, 7436–7447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holland, H.L.; Lakshmaiah, G.; Ruddock, P.L. Microbial hydroxylation of acetylaminosteroids. Steroids 1998, 63, 484–495. [Google Scholar] [CrossRef]
- Dieltiens, N.; Claeys, D.D.; Zhdankin, V.V.; Nemykin, V.N.; Allaert, B.; Verpoort, F.; Stevens, C.V. The Pyroglutamate Hydantoin Rearrangement. Eur. J. Org. Chem. 2018, 3372. [Google Scholar] [CrossRef] [Green Version]
- Krivdin, L.B. Computational protocols for calculating 13C NMR chemical shifts. Prog. Nucl. Magn. Reson. Spectrosc. 2019, 112–113, 103–156. [Google Scholar] [CrossRef] [PubMed]
- Semenov, V.A.; Krivdin, L.B. Computational 1H and 13C NMR of strychnobaillonine: On the way to larger molecules calculated at lower computational costs. Magn. Reson. Chem. 2021, 59, 108–116. [Google Scholar] [CrossRef]







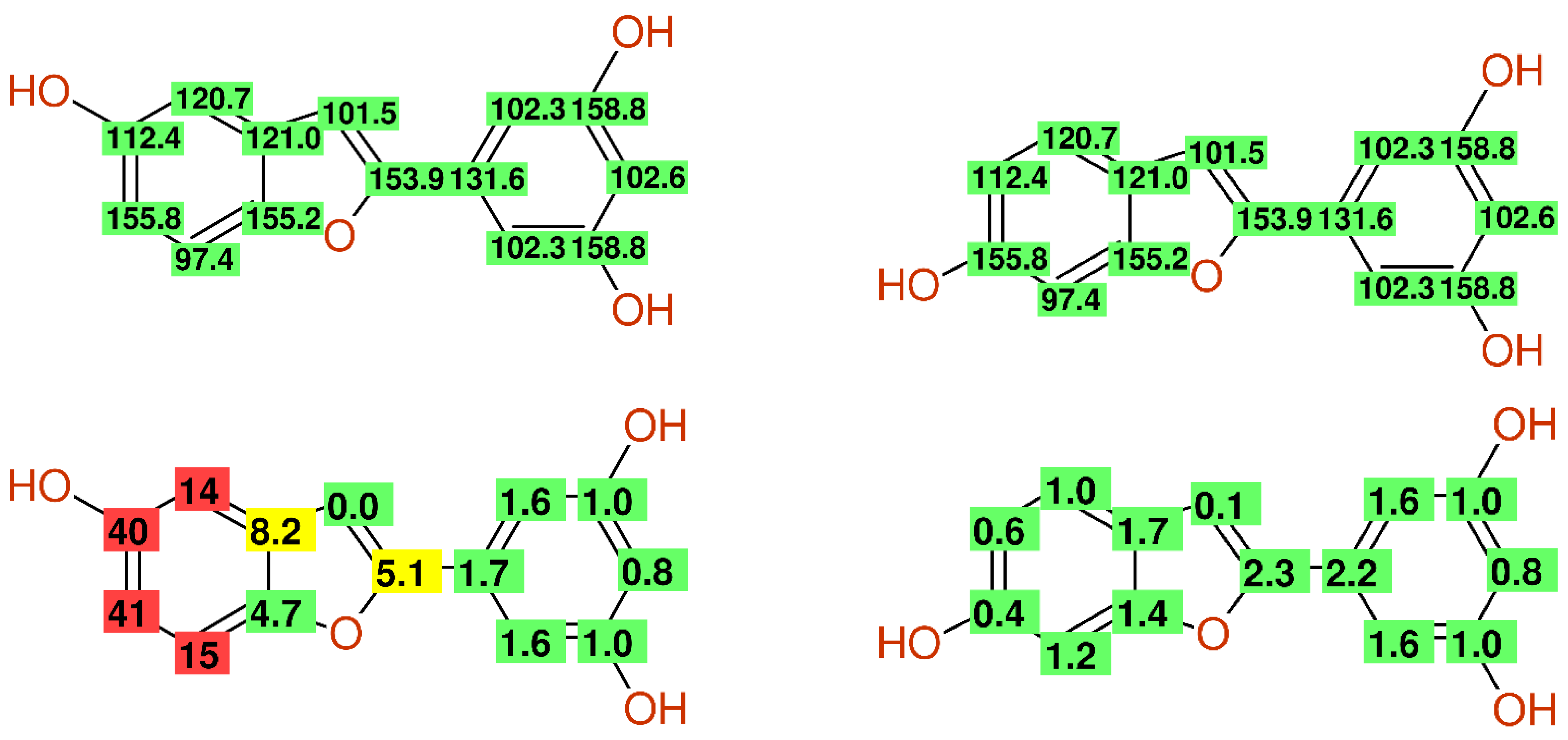
| Journal | MOL | CPB | FT | JNP | PC | PM |
|---|---|---|---|---|---|---|
| Entries | 10,039 | 17,863 | 1568 | 34,933 | 38,379 | 3621 |
| Period | 1996–2015 | 1977–2016 | 1998–2012 | 1979–2013 | 1976–2015 | 1977–2006 |
| Average MWT (amu) | 431 | 520 | 497 | 481 | 483 | 479 |
| ΔδC (ppm) | 2.07 | 1.61 | 1.85 | 1.79 | 1.66 | 1.72 |
| ΔδC > 20 ppm | 341/3.40% | 239/1.34% | 42/2.68% | 542/1.55% | 533/1.39% | 64/1.77% |
| ΔδC (ppm) Chromones | 1.59 | 1.40 | 1.74 | 1.68 | 1.45 | 1.57 |
| ΔδC (ppm) Steroids | 1.60 | 1.27 | 1.54 | 1.47 | 1.37 | 1.36 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robien, W. The Advantage of Automatic Peer-Reviewing of 13C-NMR Reference Data Using the CSEARCH-Protocol. Molecules 2021, 26, 3413. https://doi.org/10.3390/molecules26113413
Robien W. The Advantage of Automatic Peer-Reviewing of 13C-NMR Reference Data Using the CSEARCH-Protocol. Molecules. 2021; 26(11):3413. https://doi.org/10.3390/molecules26113413
Chicago/Turabian StyleRobien, Wolfgang. 2021. "The Advantage of Automatic Peer-Reviewing of 13C-NMR Reference Data Using the CSEARCH-Protocol" Molecules 26, no. 11: 3413. https://doi.org/10.3390/molecules26113413






