Electrochemical Resistive DNA Biosensor for the Detection of HPV Type 16
Abstract
:1. Introduction
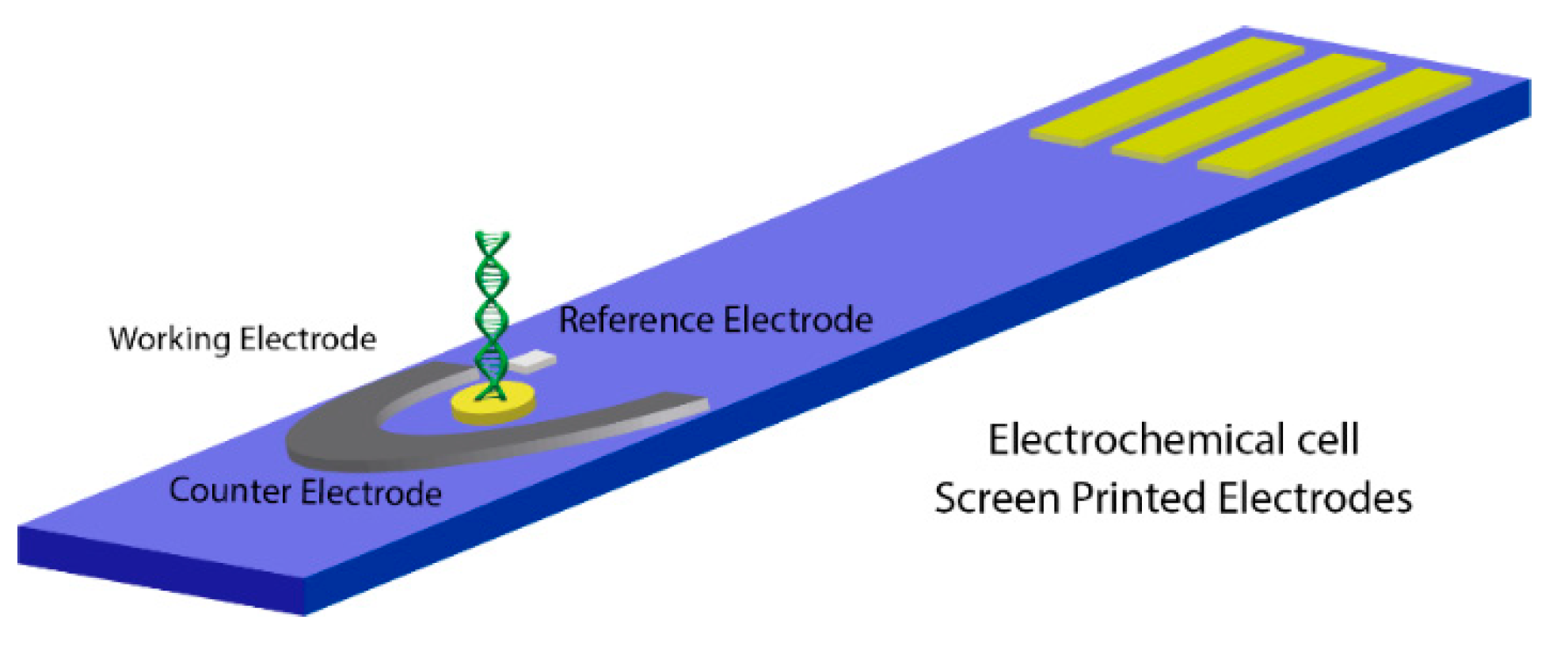
2. Materials and Methods
2.1. Chemicals and Reagents
- DNA probe = (5′-HS(CH2)6GTCATTATGTGCTGCCATATCTACTT-CAGA-3′);
- DNA complementary target = (5′-TCTGAAGTAGATATGGCAGCACATAATGAC-3′);
- DNA single-based mismatch target = (5′-TCTGAAATAGATATGGCAGCACATAATGAC-3′).
2.2. DNA Probe Immobilization and Hybridization with DNA Target
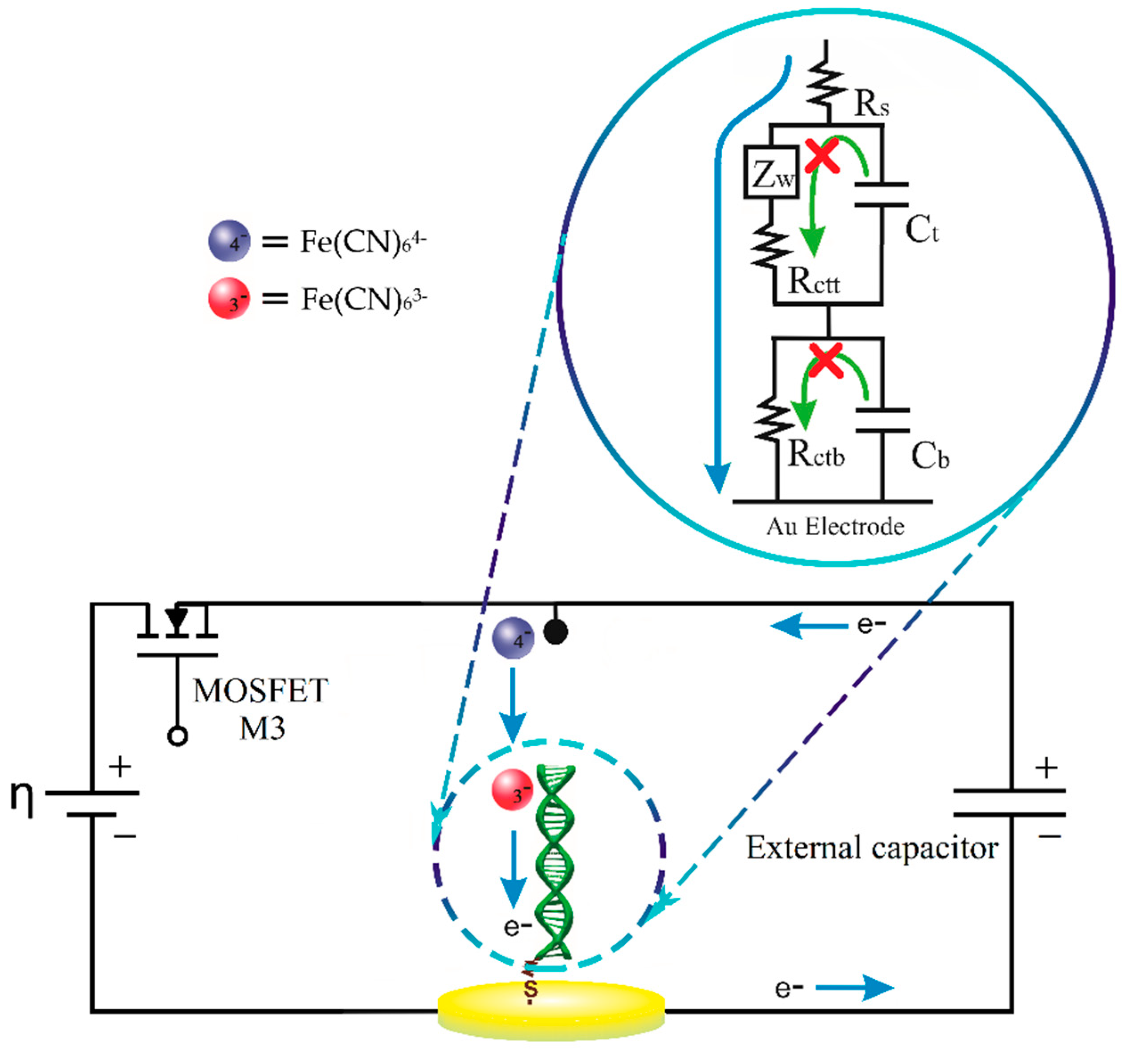
2.3. Electrochemical Measurements
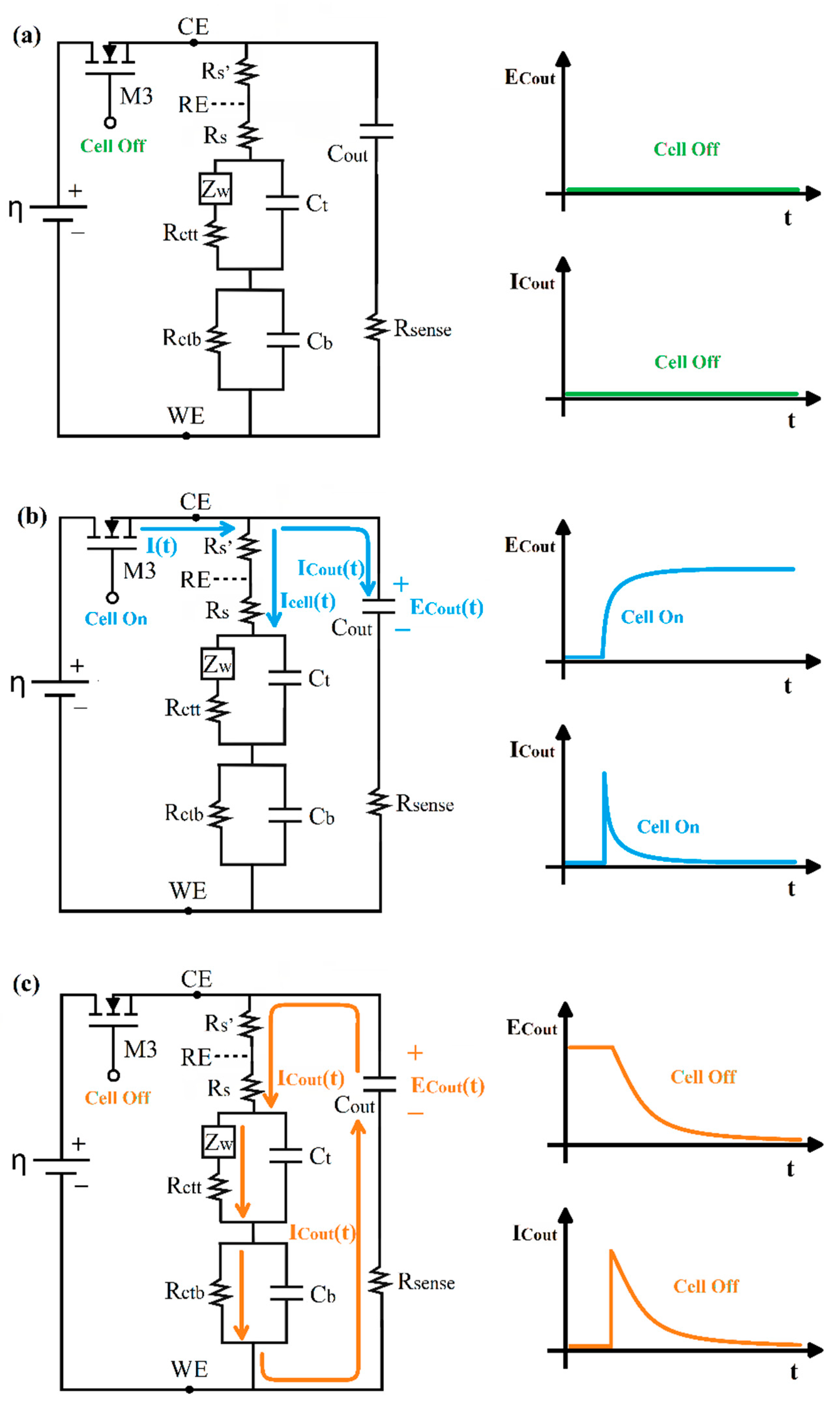
2.4. The Current Relaxation Method
2.5. Sensing Circuit
3. Results and Discussion
3.1. Step Potential
3.2. Analytical Performance
3.3. Specificity of the DNA Biosensor
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Valencia, D.; Dantas, L.; Lara, A.; García, J.; Rivera, Z.; Rosas, J.; Bertotti, M. Development of a bio-electrochemical immunosensor based on the immobilization of SPINNTKPHEAR peptide derived from HPV-L1 protein on a gold electrode surface. J. Electroanal. Chem. 2016, 770, 50–55. [Google Scholar] [CrossRef]
- He, Y.; Cheng, L.; Yang, Y.; Chen, P.; Qiu, B.; Guo, L.; Wang, Y.; Lin, Z.; Hong, G. Label-free homogeneous electrochemical biosensor for HPV DNA based on entropy-driven target recycling and hyperbranched rolling circle amplification. Sens. Actuators B Chem. 2020, 320, 128407. [Google Scholar] [CrossRef]
- De Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef] [Green Version]
- Franceschi, S.; Herrero, R.; Clifford, G.M.; Snijders, P.J.; Arslan, A.; Anh, P.T.H.; Bosch, F.X.; Ferreccio, C.; Hieu, N.T.; Lazcano-Ponce, E.; et al. Variations in the age-specific curves of human papillomavirus prevalence in women worldwide. Int. J. Cancer 2006, 119, 2677–2684. [Google Scholar] [CrossRef]
- Dondog, B.; Clifford, G.M.; Vaccarella, S.; Waterboer, T.; Unurjargal, D.; Avirmed, D.; Enkhtuya, S.; Kommoss, F.; Wentzensen, N.; Snijders, P.J.; et al. Human Papillomavirus Infection in Ulaanbaatar, Mongolia: A Population-Based Study. Cancer Epidemiol. Biomark. Prev. 2008, 17, 1731–1738. [Google Scholar] [CrossRef] [Green Version]
- Zhao, F.-H.; Lewkowitz, A.K.; Hu, S.-Y.; Chen, F.; Li, L.-Y.; Zhang, Q.-M.; Wu, R.-F.; Li, C.-Q.; Wei, L.-H.; Xu, A.-D.; et al. Prevalence of human papillomavirus and cervical intraepithelial neoplasia in China: A pooled analysis of 17 population-based studies. Int. J. Cancer 2012, 131, 2929–2938. [Google Scholar] [CrossRef] [Green Version]
- Ferlay, J.; Shin, H.-R.; Bray, F.; Forman, D.; Mathers, C.; Parkin, D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 2010, 127, 2893–2917. [Google Scholar] [CrossRef]
- Hammouda, D.; Clifford, G.M.; Pallardy, S.; Ayyach, G.; Chékiri, A.; Boudrich, A.; Snijders, P.J.; Van Kemenade, F.J.; Meijer, C.J.; Bouhadef, A.; et al. Human papillomavirus infection in a population-based sample of women in Algiers, Algeria. Int. J. Cancer 2010, 128, 2224–2229. [Google Scholar] [CrossRef] [PubMed]
- Human Papillomavirus and Related Diseases in the World. Available online: http://hpvcentre.net (accessed on 25 May 2021).
- Ozbun, M.A.; Bondu, V.; Patterson, N.A.; Sterk, R.T.; Waxman, A.G.; Bennett, E.C.; McKee, R.; Sharma, A.; Yarwood, J.; Rogers, M.; et al. Infectious titres of human papillomaviruses (HPVs) in patient lesions, methodological considerations in evaluating HPV infectivity and implications for the efficacy of high-level disinfectants. EBioMedicine 2021, 63, 103165. [Google Scholar] [CrossRef]
- Jampasa, S.; Siangproh, W.; Laocharoensuk, R.; Yanatatsaneejit, P.; Vilaivan, T.; Chailapakul, O. A new DNA sensor design for the simultaneous detection of HPV type 16 and 18 DNA. Sens. Actuators B Chem. 2018, 265, 514–521. [Google Scholar] [CrossRef]
- Gopinath, P.; Anitha, V.; Mastani, S.A. Design of biosensor array with current boost and signal conditioning circuits for HPV detection. Alex. Eng. J. 2018, 57, 671–681. [Google Scholar] [CrossRef]
- Hai, X.; Li, Y.; Zhu, C.; Song, W.; Cao, J.; Bi, S. DNA-based label-free electrochemical biosensors: From principles to applications. TrAC Trends Anal. Chem. 2020, 133, 116098. [Google Scholar] [CrossRef]
- Paleček, E.; Dorčák, V. Label-free electrochemical analysis of biomacromolecules. Appl. Mater. Today 2017, 9, 434–450. [Google Scholar] [CrossRef]
- Phan, A.D.; Viet, N.A. A new type of optical biosensor from DNA wrapped semiconductor graphene ribbons. J. Appl. Phys. 2012, 111, 114703. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Zhao, W.; Su, B.; Wu, J. A label-free optical sensor based on nanoporous gold arrays for the detection of oligodeoxynucleotides. Biosens. Bioelectron. 2011, 30, 21–27. [Google Scholar] [CrossRef]
- Singh, R.R.; Ho, D.; Nilchi, A.; Gulak, G.; Yau, P.; Genov, R. A CMOS/Thin-Film Fluorescence Contact Imaging Microsystem for DNA Analysis. IEEE Trans. Circuits Syst. I Regul. Pap. 2010, 57, 1029–1038. [Google Scholar] [CrossRef]
- Dolatabadi, J.E.N.; Mashinchian, O.; Ayoubi, B.; Jamali, A.A.; Mobed, A.; Losic, D.; Omidi, Y.; de la Guardia, M. Optical and electrochemical DNA nanobiosensors. TrAC Trends Anal. Chem. 2011, 30, 459–472. [Google Scholar] [CrossRef]
- Ianeselli, L.; Grenci, G.; Callegari, C.; Tormen, M.; Casalis, L. Development of stable and reproducible biosensors based on electrochemical impedance spectroscopy: Three-electrode versus two-electrode setup. Biosens. Bioelectron. 2014, 55, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Cesewski, E.; Johnson, B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020, 159, 112214. [Google Scholar] [CrossRef]
- Sankoh, S.; Samanman, S.; Thipmanee, O.; Numnuam, A.; Limbut, W.; Kanatharana, P.; Vilaivan, T.; Thavarungkul, P. A comparative study of a label-free DNA capacitive sensor using a pyrrolidinyl peptide nucleic acid probe immobilized through polyphenylenediamine and polytyramine non-conducting polymers. Sens. Actuators B Chem. 2013, 177, 543–554. [Google Scholar] [CrossRef]
- Thipmanee, O.; Samanman, S.; Sankoh, S.; Numnuam, A.; Limbut, W.; Kanatharana, P.; Vilaivan, T.; Thavarungkul, P. Label-free capacitive DNA sensor using immobilized pyrrolidinyl PNA probe: Effect of the length and terminating head group of the blocking thiols. Biosens. Bioelectron. 2012, 38, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Rasouli, E.; Shahnavaz, Z.; Basirun, W.J.; Rezayi, M.; Avan, A.; Ghayour-Mobarhan, M.; Khandanlou, R.; Johan, M.R. Advancements in electrochemical DNA sensor for detection of human papilloma virus—A review. Anal. Biochem. 2018, 556, 136–144. [Google Scholar] [CrossRef]
- Khan, M.; Hasan, M.; Hossain, S.; Ahommed, M.; Daizy, M. Ultrasensitive detection of pathogenic viruses with electrochemical biosensor: State of the art. Biosens. Bioelectron. 2020, 166, 112431. [Google Scholar] [CrossRef] [PubMed]
- Rashid, J.I.A.; Yusof, N.A. The strategies of DNA immobilization and hybridization detection mechanism in the construction of electrochemical DNA sensor: A review. Sens. Bio-Sens. Res. 2017, 16, 19–31. [Google Scholar] [CrossRef]
- Chang, B.-Y.; Park, S.-M. Electrochemical impedance spectroscopy. Annu. Rev. Anal. Chem. 2010, 3, 207–229. [Google Scholar] [CrossRef]
- Uygun, Z.O.; Uygun, H.D.E. A short footnote: Circuit design for faradaic impedimetric sensors and biosensors. Sens. Actuators B Chem. 2014, 202, 448–453. [Google Scholar] [CrossRef]
- Mahmoud, A.M.; Tang, T.; Harrison, D.J.; Lee, W.E.; Jemere, A.B. A regenerating self-assembled gold nanoparticle-containing electrochemical impedance sensor. Biosens. Bioelectron. 2014, 56, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Kong, R.-M.; Song, Z.-L.; Meng, H.-M.; Zhang, X.-B.; Shen, G.-L.; Yu, R.-Q. A label-free electrochemical biosensor for highly sensitive and selective detection of DNA via a dual-amplified strategy. Biosens. Bioelectron. 2014, 54, 442–447. [Google Scholar] [CrossRef]
- Galán, T.; Prieto-Simón, B.; Alvira, M.; Eritja, R.; Götz, G.; Bäuerle, P.; Samitier, J. Label-free electrochemical DNA sensor using “click”-functionalized PEDOT electrodes. Biosens. Bioelectron. 2015, 74, 751–756. [Google Scholar] [CrossRef] [Green Version]
- Shi, A.; Wang, J.; Han, X.; Fang, X.; Zhang, Y. A sensitive electrochemical DNA biosensor based on gold nanomaterial and graphene amplified signal. Sens. Actuators B Chem. 2014, 200, 206–212. [Google Scholar] [CrossRef]
- Zheng, D.; Wang, Q.; Gao, F.; Wang, Q.; Qiu, W.; Gao, F. Development of a novel electrochemical DNA biosensor based on elongated hexagonal-pyramid CdS and poly-isonicotinic acid composite film. Biosens. Bioelectron. 2014, 60, 167–174. [Google Scholar] [CrossRef]
- Miao, X.; Guo, X.; Xiao, Z.; Ling, L. Electrochemical molecular beacon biosensor for sequence-specific recognition of double-stranded DNA. Biosens. Bioelectron. 2014, 59, 54–57. [Google Scholar] [CrossRef]
- Goda, T.; Singi, A.B.; Maeda, Y.; Matsumoto, A.; Torimura, M.; Aoki, H.; Miyahara, Y. Label-Free Potentiometry for Detecting DNA Hybridization Using Peptide Nucleic Acid and DNA Probes. Sensors 2013, 13, 2267–2278. [Google Scholar] [CrossRef]
- Yi, Z.; Wang, H.-B.; Chen, K.; Gao, Q.; Tang, H.; Yu, R.-Q.; Chu, X. A novel electrochemical biosensor for sensitive detection of telomerase activity based on structure-switching DNA. Biosens. Bioelectron. 2014, 53, 310–315. [Google Scholar] [CrossRef]
- Langer, A.; Hampel, P.A.; Kaiser, W.A.; Knezevic, J.; Welte, T.; Villa, V.; Maruyama, M.; Svejda, M.; Jähner, S.; Fischer, F.; et al. Protein analysis by time-resolved measurements with an electro-switchable DNA chip. Nat. Commun. 2013, 4, 2099. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Cui, C.; Higgins, D.A.; Li, J. Fluorescence Quenching Studies of Potential-Dependent DNA Reorientation Dynamics at Glassy Carbon Electrode Surfaces. J. Am. Chem. Soc. 2012, 134, 14467–14475. [Google Scholar] [CrossRef]
- Espinosa, J.R.; Galván, M.; Quiñones, A.S.; Ayala, J.L.; Durón, S.M. DNA Biosensor Based on Double-Layer Discharge for the Detection of HPV Type 16. Sensors 2019, 19, 3956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobs, M.V.; Husman, A.M.D.R.; Brule, A.J.V.D.; Snijders, P.J.; Meijer, C.J.; Walboomers, J.M. Group-specific differentiation between high- and low-risk human papillomavirus genotypes by general primer-mediated PCR and two cocktails of oligonucleotide probes. J. Clin. Microbiol. 1995, 33, 901–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holtan, M.D.; Somasundaram, S.; Khuda, N.; Easley, C.J. Nonfaradaic Current Suppression in DNA-Based Electrochemical Assays with a Differential Potentiostat. Anal. Chem. 2019, 91, 15833–15839. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Currás, N.; Dauphin-Ducharme, P.; Ortega, G.; Ploense, K.L.; Kippin, T.E.; Plaxco, K.W. Subsecond-Resolved Molecular Measurements in the Living Body Using Chronoamperometrically Interrogated Aptamer-Based Sensors. ACS Sens. 2018, 3, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Pellitero, M.A.; Shaver, A.; Arroyo-Currás, N. Critical Review—Approaches for the Electrochemical Interrogation of DNA-Based Sensors: A Critical Review. J. Electrochem. Soc. 2020, 167, 037529. [Google Scholar] [CrossRef]
- Souza, E.; Nascimento, G.; Santana, N.; Campos-Ferreira, D.; Bibiano, J.; Arruda, M.; Bruneska, D.; Lima-Filho, J.L. Electrochemical DNA biosensor for sequences related to the human papillomavirus type 16 using methylene blue. Biosens. J. 2014, 3, 107. [Google Scholar]
- Jampasa, S.; Wonsawat, W.; Rodthongkum, N.; Siangproh, W.; Yanatatsaneejit, P.; Vilaivan, T.; Chailapakul, O. Electrochemical detection of human papillomavirus DNA type 16 using a pyrrolidinyl peptide nucleic acid probe immobilized on screen-printed carbon electrodes. Biosens. Bioelectron. 2014, 54, 428–434. [Google Scholar] [CrossRef]
- Teengam, P.; Siangproh, W.; Tuantranont, A.; Henry, C.S.; Vilaivan, T.; Chailapakul, O. Electrochemical paper-based peptide nucleic acid biosensor for detecting human papillomavirus. Anal. Chim. Acta 2017, 952, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Bartolome, J.P.; Echegoyen, L.; Fragoso, A. Reactive Carbon Nano-Onion Modified Glassy Carbon Surfaces as DNA Sensors for Human Papillomavirus Oncogene Detection with Enhanced Sensitivity. Anal. Chem. 2015, 87, 6744–6751. [Google Scholar] [CrossRef] [PubMed]
- Bartosik, M.; Durikova, H.; Vojtesek, B.; Anton, M.; Jandakova, E.; Hrstka, R. Electrochemical chip-based genomagnetic assay for detection of high-risk human papillomavirus DNA. Biosens. Bioelectron. 2016, 83, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Karimizefreh, A.; Mahyari, F.A.; Vaezjalali, M.; Mohammadpour, R.; Sasanpour, P. Impedimetic biosensor for the DNA of the human papilloma virus based on the use of gold nanosheets. Microchim. Acta 2017, 184, 1729–1737. [Google Scholar] [CrossRef]
- Kowalczyk, A.; Nowicka, A.M. Application of mercury–mediated thymine–base pairs for successful voltammetric detection of HPV 18. Sens. Actuators B Chem. 2016, 237, 810–816. [Google Scholar] [CrossRef]


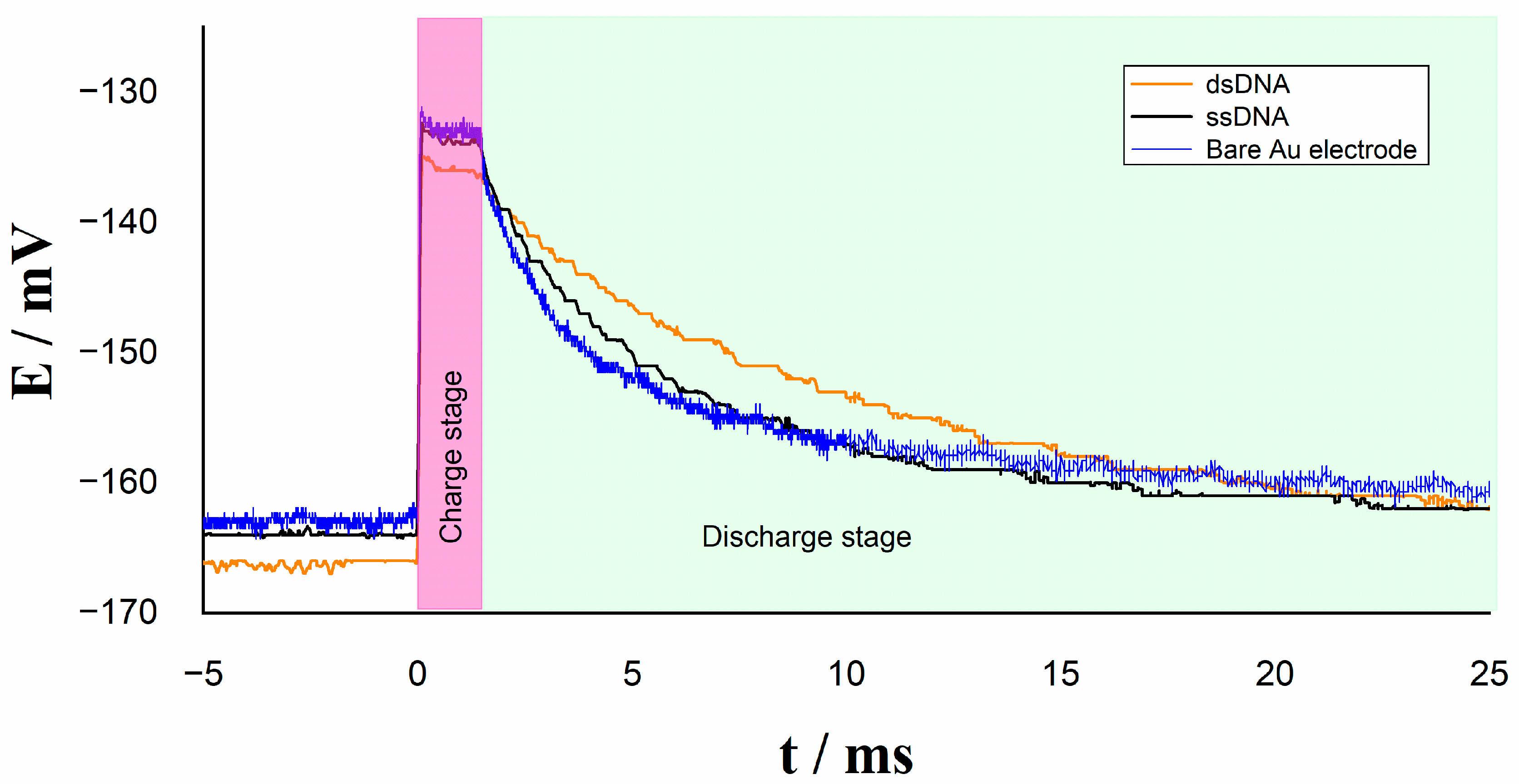
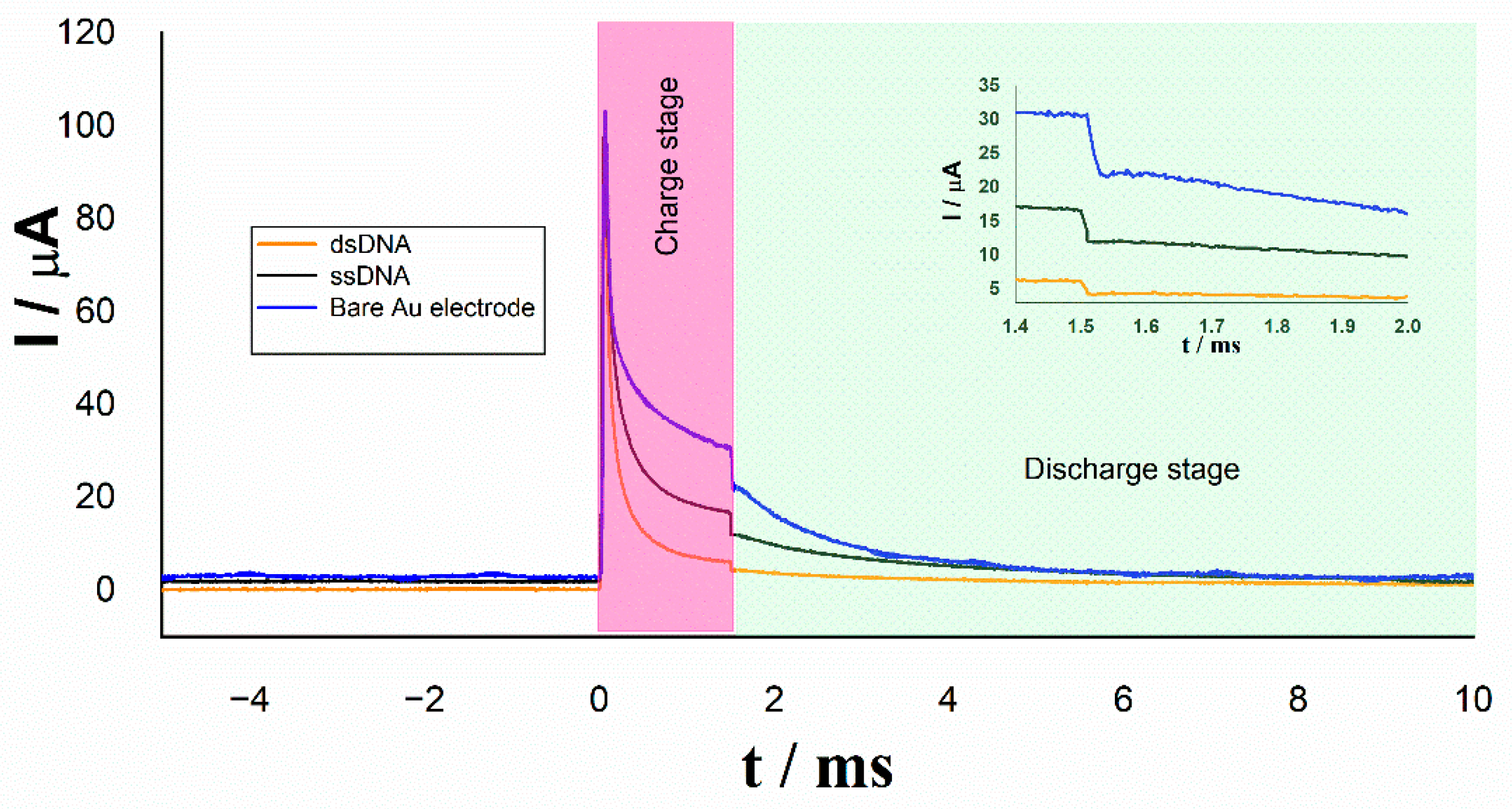
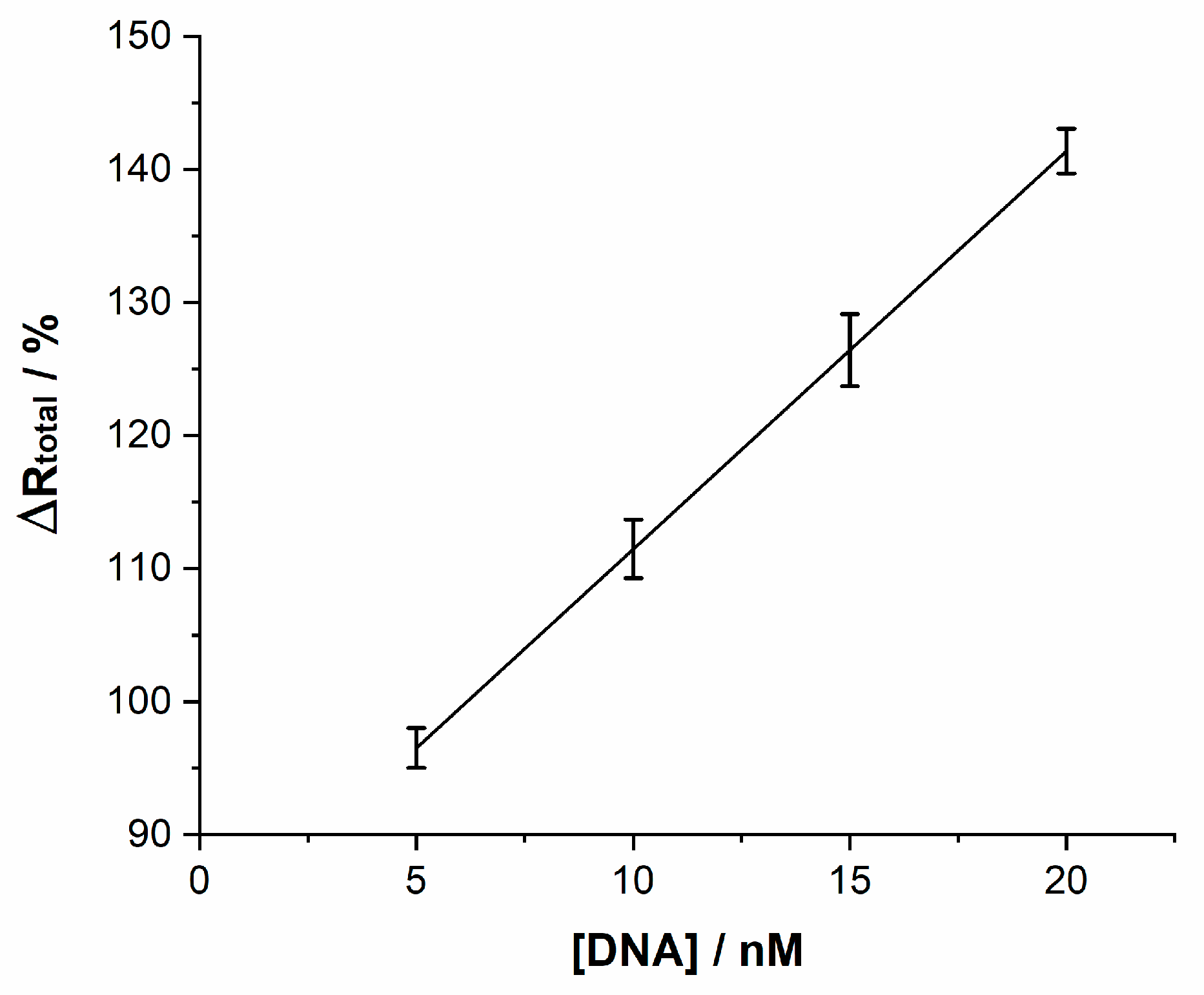
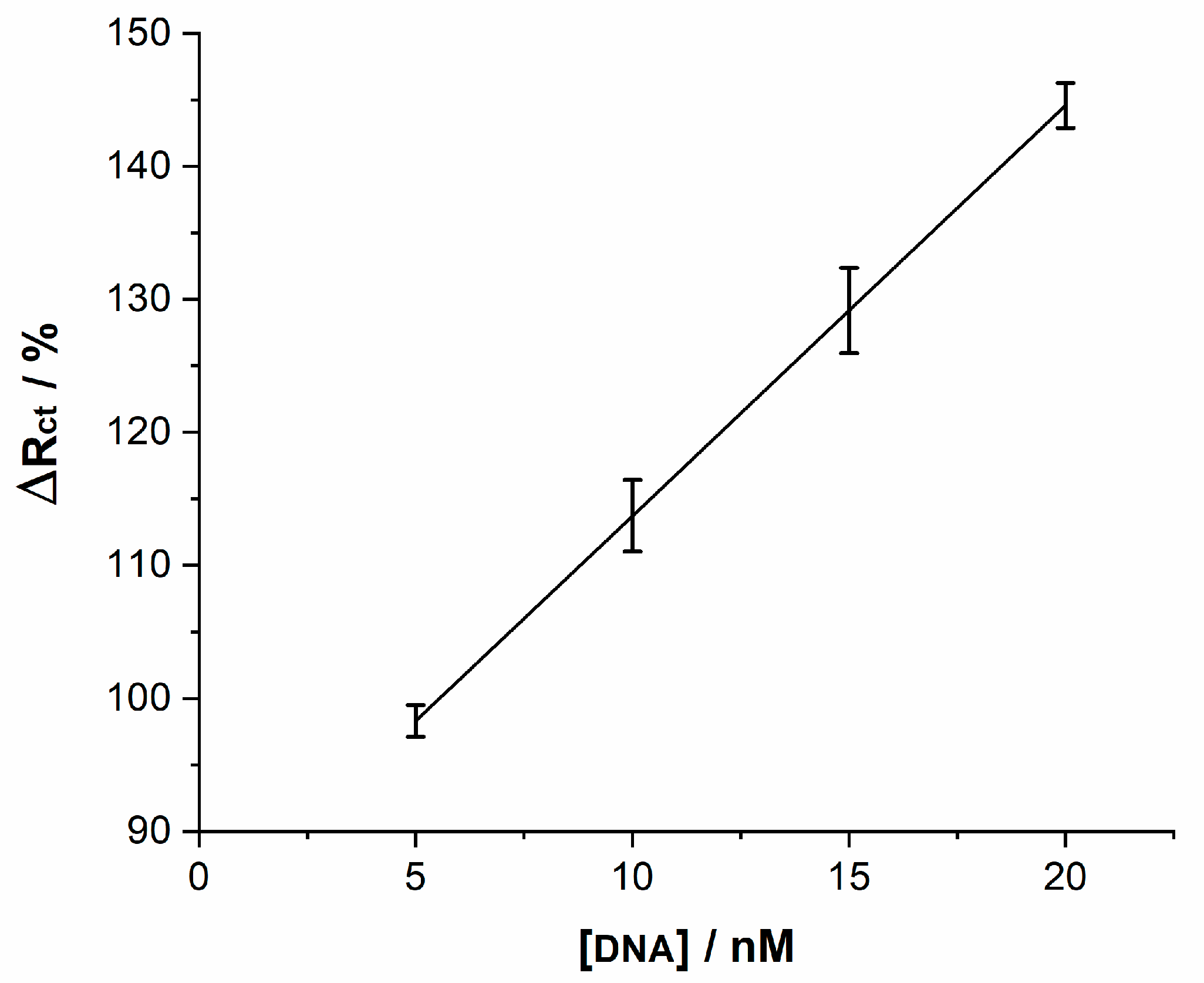
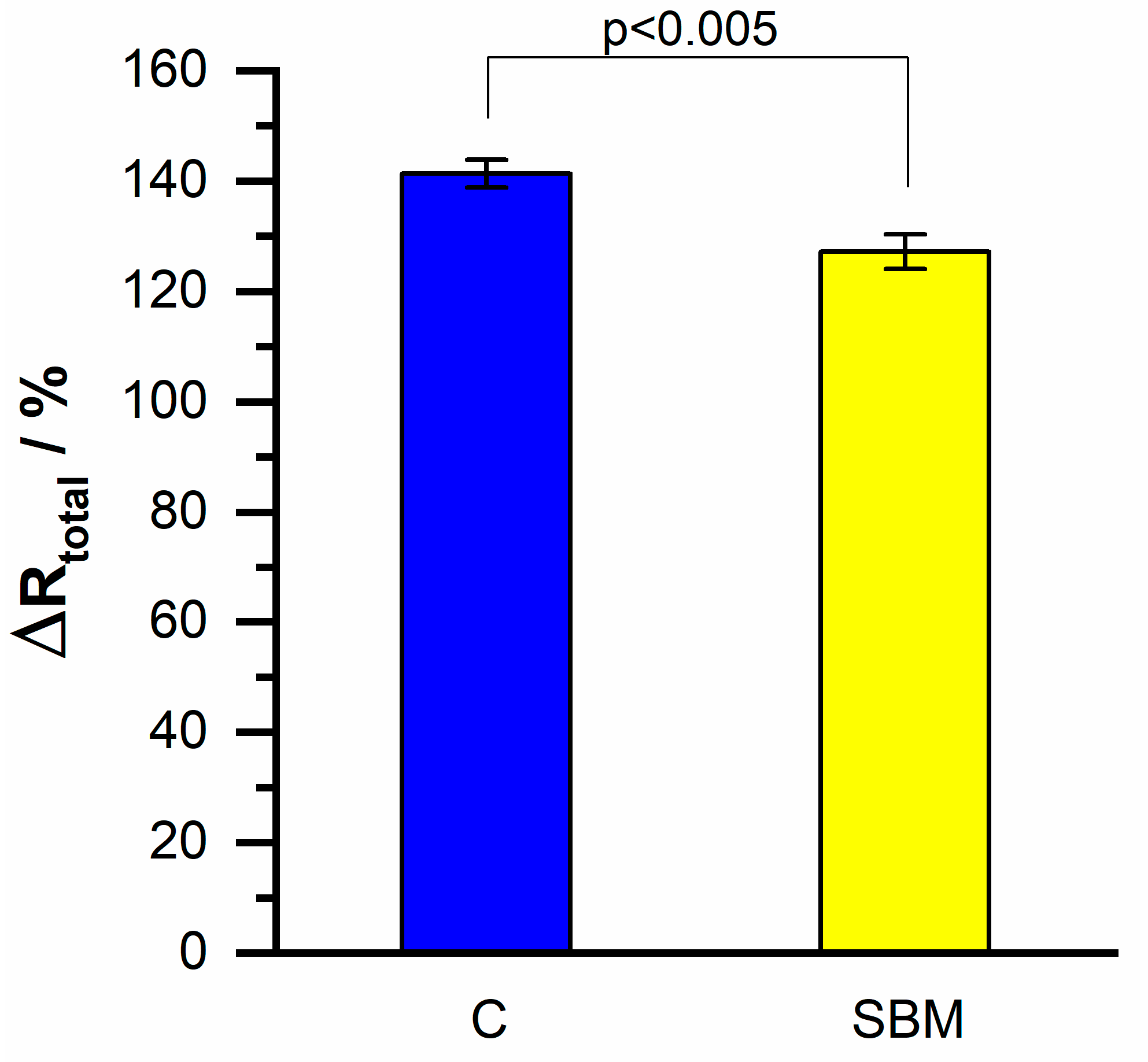
| Irelax/µA | Rtotal/kΩ | |
|---|---|---|
| Bare Au electrode | 22.2 | 1.3 |
| ssDNA | 11.9 | 2.4 |
| dsDNA | 4.3 | 6.6 |
| HPV Type | Technique | Sensor Platform | Detection Limit | Response Time | Ref. |
|---|---|---|---|---|---|
| HPV-16 | DPV | PGE | 1.49 nM | 40 s | [43] |
| HPV-16 | SWV | Carbon surface/chitosan | 4 nM | 10 s | [44] |
| HPV-16 | EIS, SWV | Paper base/G-PANI | 2.3 nM | 17 min, 15 s | [45] |
| HPV-16 | CV | GCE/CNO | 0.50 nM | 7 min | [46] |
| HPV-45 | CA | Gold surface | 110 pM | 60 s | [47] |
| HPV-16 | EIS | GCE/gold nanosheet | 0.15 pM | 17 min | [48] |
| HPV-18 | SWV | GCE/carboxyphenyl layer | 1.2 × 10−5 nM | 10 s | [49] |
| HPV-16 | CA | Gold surface | 2.39 nM | 750 µs | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Espinosa, J.R.; Galván, M.; Quiñones, A.S.; Ayala, J.L.; Ávila, V.; Durón, S.M. Electrochemical Resistive DNA Biosensor for the Detection of HPV Type 16. Molecules 2021, 26, 3436. https://doi.org/10.3390/molecules26113436
Espinosa JR, Galván M, Quiñones AS, Ayala JL, Ávila V, Durón SM. Electrochemical Resistive DNA Biosensor for the Detection of HPV Type 16. Molecules. 2021; 26(11):3436. https://doi.org/10.3390/molecules26113436
Chicago/Turabian StyleEspinosa, José R., Marisol Galván, Arturo S. Quiñones, Jorge L. Ayala, Verónica Ávila, and Sergio M. Durón. 2021. "Electrochemical Resistive DNA Biosensor for the Detection of HPV Type 16" Molecules 26, no. 11: 3436. https://doi.org/10.3390/molecules26113436






