Epigenetic and Transcriptional Control of the Opioid Prodynorphine Gene: In-Depth Analysis in the Human Brain
Abstract
:1. Introduction
2. Prodynorphin Transcripts and Proteins in the Human Brain
3. PDYN Promoter Mapping and Identification of Transcription Factors
4. Genetic Factors Contributing to PDYN Regulation
5. PDYN Epigenetic Mechanisms
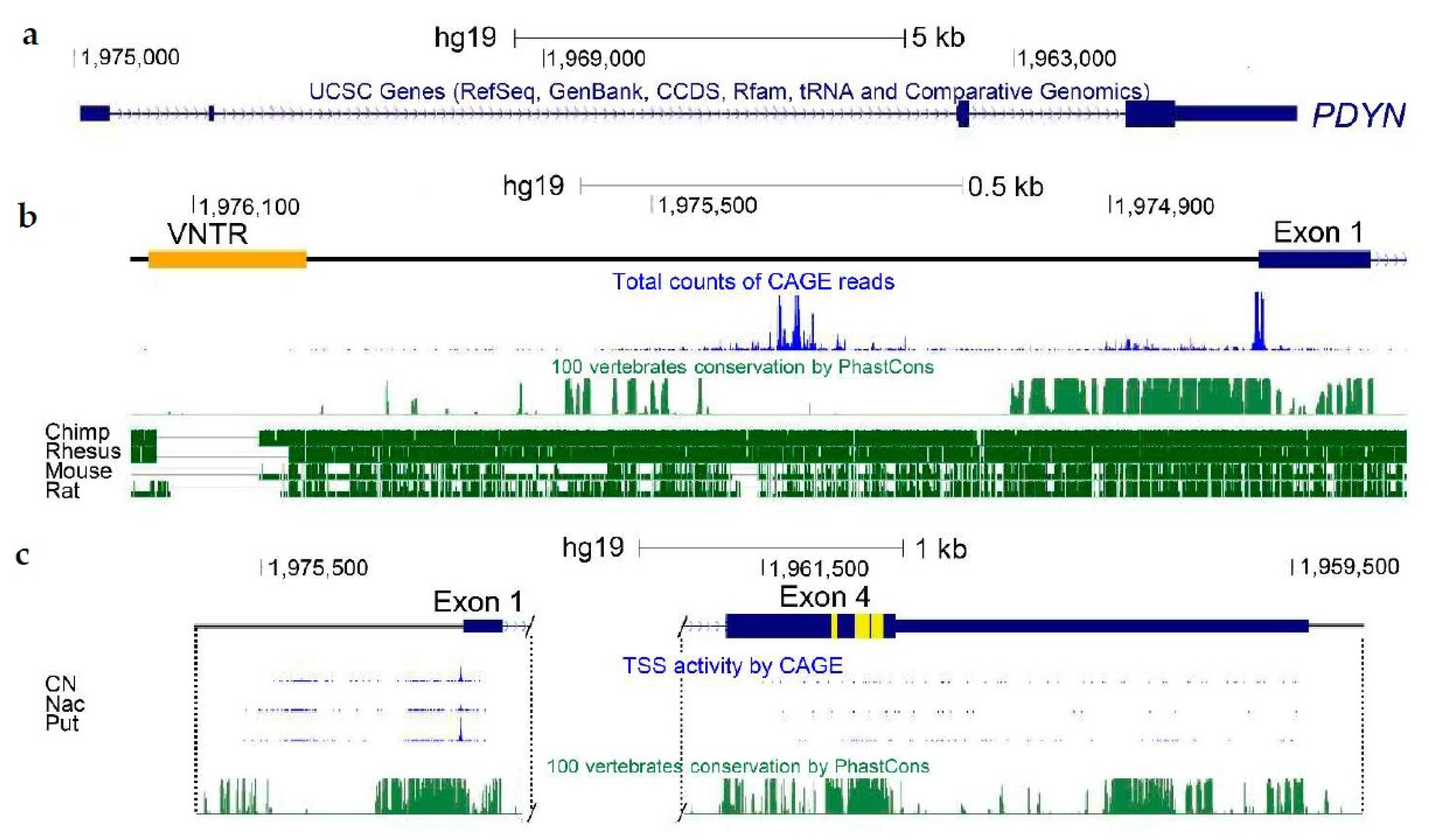
5.1. PDYN Regulation in Chromosomal Context
5.2. DNA Methylation
5.3. The CpG-SNP Hypothesis: Epialleles of PDYN SNPs Associated with Alcoholism
5.4. PDYN Regulation by REST
5.5. Dual Epigenetic and Transcriptional Mechanism Controls Neuronal PDYN Expression
6. PDYN Transcriptional Adaptations Concomitant with Neuronal Decline in Human Alcoholics
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nusbaum, M.P.; Blitz, D.M.; Marder, E. Functional consequences of neuropeptide and small-molecule co-transmission. Nat. Rev. Neurosci. 2017, 18, 389–403. [Google Scholar] [CrossRef] [Green Version]
- Chavkin, C. Dynorphin—Still an extraordinarily potent opioid peptide. Mol. Pharmacol. 2013, 83, 729–736. [Google Scholar] [CrossRef] [Green Version]
- Schwarzer, C. 30 years of dynorphins—New insights on their functions in neuropsychiatric diseases. Pharmacol. Ther. 2009, 123, 353–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruchas, M.R.; Chavkin, C. Kinase cascades and ligand-directed signaling at the kappa opioid receptor. Psychopharmacology 2010, 210, 137–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurd, Y.L. Differential messenger RNA expression of prodynorphin and proenkephalin in the human brain. Neuroscience 1996, 72, 767–783. [Google Scholar] [CrossRef]
- Nikoshkov, A.; Drakenberg, K.; Wang, X.; Horvath, M.C.; Keller, E.; Hurd, Y.L. Opioid neuropeptide genotypes in relation to heroin abuse: Dopamine tone contributes to reversed mesolimbic proenkephalin expression. Proc. Natl. Acad. Sci. USA 2008, 105, 786–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knoll, A.T.; Carlezon, W.A., Jr. Dynorphin, stress, and depression. Brain Res. 2010, 1314, 56–73. [Google Scholar] [CrossRef] [Green Version]
- Negrete, R.; García Gutiérrez, M.S.; Manzanares, J.; Maldonado, R. Involvement of the dynorphin/KOR system on the nociceptive, emotional and cognitive manifestations of joint pain in mice. Neuropharmacology 2017, 116, 315–327. [Google Scholar] [CrossRef] [Green Version]
- Nogueiras, R.; Romero-Picó, A.; Vazquez, M.J.; Novelle, M.G.; López, M.; Diéguez, C. The opioid system and food intake: Homeostatic and hedonic mechanisms. Obes. Facts. 2012, 5, 196–207. [Google Scholar] [CrossRef]
- Przewlocki, R.; Przewlocka, B. Opioids in chronic pain. Eur. J. Pharmacol. 2001, 429, 79–91. [Google Scholar] [CrossRef]
- Zangrandi, L.; Schwarzer, C. The Kappa Opioid Receptor System in Temporal Lobe Epilepsy. Handb. Exp. Pharmacol. 2021. [Google Scholar]
- Shippenberg, T.S.; Zapata, A.; Chefer, V.I. Dynorphin and the pathophysiology of drug addiction. Pharmacol. Ther. 2007, 116, 306–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bazov, I.; Kononenko, O.; Watanabe, H.; Kuntić, V.; Sarkisyan, D.; Taqi, M.M.; Hussain, M.Z.; Nyberg, F.; Yakovleva, T.; Bakalkin, G. The endogenous opioid system in human alcoholics: Molecular adaptations in brain areas involved in cognitive control of addiction. Addict. Biol. 2013, 18, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.D.; Abi-Dargham, A. The Role of Dynorphin and the Kappa Opioid Receptor in the Symptomatology of Schizophrenia: A Review of the Evidence. Biol. Psychiatry 2019, 86, 502–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Marchant, N.J.; Shaham, Y. Opposing roles of cotransmission of dynorphin and hypocretin on reward and motivation. Proc. Natl. Acad. Sci. USA 2014, 111, 5765–5766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muschamp, J.W.; Hollander, J.A.; Thompson, J.L.; Voren, G.; Hassinger, L.C.; Onvani, S.; Kamenecka, T.M.; Borgland, S.L.; Kenny, P.J.; Carlezon, W.A., Jr. Hypocretin (orexin) facilitates reward by attenuating the antireward effects of its cotransmitter dynorphin in ventral tegmental area. Proc. Natl. Acad. Sci. USA 2014, 111, 1648–1655. [Google Scholar] [CrossRef] [Green Version]
- Tejeda, H.A.; Shippenberg, T.S.; Henriksson, R. The dynorphin/kappa-opioid receptor system and its role in psychiatric disorders. Cell Mol. Life Sci. 2012, 69, 857–896. [Google Scholar] [CrossRef] [PubMed]
- Bakalkin, G.; Watanabe, H.; Jezierska, J.; Depoorter, C.; Verschuuren-Bemelmans, C.; Bazov, I.; Artemenko, K.A.; Yakovleva, T.; Dooijes, D.; Van de Warrenburg, B.P.; et al. Prodynorphin mutations cause the neurodegenerative disorder spinocerebellar ataxia type 23. Am. J. Hum. Genet. 2010, 87, 593–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smeets, C.J.; Jezierska, J.; Watanabe, H.; Duarri, A.; Fokkens, M.R.; Meijer, M.; Zhou, Q.; Yakovleva, T.; Boddeke, E.; den Dunnen, W.; et al. Elevated mutant dynorphin A causes Purkinje cell loss and motor dysfunction in spinocerebellar ataxia type 23. Brain 2015, 138, 2537–2552. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Wang, X.; Li, X.; Teng, H.; Tian, T.; Bai, J. Spinocerebellar ataxia type 23 (SCA23): A review. J. Neurol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Mague, S.D.; Pliakas, A.M.; Todtenkopf, M.S.; Tomasiewicz, H.C.; Zhang, Y.; Stevens, W.C., Jr.; Jones, R.M.; Portoghese, P.S.; Carlezon, W.A., Jr. Antidepressant-like effects of kappa-opioid receptor antagonists in the forced swim test in rats. J. Pharmacol. Exp. Ther. 2003, 305, 323–330. [Google Scholar] [CrossRef] [Green Version]
- Pfeiffer, A.; Brantl, V.; Herz, A.; Emrich, H.M. Psychotomimesis mediated by kappa opiate receptors. Science 1986, 233, 774–776. [Google Scholar] [CrossRef]
- Land, B.B.; Bruchas, M.R.; Lemos, J.C.; Xu, M.; Melief, E.J.; Chavkin, C. The dysphoric component of stress is encoded by activation of the dynorphin kappa-opioid system. J. Neurosci. 2008, 28, 407–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todtenkopf, M.S.; Marcus, J.F.; Portoghese, P.S.; Carlezon, W.A., Jr. Effects of kappa-opioid receptor ligands on intracranial self-stimulation in rats. Psychopharmacology 2004, 172, 463–470. [Google Scholar] [CrossRef]
- De Kloet, E.R.; Joels, M.; Holsboer, F. Stress and the brain: From adaptation to disease. Nat. Rev. Neurosci. 2005, 6, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Shaham, Y.; Erb, S.; Stewart, J. Stress-induced relapse to heroin and cocaine seeking in rats: A review. Brain Res. 2000, 33, 13–33. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, F.; Qin, W.; Jiang, T.; Yu, C. Multiscale neurobiological correlates of human neuroticism. Hum. Brain Mapp. 2020. [Google Scholar] [CrossRef]
- Altshuler, H.L.; Phillips, P.E.; Feinhandler, D.A. Alteration of ethanol self-administration by naltrexone. Life Sci. 1980, 26, 679–688. [Google Scholar] [CrossRef]
- Cichelli, M.J.; Lewis, M.J. Naloxone nonselective suppression of drinking of ethanol, sucrose, saccharin, and water by rats. Pharmacol. Biochem. Behav. 2002, 72, 699–706. [Google Scholar] [CrossRef]
- Walker, B.M.; Zorrilla, E.P.; Koob, G.F. Systemic kappa-opioid receptor antagonism by nor-binaltorphimine reduces dependence-induced excessive alcohol self-administration in rats. Addict. Biol. 2011, 16, 116–119. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez-Cuesta, J.; Burokas, A.; Mancino, S.; Kummer, S.; Martín-García, E.; Maldonado, R. Effects of genetic deletion of endogenous opioid system components on the reinstatement of cocaine-seeking behavior in mice. Neuropsychopharmacology 2014, 39, 2974–2988. [Google Scholar] [CrossRef] [Green Version]
- Edenberg, H.J.; Wang, J.; Tian, H.; Pochareddy, S.; Xuei, X.; Wetherill, L.; Goate, A.; Hinrichs, T.; Kuperman, S.; Nurnberger, J.I.; et al. A regulatory variation in OPRK1, the gene encoding the kappa-opioid receptor, is associated with alcohol dependence. Hum. Mol. Genet. 2008, 17, 1783–1789. [Google Scholar] [CrossRef] [Green Version]
- Egervari, G.; Jutras-Aswad, D.; Landry, J.; Miller, M.L.; Anderson, S.A.; Michaelides, M.; Jacobs, M.M.; Peter, C.; Yiannoulos, G.; Liu, X.; et al. A Functional 3′UTR Polymorphism (rs2235749) of Prodynorphin Alters microRNA-365 Binding in Ventral Striatonigral Neurons to Influence Novelty Seeking and Positive Reward Traits. Neuropsychopharmacology 2016, 41, 2512–2520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramchandani, V.A.; Umhau, J.; Pavon, F.J.; Ruiz-Velasco, V.; Margas, W.; Sun, H.; Damadzic, R.; Eskay, R.; Schoor, M.; Thorsell, A.; et al. A genetic determinant of the striatal dopamine response to alcohol in men. Mol Psychiatry 2011, 16, 809–817. [Google Scholar] [CrossRef]
- Xuei, X.; Dick, D.; Flury-Wetherill, L.; Tian, H.J.; Agrawal, A.; Bierut, L.; Goate, A.; Bucholz, K.; Schuckit, M.; Nurnberger, J.; et al. Association of the kappa-opioid system with alcohol dependence. Mol. Psychiatry 2006, 11, 1016–1024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xuei, X.; Flury-Wetherill, L.; Bierut, L.; Dick, D.; Nurnberger, J.; Nurnberger, J., Jr.; Foroud, T.; Edenberg, H.J. The opioid system in alcohol and drug dependence: Family-based association study. Am. J. Med. Gene.t B. Neuropsychiatr. Genet. 2007, 144, 877–884. [Google Scholar] [CrossRef]
- Ji, Y.; Su, R.; Tang, H.; Cui, J.; Deji, C.; Shi, Y.; Wei, S. Genetic association analyses and meta-analysis of Dynorphin-Kappa Opioid system potential functional variants with heroin dependence. Neurosci. Lett. 2018, 685, 75–82. [Google Scholar] [CrossRef]
- Karpyak, V.M.; Winham, S.J.; Preuss, U.W.; Zill, P.; Cunningham, J.M.; Walker, D.L.; Lewis, K.A.; Geske, J.R.; Colby, C.L.; Abulseoud, O.A.; et al. Association of the PDYN gene with alcohol dependence and the propensity to drink in negative emotional states. Int. J. Neuropsychopharmacol. 2013, 16, 975–985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, R.I.; Lopez, M.F.; Griffin, W.C.; Haun, H.L.; Bloodgood, D.W.; Pati, D.; Boyt, K.M.; Kash, T.L.; Becker, H.C. Dynorphin-kappa opioid receptor activity in the central amygdala modulates binge-like alcohol drinking in mice. Neuropsychopharmacology 2019, 44, 1084–1092. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zheng, M.Q.; Naganawa, M.; Gao, H.; Pracitto, R.; Shirali, A.; Lin, S.F.; Teng, J.K.; Ropchan, J.; Huang, Y. Novel Kappa Opioid Receptor Agonist as Improved PET Radiotracer: Development and in Vivo Evaluation. Mol. Pharm. 2019, 16, 1523–1531. [Google Scholar] [CrossRef]
- Bazov, I.; Bakalkin, G. Prodynorphin Epialleles, in Epigenetics and Human Health, Clinical Focus on Psychiatry. Springer 2016, 2, 43–76. [Google Scholar]
- Nikoshkov, A.; Hurd, Y.L.; Yakovleva, T.; Bazov, I.; Marinova, Z.; Cebers, G.; Pasikova, N.; Gharibyan, A.; Terenius, L.; Bakalkin, G. Prodynorphin transcripts and proteins differentially expressed and regulated in the adult human brain. FASEB J. 2005, 19, 1543–1545. [Google Scholar] [CrossRef]
- Lizio, M.; Harshbarger, J.; Shimoji, H.; Severin, J.; Kasukawa, T.; Sahin, S.; Abugessaisa, I.; Fukuda, S.; Hori, F.; Ishikawa-Kato, S.; et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome. Biol. 2015, 16, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Telkov, M.; Geijer, T.; Terenius, L. Human prodynorphin gene generates several tissue-specific transcripts. Brain Res. 1998, 804, 284–295. [Google Scholar] [CrossRef]
- Carninci, P.; Sandelin, A.; Lenhard, B.; Katayama, S.; Shimokawa, K.; Ponjavic, J.; Semple, C.A.; Taylor, M.S.; Engström, P.G.; Frith, M.C.; et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006, 38, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Kononenko, O.; Bazov, I.; Watanabe, H.; Gerashchenko, G.; Dyachok, O.; Verbeek, D.S.; Alkass, K.; Druid, H.; Andersson, M.; Mulder, J.; et al. Opioid precursor protein isoform is targeted to the cell nuclei in the human brain. Biochim. Biophys. Acta. Gen. Subj. 2017, 1861, 246–255. [Google Scholar] [CrossRef] [Green Version]
- Hara, Y.; Yakovleva, T.; Bakalkin, G.; Pickel, V.M. Dopamine D1 receptors have subcellular distributions conducive to interactions with prodynorphin in the rat nucleus accumbens shell. Synapse 2006, 60, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Bottger, A.; Spruce, B.A. Proenkephalin is a nuclear protein responsive to growth arrest and differentiation signals. J. Cell Biol. 1995, 130, 1251–1262. [Google Scholar] [CrossRef]
- McTavish, N.; Copeland, L.A.; Saville, M.K.; Perkins, N.D.; Spruce, B.A. Proenkephalin assists stress-activated apoptosis through transcriptional repression of NF-kappaB- and p53-regulated gene targets. Cell Death. Differ. 2007, 14, 1700–1710. [Google Scholar] [CrossRef] [PubMed]
- Bakalkin, G.; Ponomariev, D.; Sarkisyan, R.A.; Terenius, L. Sequence similarity between opioid peptide precursors and DNA-binding proteins. FEBS Lett. 1991, 282, 175–177. [Google Scholar] [CrossRef] [Green Version]
- Ota, T.; Suzuki, Y.; Nishikawa, T.; Otsuki, T.; Sugiyama, T.; Irie, R.; Wakamatsu, A.; Hayashi, K.; Sato, H.; Nagai, K.; et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004, 36, 40–45. [Google Scholar] [CrossRef]
- Rockman, M.V.; Hahn, M.W.; Soranzo, N.; Zimprich, F.; Goldstein, D.B.; Wray, G.A. Ancient and recent positive selection transformed opioid cis-regulation in humans. PLoS. Biol. 2005, 3, 387. [Google Scholar] [CrossRef] [PubMed]
- Carrion, A.M.; Link, W.A.; Ledo, F.; Mellström, B.; Naranjo, J.R. DREAM is a Ca2+-regulated transcriptional repressor. Nature 1999, 398, 80–84. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Pitcher, G.M.; Laviolette, S.R.; Whishaw, I.Q.; Tong, K.I.; Kockeritz, L.K.; Wada, T.; Joza, N.A.; Crackower, M.; Goncalves, J.; et al. DREAM is a critical transcriptional repressor for pain modulation. Cell 2002, 108, 31–43. [Google Scholar] [CrossRef] [Green Version]
- Ledo, F.; Carrión, A.M.; Link, W.A.; Mellström, B.; Naranjo, J.R. DREAM-alphaCREM interaction via leucine-charged domains derepresses downstream regulatory element-dependent transcription. Mol. Cell Biol. 2000, 20, 9120–9126. [Google Scholar] [CrossRef] [Green Version]
- Campos, D.; Jimenez-Diaz, L.; Carrion, A.M. Ca(2+)-dependent prodynorphin transcriptional derepression in neuroblastoma cells is exerted through DREAM protein activity in a kinase-independent manner. Mol. Cell Neurosci. 2003, 22, 135–145. [Google Scholar] [CrossRef]
- Bakalkin, G.; Telkov, M.; Yakovleva, T.; Terenius, L. [Leu5]enkephalin-encoding sequences are targets for a specific DNA-binding factor. Proc. Natl. Acad. Sci. USA 1995, 92, 9024–9028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakalkin, G.; Yakovleva, T.; Terenius, L. Prodynorphin gene expression relates to NF-kappa B factors. Brain Res. Mol. Brain Res. 1994, 24, 301–312. [Google Scholar] [CrossRef]
- Bakalkin, G.; Yakovleva, T.; Terenius, L. The Leu-enkephalin-encoding sequence DNA-binding factor (LEF) is the transcription factor YY1. Biochem. Biophys. Res. Commun. 1997, 231, 135–139. [Google Scholar] [CrossRef] [PubMed]
- McClung, C.A.; Ulery, P.G.; Perrotti, L.I.; Zachariou, V.; Berton, O.; Nestler, E.J. DeltaFosB: A molecular switch for long-term adaptation in the brain. Brain Res. Mol. Brain Res. 2004, 132, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Zachariou, V.; Bolanos, C.A.; Selley, D.E.; Theobald, D.; Cassidy, M.P.; Kelz, M.B.; Shaw-Lutchman, T.; Berton, O.; Sim-Selley, L.J.; Dileone, R.J.; et al. An essential role for DeltaFosB in the nucleus accumbens in morphine action. Nat. Neurosci. 2006, 9, 205–211. [Google Scholar] [CrossRef]
- Taqi, M.M.; Bazov, I.; Watanabe, H.; Nyberg, F.; Yakovleva, T.; Bakalkin, G. Prodynorphin promoter SNP associated with alcohol dependence forms noncanonical AP-1 binding site that may influence gene expression in human brain. Brain Res. 2011, 1385, 18–25. [Google Scholar] [CrossRef]
- Yuferov, V.; Ji, F.; Nielsen, D.A.; Levran, O.; Ho, A.; Morgello, S.; Shi, R.; Ott, J.; Kreek, M.J. A functional haplotype implicated in vulnerability to develop cocaine dependence is associated with reduced PDYN expression in human brain. Neuropsychopharmacology 2009, 34, 1185–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preuss, U.W.; Winham, S.J.; Biernacka, J.M.; Geske, J.R.; Bakalkin, G.; Koller, G.; Zill, P.; Soyka, M.; Karpyak, V.M. PDYN rs2281285 variant association with drinking to avoid emotional or somatic discomfort. PLoS ONE 2013, 8, 78688. [Google Scholar] [CrossRef] [Green Version]
- Zimprich, A.; Kraus, J.; Wöltje, M.; Mayer, P.; Rauch, E.; Höllt, V. An allelic variation in the human prodynorphin gene promoter alters stimulus-induced expression. J. Neurochem. 2000, 74, 472–477. [Google Scholar] [CrossRef]
- Rouault, M.; Nielsen, D.A.; Ho, A.; Kreek, M.J.; Yuferov, V. Cell-specific effects of variants of the 68-base pair tandem repeat on prodynorphin gene promoter activity. Addict. Biol. 2011, 16, 334–346. [Google Scholar] [CrossRef] [Green Version]
- Stogmann, E.; Zimprich, A.; Baumgartner, C.; Aull-Watschinger, S.; Höllt, V.; Zimprich, F. A functional polymorphism in the prodynorphin gene promotor is associated with temporal lobe epilepsy. Ann. Neurol. 2002, 51, 260–263. [Google Scholar] [CrossRef]
- Chen, A.C.; LaForge, K.S.; Ho, A.; McHugh, P.F.; Kellogg, S.; Bell, K.; Schluger, R.P.; Leal, S.M.; Kreek, M.J. Potentially functional polymorphism in the promoter region of prodynorphin gene may be associated with protection against cocaine dependence or abuse. Am. J. Med. Genet. 2002, 114, 429–435. [Google Scholar] [CrossRef]
- Zhang, C.S.; Tan, Z.; Lu, L.; Wu, S.N.; He, Y.; Gu, N.F.; Feng, G.Y.; He, L. Polymorphism of Prodynorphin promoter is associated with schizophrenia in Chinese population. Acta. Pharmacol. Sin. 2004, 25, 1022–1026. [Google Scholar]
- Saify, K.; Saadat, I.; Saadat, M. Association between VNTR polymorphism in promoter region of prodynorphin (PDYN) gene and heroin dependence. Psychiatry Res. 2014, 219, 690–692. [Google Scholar] [CrossRef]
- Ray, R.; Doyle, G.A.; Crowley, J.J.; Buono, R.J.; Oslin, D.W.; Patkar, A.A.; Mannelli, P.; DeMaria, P.A., Jr.; O’Brien, C.P.; Berrettini, W.H. A functional prodynorphin promoter polymorphism and opioid dependence. Psychiatr Genet. 2005, 15, 295–298. [Google Scholar] [CrossRef]
- Nomura, A.; Ujike, H.; Tanaka, Y.; Otani, K.; Morita, Y.; Kishimoto, M.; Morio, A.; Harano, M.; Inada, T.; Yamada, M.; et al. Genetic variant of prodynorphin gene is risk factor for methamphetamine dependence. Neurosci Lett. 2006, 400, 158–162. [Google Scholar] [CrossRef] [Green Version]
- Saify, K.; Saadat, M. Association between VNTR Polymorphism in Promoter Region of Prodynorphin (PDYN) Gene and Methamphetamine Dependence. J. Med. Sci. 2015, 3, 371–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, T.J.; LaForge, K.S.; Gordon, D.; Bart, G.; Kellogg, S.; Ott, J.; Kreek, M.J. Prodynorphin gene promoter repeat associated with cocaine/alcohol codependence. Addict. Biol. 2007, 12, 496–502. [Google Scholar] [CrossRef]
- Bovo, G.; Diani, E.; Bisulli, F.; Di Bonaventura, C.; Striano, P.; Gambardella, A.; Ferlazzo, E.; Egeo, G.; Mecarelli, O.; Elia, M.; et al. Analysis of LGI1 promoter sequence, PDYN and GABBR1 polymorphisms in sporadic and familial lateral temporal lobe epilepsy. Neurosci. Lett. 2008, 436, 23–26. [Google Scholar] [CrossRef] [PubMed]
- Dahl, J.P.; Weller, A.E.; Kampman, K.M.; Oslin, D.W.; Lohoff, F.W.; Ferraro, T.N.; O’Brien, C.P.; Berrettini, W.H. Confirmation of the association between a polymorphism in the promoter region of the prodynorphin gene and cocaine dependence. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2005, 139, 106–108. [Google Scholar] [CrossRef]
- Hou, C.; Zhao, H.; Tanimoto, K.; Dean, A. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc. Natl. Acad. Sci. USA 2008, 105, 20398–20403. [Google Scholar] [CrossRef] [Green Version]
- Majumder, P.; Gomez, J.A.; Chadwick, B.P.; Boss, J.M. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J. Exp. Med. 2008, 205, 785–798. [Google Scholar] [CrossRef] [Green Version]
- Bazov, I.; Sarkisyan, D.; Kononenko, O.; Watanabe, H.; Taqi, M.M.; Stålhandske, L.; Verbeek, D.S.; Mulder, J.; Rajkowska, G.; Sheedy, D.; et al. Neuronal Expression of Opioid Gene is Controlled by Dual Epigenetic and Transcriptional Mechanism in Human Brain. Cereb. Cortex. 2018, 28, 3129–3142. [Google Scholar] [CrossRef]
- Yuferov, V.; Nielsen, D.A.; Levran, O.; Randesi, M.; Hamon, S.; Ho, A.; Morgello, S.; Kreek, M.J. Tissue-specific DNA methylation of the human prodynorphin gene in post-mortem brain tissues and PBMCs. Pharm. Genomics. 2011, 21, 185–196. [Google Scholar] [CrossRef] [Green Version]
- Doi, A.; Park, I.H.; Wen, B.; Murakami, P.; Aryee, M.J.; Irizarry, R.; Herb, B.; Ladd-Acosta, C.; Rho, J.; Loewer, S.; et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009, 41, 1350–1353. [Google Scholar] [CrossRef] [Green Version]
- Ciernia, A.V.; LaSalle, J. The landscape of DNA methylation amid a perfect storm of autism aetiologies. Nat. Rev. Neurosci. 2016, 17, 411–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Li, X.; Aryee, M.J.; Ekström, T.J.; Padyukov, L.; Klareskog, L.; Vandiver, A.; Moore, A.Z.; Tanaka, T.; Ferrucci, L.; et al. GeMes, clusters of DNA methylation under genetic control, can inform genetic and epigenetic analysis of disease. Am. J. Hum. Genet. 2014, 94, 485–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naranjo, J.R.; Mellstrom, B. Ca2+-dependent transcriptional control of Ca2+ homeostasis. J. Biol. Chem. 2012, 287, 31674–31680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, C.; Liang, G.; Nguyen, T.T.; Tsao-Wei, D.; Groshen, S.; Lübbert, M.; Zhou, J.H.; Benedict, W.F.; Jones, P.A. Susceptibility of nonpromoter CpG islands to de novo methylation in normal and neoplastic cells. J. Natl. Cancer Inst. 2001, 93, 1465–1472. [Google Scholar] [CrossRef] [Green Version]
- Ball, M.P.; Li, J.B.; Gao, Y.; Lee, J.H.; LeProust, E.M.; Park, I.H.; Xie, B.; Daley, G.Q.; Church, G.M. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009, 27, 361–368. [Google Scholar] [CrossRef] [Green Version]
- Moen, E.L.; Zhang, X.; Mu, W.; Delaney, S.M.; Wing, C.; McQuade, J.; Myers, J.; Godley, L.A.; Dolan, M.E.; Zhang, W. Genome-wide variation of cytosine modifications between European and African populations and the implications for complex traits. Genetics 2013, 194, 987–996. [Google Scholar] [CrossRef] [Green Version]
- Tomso, D.J.; Bell, D.A. Sequence context at human single nucleotide polymorphisms: Overrepresentation of CpG dinucleotide at polymorphic sites and suppression of variation in CpG islands. J. Mol. Biol. 2003, 327, 303–308. [Google Scholar] [CrossRef]
- Taqi, M.M.; Bazov, I.; Watanabe, H.; Sheedy, D.; Harper, C.; Alkass, K.; Druid, H.; Wentzel, P.; Nyberg, F.; Yakovleva, T.; et al. Prodynorphin CpG-SNPs associated with alcohol dependence: Elevated methylation in the brain of human alcoholics. Addict. Biol. 2011, 16, 499–509. [Google Scholar] [CrossRef] [Green Version]
- Oertel, B.G.; Doehring, A.; Roskam, B.; Kettner, M.; Hackmann, N.; Ferreirós, N.; Schmidt, P.H.; Lötsch, J. Genetic-epigenetic interaction modulates mu-opioid receptor regulation. Hum. Mol. Genet. 2012, 21, 4751–4760. [Google Scholar] [CrossRef]
- Pun, F.W.; Zhao, C.; Lo, W.S.; Ng, S.K.; Tsang, S.Y.; Nimgaonkar, V.; Chung, W.S.; Ungvari, G.S.; Xue, H. Imprinting in the schizophrenia candidate gene GABRB2 encoding GABA(A) receptor beta(2) subunit. Mol. Psychiatry. 2011, 16, 557–568. [Google Scholar] [CrossRef] [Green Version]
- Ursini, G.; Bollati, V.; Fazio, L.; Porcelli, A.; Iacovelli, L.; Catalani, A.; Sinibaldi, L.; Gelao, B.; Romano, R.; Rampino, A.; et al. Stress-related methylation of the catechol-O-methyltransferase Val 158 allele predicts human prefrontal cognition and activity. J. Neurosci. 2011, 31, 6692–6698. [Google Scholar] [CrossRef] [Green Version]
- Sigurdsson, M.I.; Smith, A.V.; Bjornsson, H.T.; Jonsson, J.J. HapMap methylation-associated SNPs, markers of germline DNA methylation, positively correlate with regional levels of human meiotic recombination. Genome. Res. 2009, 19, 581–589. [Google Scholar] [CrossRef] [Green Version]
- Hellman, A.; Chess, A. Extensive sequence-influenced DNA methylation polymorphism in the human genome. Epigenetics Chromatin. 2010, 3, 11. [Google Scholar] [CrossRef] [Green Version]
- Xie, H.; Wang, M.; Bischof, J.; Bonaldo, M.; Soares, M.B. SNP-based prediction of the human germ cell methylation landscape. Genomics 2009, 93, 434–440. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.A.; Tapia-Ramírez, J.; Kim, S.; Toledo-Aral, J.J.; Zheng, Y.; Boutros, M.C.; Altshuller, Y.M.; Frohman, M.A.; Kraner, S.D.; Mandel, G. REST: A mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 1995, 80, 949–957. [Google Scholar] [CrossRef] [Green Version]
- Schoenherr, C.J.; Anderson, D.J. The neuron-restrictive silencer factor (NRSF): A coordinate repressor of multiple neuron-specific genes. Science 1995, 267, 1360–1363. [Google Scholar] [CrossRef]
- Gerstein, M.B.; Kundaje, A.; Hariharan, M.; Landt, S.G.; Yan, K.K.; Cheng, C.; Mu, X.J.; Khurana, E.; Rozowsky, J.; Alexander, R.; et al. Architecture of the human regulatory network derived from ENCODE data. Nature 2012, 489, 91–100. [Google Scholar] [CrossRef]
- Wang, J.; Zhuang, J.; Iyer, S.; Lin, X.; Whitfield, T.W.; Greven, M.C.; Pierce, B.G.; Dong, X.; Kundaje, A.; Cheng, Y.; et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome. Res. 2012, 22, 1798–1812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruce, A.W.; Donaldson, I.J.; Wood, I.C.; Yerbury, S.A.; Sadowski, M.I.; Chapman, M.; Göttgens, B.; Buckley, N.J. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc. Natl. Acad. Sci. USA 2004, 101, 10458–10463. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.S.; Mortazavi, A.; Myers, R.M.; Wold, B. Genome-wide mapping of in vivo protein-DNA interactions. Science 2007, 316, 1497–1502. [Google Scholar] [CrossRef] [Green Version]
- Rockowitz, S.; Lien, W.H.; Pedrosa, E.; Wei, G.; Lin, M.; Zhao, K.; Lachman, H.M.; Fuchs, E.; Zheng, D. Comparison of REST cistromes across human cell types reveals common and context-specific functions. PLoS. Comput. Biol. 2014, 10, 1003671. [Google Scholar] [CrossRef]
- Tapia-Ramirez, J.; Eggen, B.J.; Peral-Rubio, M.J.; Toledo-Aral, J.J.; Mandel, G. A single zinc finger motif in the silencing factor REST represses the neural-specific type II sodium channel promoter. Proc. Natl. Acad. Sci. USA 1997, 94, 1177–1182. [Google Scholar] [CrossRef] [Green Version]
- Henriksson, R.; Bäckman, C.M.; Harvey, B.K.; Kadyrova, H.; Bazov, I.; Shippenberg, T.S.; Bakalkin, G. PDYN, a gene implicated in brain/mental disorders, is targeted by REST in the adult human brain. Biochim Biophys Acta. 2014, 1839, 1226–1232. [Google Scholar] [CrossRef]
- Packer, A.N.; Xing, Y.; Harper, S.Q.; Jones, L.; Davidson, B.L. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J. Neurosci. 2008, 28, 14341–14346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, A.S.; Staahl, B.T.; Chen, L.; Crabtree, G.R. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 2009, 460, 642–646. [Google Scholar] [CrossRef] [Green Version]
- Brohl, D.; Strehle, M.; Wende, H.; Hori, K.; Bormuth, I.; Nave, K.A.; Müller, T.; Birchmeier, C. A transcriptional network coordinately determines transmitter and peptidergic fate in the dorsal spinal cord. Dev. Biol. 2008, 322, 381–393. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Huang, T.; Xiang, Y.; Xie, Z.; Chen, Y.; Yan, R.; Xu, J.; Cheng, L. Ptf1a, Lbx1 and Pax2 coordinate glycinergic and peptidergic transmitter phenotypes in dorsal spinal inhibitory neurons. Dev. Biol. 2008, 322, 394–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wildner, H.; Das Gupta, R.; Bröhl, D.; Heppenstall, P.A.; Zeilhofer, H.U.; Birchmeier, C. Genome-wide expression analysis of Ptf1a- and Ascl1-deficient mice reveals new markers for distinct dorsal horn interneuron populations contributing to nociceptive reflex plasticity. J. Neurosci. 2013, 33, 7299–7307. [Google Scholar] [CrossRef]
- Lu, K.M.; Evans, S.M.; Hirano, S.; Liu, F.C. Dual role for Islet-1 in promoting striatonigral and repressing striatopallidal genetic programs to specify striatonigral cell identity. Proc. Natl. Acad. Sci. USA 2014, 111, 168–177. [Google Scholar] [CrossRef] [Green Version]
- Kardon, A.P.; Polgár, E.; Hachisuka, J.; Snyder, L.M.; Cameron, D.; Savage, S.; Cai, X.; Karnup, S.; Fan, C.R.; Hemenway, G.M.; et al. Dynorphin acts as a neuromodulator to inhibit itch in the dorsal horn of the spinal cord. Neuron 2014, 82, 573–586. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.; Huang, D.; Grady, F.S.; Peltekian, L.; Geerling, J.C. Efferent projections of Vglut2, Foxp2, and Pdyn parabrachial neurons in mice. J. Comp. Neurol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.K.; Owyang, V.V.; Hong, J.S.; Gallagher, M. Elevated dynorphin in the hippocampal formation of aged rats: Relation to cognitive impairment on a spatial learning task. Proc. Natl. Acad. Sci. USA 1989, 86, 2948–2951. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, X.V.; Masse, J.; Kumar, A.; Vijitruth, R.; Kulik, C.; Liu, M.; Choi, D.Y.; Foster, T.C.; Usynin, I.; Bakalkin, G.; et al. Prodynorphin knockout mice demonstrate diminished age-associated impairment in spatial water maze performance. Behav. Brain Res. 2005, 161, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Carey, A.N.; Lyons, A.M.; Shay, C.F.; Dunton, O.; McLaughlin, J.P. Endogenous kappa opioid activation mediates stress-induced deficits in learning and memory. J. Neurosci. 2009, 29, 4293–4300. [Google Scholar] [CrossRef] [PubMed]
- Kolsch, H.; Wagner, M.; Bilkei-Gorzó, A.; Toliat, M.R.; Pentzek, M.; Fuchs, A.; Kaduszkiewicz, H.; van den Bussche, H.; Riedel-Heller, S.G.; Angermeyer, M.C.; et al. Gene polymorphisms in prodynorphin (PDYN) are associated with episodic memory in the elderly. J. Neural Transm. 2009, 116, 897–903. [Google Scholar] [CrossRef]
- Yakovleva, T.; Marinova, Z.; Kuzmin, A.; Seidah, N.G.; Haroutunian, V.; Terenius, L.; Bakalkin, G. Dysregulation of dynorphins in Alzheimer disease. Neurobiol. Aging. 2007, 28, 1700–1708. [Google Scholar] [CrossRef]
- Sandin, J.; Nylander, I.; Georgieva, J.; Schött, P.A.; Ogren, S.O.; Terenius, L. Hippocampal dynorphin B injections impair spatial learning in rats: A kappa-opioid receptor-mediated effect. Neuroscience 1998, 85, 375–382. [Google Scholar] [CrossRef]
- Cippitelli, A.; Damadzic, R.; Frankola, K.; Goldstein, A.; Thorsell, A.; Singley, E.; Eskay, R.L.; Heilig, M. Alcohol-induced neurodegeneration, suppression of transforming growth factor-beta, and cognitive impairment in rats: Prevention by group II metabotropic glutamate receptor activation. Biol. Psychiatry. 2010, 67, 823–830. [Google Scholar] [CrossRef]
- Kuzmin, A.; Chefer, V.; Bazov, I.; Meis, J.; Ögren, S.O.; Shippenberg, T.; Bakalkin, G. Upregulated dynorphin opioid peptides mediate alcohol-induced learning and memory impairment. Transl. Psychiatry. 2013, 3, 310. [Google Scholar] [CrossRef] [Green Version]
- Kuzmin, A.; Liljequist, S.; Meis, J.; Chefer, V.; Shippenberg, T.; Bakalkin, G. Repeated moderate-dose ethanol bouts impair cognitive function in Wistar rats. Addict. Biol. 2012, 17, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Chefer, V.; Meis, J.; Wang, G.; Kuzmin, A.; Bakalkin, G.; Shippenberg, T. Repeated exposure to moderate doses of ethanol augments hippocampal glutamate neurotransmission by increasing release. Addict. Biol. 2011, 16, 229–237. [Google Scholar] [CrossRef] [Green Version]
- Walker, B.M.; Koob, G.F. Pharmacological evidence for a motivational role of kappa-opioid systems in ethanol dependence. Neuropsychopharmacology 2008, 33, 643–652. [Google Scholar] [CrossRef] [Green Version]
- Schlosburg, J.E.; Whitfield, T.W., Jr.; Park, P.E.; Crawford, E.F.; George, O.; Vendruscolo, L.F.; Koob, G.F. Long-term antagonism of kappa opioid receptors prevents escalation of and increased motivation for heroin intake. J. Neurosci. 2013, 33, 19384–19392. [Google Scholar] [CrossRef]
- Nealey, K.A.; Smith, A.W.; Davis, S.M.; Smith, D.G.; Walker, B.M. kappa-opioid receptors are implicated in the increased potency of intra-accumbens nalmefene in ethanol-dependent rats. Neuropharmacology 2011, 61, 35–42. [Google Scholar] [CrossRef] [Green Version]
- Kissler, J.L.; Sirohi, S.; Reis, D.J.; Jansen, H.T.; Quock, R.M.; Smith, D.G.; Walker, B.M. The one-two punch of alcoholism: Role of central amygdala dynorphins/kappa-opioid receptors. Biol. Psychiatry 2014, 75, 774–782. [Google Scholar] [CrossRef] [Green Version]
- Bazov, I.; Sarkisyan, D.; Kononenko, O.; Watanabe, H.; Karpyak, V.M.; Yakovleva, T.; Bakalkin, G. Downregulation of the neuronal opioid gene expression concomitantly with neuronal decline in dorsolateral prefrontal cortex of human alcoholics. Transl. Psychiatry 2018, 8, 122. [Google Scholar] [CrossRef] [Green Version]
- Bazov, I.; Sarkisyan, D.; Kononenko, O.; Watanabe, H.; Yakovleva, T.; Hansson, A.C.; Sommer, W.H.; Spanagel, R.; Bakalkin, G. Dynorphin and kappa-Opioid Receptor Dysregulation in the Dopaminergic Reward System of Human. Alcoholics. Mol. Neurobiol. 2018, 55, 7049–7061. [Google Scholar] [CrossRef] [Green Version]
- Sarkisyan, D.; Hussain, M.Z.; Watanabe, H.; Kononenko, O.; Bazov, I.; Zhou, X.; Yamskova, O.; Krishtal, O.; Karpyak, V.M.; Yakovleva, T.; et al. Downregulation of the endogenous opioid peptides in the dorsal striatum of human alcoholics. Front. Cell Neurosci. 2015, 9, 187. [Google Scholar] [CrossRef]
- De la Monte, S.M.; Kril, J.J. Human alcohol-related neuropathology. Acta. Neuropathol. 2014, 127, 71–90. [Google Scholar] [CrossRef]
- Zahr, N.M.; Kaufman, K.L.; Harper, C.G. Clinical and pathological features of alcohol-related brain damage. Nat. Rev. Neurol. 2011, 7, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Sarkisyan, D.; Bazov, I.; Watanabe, H.; Kononenko, O.; Syvänen, A.C.; Schumann, G.; Yakovleva, T.; Bakalkin, G. Damaged reward areas in human alcoholics: Neuronal proportion decline and astrocyte activation. Acta. Neuropathol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Robison, A.J.; Nestler, E.J. Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci. 2011, 12, 623–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nassel, D.R. Neuropeptide signaling near and far: How localized and timed is the action of neuropeptides in brain circuits? Invert. Neurosci. 2009, 9, 57–75. [Google Scholar] [CrossRef] [PubMed]
- Gohler, T.; Reimann, M.; Cherny, D.; Walter, K.; Warnecke, G.; Kim, E.; Deppert, W. Specific interaction of p53 with target binding sites is determined by DNA conformation and is regulated by the C-terminal domain. J. Biol. Chem. 2002, 277, 41192–41203. [Google Scholar] [CrossRef] [Green Version]
- Lai, J.; Ossipov, M.H.; Vanderah, T.W.; Malan, T.P.; Porreca, F. Neuropathic pain: The paradox of dynorphin. Mol. Interv. 2001, 1, 160–167. [Google Scholar]
- Bazov, I.; Kononenko, O.; Watanabe, H.; Taqi, M.M.; Gerashchenko, G.; Yakovleva, T.; Bakalkin, G. Epigenetic mechanism of endogenous opioid peptide precursor prodynorphin upregulation in the brain of human alcoholics: Methylation of DNA in a single promoter nucleosome mediates USF2 effects. Soc. Neurosci. Meet. 2011. [Google Scholar]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nosova, O.; Bazov, I.; Karpyak, V.; Hallberg, M.; Bakalkin, G. Epigenetic and Transcriptional Control of the Opioid Prodynorphine Gene: In-Depth Analysis in the Human Brain. Molecules 2021, 26, 3458. https://doi.org/10.3390/molecules26113458
Nosova O, Bazov I, Karpyak V, Hallberg M, Bakalkin G. Epigenetic and Transcriptional Control of the Opioid Prodynorphine Gene: In-Depth Analysis in the Human Brain. Molecules. 2021; 26(11):3458. https://doi.org/10.3390/molecules26113458
Chicago/Turabian StyleNosova, Olga, Igor Bazov, Victor Karpyak, Mathias Hallberg, and Georgy Bakalkin. 2021. "Epigenetic and Transcriptional Control of the Opioid Prodynorphine Gene: In-Depth Analysis in the Human Brain" Molecules 26, no. 11: 3458. https://doi.org/10.3390/molecules26113458
APA StyleNosova, O., Bazov, I., Karpyak, V., Hallberg, M., & Bakalkin, G. (2021). Epigenetic and Transcriptional Control of the Opioid Prodynorphine Gene: In-Depth Analysis in the Human Brain. Molecules, 26(11), 3458. https://doi.org/10.3390/molecules26113458





