Parvaxanthines D–F and Asponguanosines C and D, Racemic Natural Hybrids from the Insect Cyclopelta parva
Abstract
:1. Introduction
2. Results and Discussion
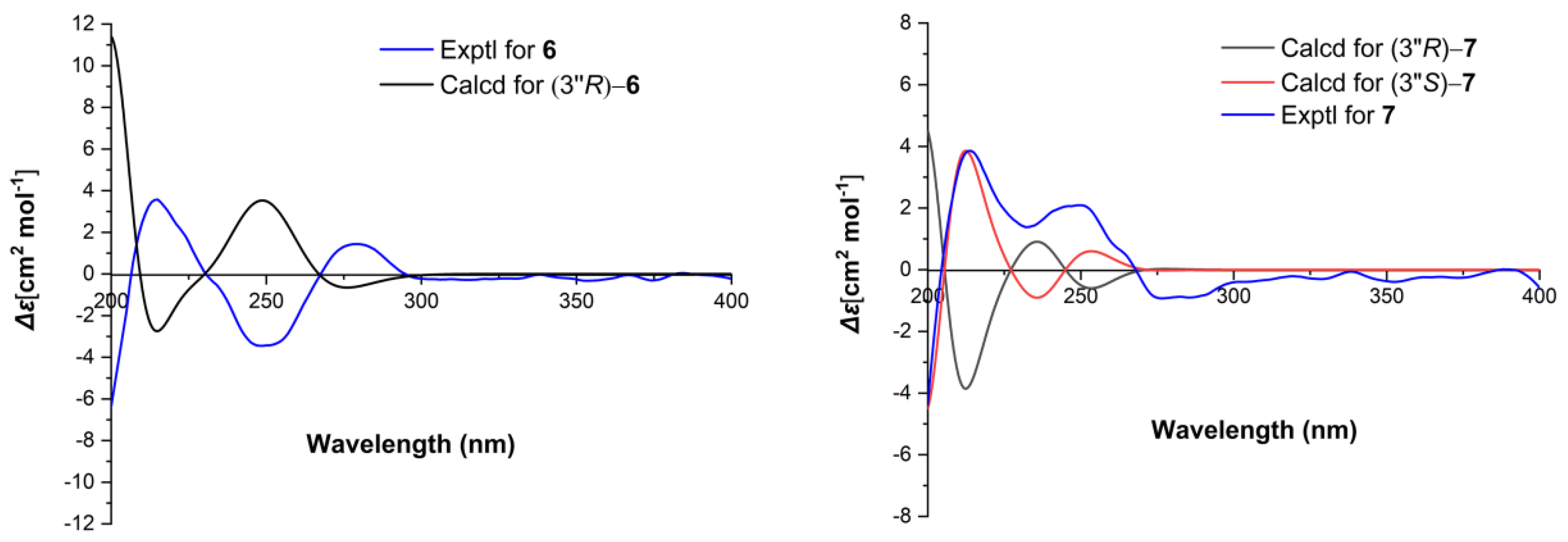
2.1. Structure Elucidation of the Compounds
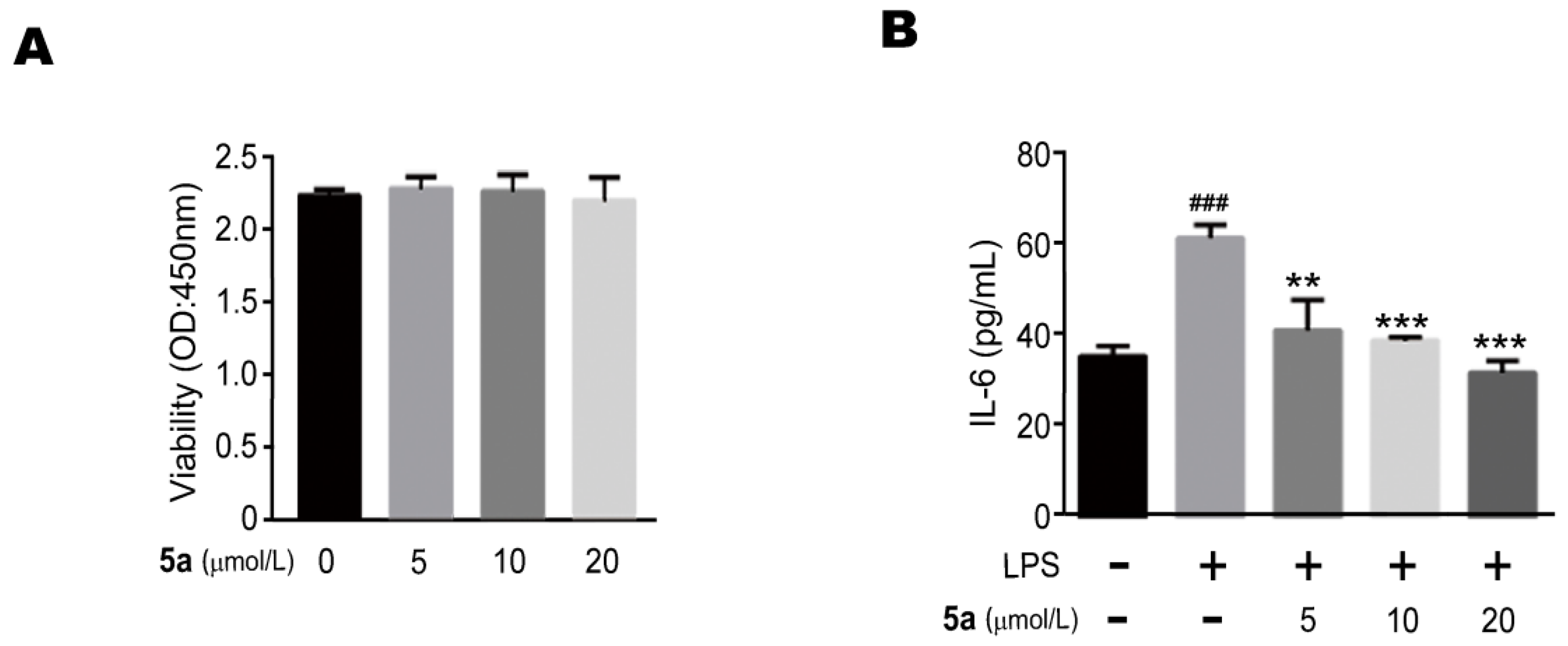
2.2. Biological Evaluation
3. Experimental Section
3.1. General Procedures
3.2. Insect Material
3.3. Extraction and Isolation
3.4. Compound Characterization Data
3.5. Acid Hydrolysis and Preparation of Sugar Derivatives of Compounds 6 and 7
3.6. Computational Methods
3.7. Antiviral Activity Assay
3.8. Immunosuppressive Activity Assay
3.9. Biological Evaluation for Human Cancer Cells (BGC-823, MDA-MB-231, HepG2, Kyse30)
3.10. Anti-Inflammatory Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Sun, G.; Hu, C.Q.; Mei, Q.; Luo, M.H.; Chen, X.; Li, Z.Y.; Liu, Y.Z.; Deng, Z.X.; Zhang, Z.Y.; Sun, Y.H. Uncovering the cytochrome P450-catalyzed methylenedioxy bridge formation in streptovaricins biosynthesis. Nat. Commun. 2020, 11, 4501. [Google Scholar] [CrossRef] [PubMed]
- Dossy, A.D. Insects and their chemical weaponry: New potential for drug discovery. Nat. Prod. Rep. 2010, 27, 1737–1757. [Google Scholar] [CrossRef]
- Seabrooks, L.; Hu, L.Q. Insects: An underrepresented resource for the discovery of biologically active natural products. Acta Pharm Sin. B. 2017, 7, 409–426. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.L.; Huang, X.J.; Wang, Y.; Jiang, R.W.; Wang, L.; Bai, L.L.; Peng, Q.L.; Song, C.L.; Zhang, D.M.; Ye, W.C. Isocoumarins from American cockroach (Periplaneta americana) and their cytotoxic activities. Fitoterapia 2014, 95, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.F.; Li, Y.P.; Qin, F.Y.; Yan, Y.M.; Wang, S.M.; Zhang, H.X.; Cheng, Y.X. Periplanetols A–F, phenolic compounds from Periplaneta americana with potent COX-2 inhibitory activity. Fitoterapia 2020, 143, 104589. [Google Scholar] [CrossRef]
- Tang, J.J.; Zhang, L.; Jiang, L.P.; Di, L.; Yan, Y.M.; Tu, Z.C.; Yang, C.P.; Zuo, Z.L.; Hou, B.; Xia, H.L.; et al. Dopamine derivatives from the insect Polyrhachis dives as inhibitors of ROCK1/2 and stimulators of neural stem cell proliferation. Tetrahedron 2014, 70, 8852–8857. [Google Scholar] [CrossRef]
- Tang, J.J.; Fang, P.; Xia, H.L.; Tu, Z.C.; Hou, B.Y.; Yan, Y.M.; Di, L.; Zhang, L.; Cheng, Y.X. Constituents from the edible Chinese black ants (Polyrhachis dives) showing protective effect on rat mesangial cells and anti-inflammatory activity. Food Res. Int. 2015, 67, 163–168. [Google Scholar] [CrossRef]
- Zhu, H.J.; Xu, T.; Yan, Y.M.; Cheng, Y.X. Nonpeptidal compounds from the insect Polyphaga plancyi and their biological evaluation. Bioorg. Chem. 2020, 104, 104258. [Google Scholar] [CrossRef]
- Yan, Y.M.; Zhu, H.J.; Zhou, F.J.; Tu, Z.C.; Cheng, Y.X. Phenolic compounds from the insect Blaps japanensis with inhibitory activities towards cancer cells, COX-2, ROCK1 and JAK3. Tetrahedron 2019, 75, 1029–1033. [Google Scholar] [CrossRef]
- Yan, Y.M.; Xu, T.; Tu, Z.C.; Zhu, H.J.; Cheng, Y.X. Sulfur and nitrogen-containing compounds from the whole bodies of Blaps Japanensis. Bioorg. Chem. 2020, 102, 104086. [Google Scholar] [CrossRef]
- Song, Z.W.; Yin, W.P.; Liu, P.; Dong, J.F.; Jiang, Y.L.; Gao, T. Advances in chemical constituents and pharmacological effects of familiar Pentatomoids. Chin. J. Appl. Entomol. 2011, 48, 753–756. [Google Scholar]
- Yang, Z.H.; Yang, X.Y.; Yuan, X.H.; Luo, K.M.; Luo, X.Y.; Zhang, Y.J.; Guo, Y.L.; Pei, Y. Purification of the antimicrobial peptide AMP-1 of Cyclopelta parva distant by HPLC. J. Southwest Agric. Univ. 2001, 23, 104–107. [Google Scholar]
- Wu, Y.F.; Cao, M.L.; Tan, J.; Guo, J.J. Comparison of the inhibitory effects between Aspongopus chinensis and Cyclopelta parva on proliferation of human gastric cancer cell SGC–7901. J. Mountain Agric. Bilo. 2018, 37, 11–14. [Google Scholar] [CrossRef]
- Di, L.; Shi, Y.N.; Yan, Y.M.; Jiang, L.P.; Hou, B.; Wang, X.L.; Zuo, Z.L.; Chen, Y.B.; Yang, C.P.; Cheng, Y.X. Nonpeptide small molecules from the insect Aspongopus chinensis and their neural stem cell proliferation stimulating properties. RSC Adv. 2015, 5, 70985–70991. [Google Scholar] [CrossRef]
- Shi, Y.N.; Tu, Z.C.; Wang, X.L.; Yan, Y.M.; Fang, P.; Zuo, Z.L.; Hou, B.; Yang, T.H.; Cheng, Y.X. Bioactive compounds from the insect Aspongopus chinensis. Bioorg. Med. Chem. Lett. 2014, 24, 5164–5169. [Google Scholar] [CrossRef]
- Yan, Y.M.; Ai, J.; Shi, Y.N.; Zuo, Z.L.; Hou, B.; Luo, J.; Cheng, Y.X. (±)-Aspongamide A, an N-acetyldopamine trimer isolated from the insect Aspongopus chinensis, is an inhibitor of p-Smad3. Org. Lett. 2014, 16, 532–535. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.P.; Qin, F.Y.; Yan, Y.M.; Zhang, H.X.; Cheng, Y.X. Racemic xanthine and dihydroxydopamine conjugates from Cyclopelta parva and their COX-2 inhibitory activity. Fitoterapia 2020, 142, 104534. [Google Scholar] [CrossRef]
- Yeh, G.C.; Henderson, J.P.; Byun, J.; Avignon, D.A.; Heineckeb, J.W. 8-Nitroxanthine, a product of myeloperoxidase, peroxynitrite, and activated human neutrophils, enhances generation of superoxide by xanthine oxidase. Arch. Biochem. Biophys. 2003, 418, 1–12. [Google Scholar] [CrossRef]
- Ding, W.Y.; Yan, Y.M.; Meng, X.H.; Nafie, L.A.; Xu, T.; Dukor, R.K.; Qin, H.B.; Cheng, Y.X. Isolation, total synthesis, and absolute configuration determination of renoprotective dimeric N-acetyldopamine-adenine hybrids from the insect Aspongopus chinensis. Org. Lett. 2020, 22, 5726–5730. [Google Scholar] [CrossRef]
- Wang, C.X.; Chen, G.D.; Feng, C.C.; He, R.R.; Qin, S.Y.; Hu, D.; Chen, H.R.; Liu, X.Z.; Yao, X.S.; Gao, H. Same data, different structures: Diastereoisomers with substantially identical NMR data from nature. Chem. Commun. 2016, 52, 1250–1253. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.P.; Zhou, Z.X.; Zhang, Y.; Li, J.; Cheng, F.; Xu, P.S.; Zhou, G.; Li, X.M.; Xu, K.P.; Tan, G.S. New adenine analogues and a pyrrole alkaloid from Selaginella delicatula. Nat. Prod. Res. 2019, 33, 1985–1991. [Google Scholar] [CrossRef]
- Tietze, L.F.; Bell, H.P.; Chandasekhar, S. Natural product hybrids as new leads for drug discovery. Angew. Chem. Int. Ed. 2003, 115, 4128–4160. [Google Scholar] [CrossRef]
- Walsh, J.J.; Coughlan, D.; Heneghan, N.; Gaynor, C.; Bell, A. A novel artemisinin-quinine hybrid with potent antimalarial activity. Bioorg. Med. Chem. Lett. 2001, 17, 3599–3602. [Google Scholar] [CrossRef]
- Tanaka, T.; Nakashima, T.; Ueda, T.; Tomii, K.; Kouno, I. Facile discrimination of aldose enantiomers by reversed-phase HPLC. Chem. Pharm. Bull. 2007, 55, 899–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.M.; Luo, Q.; Di, L.; Shi, Y.N.; Tu, Z.C.; Cheng, Y.X. Nucleoside and N-acetyldopamine derivatives from the insect Aspongopus chinensis. Fitoterapia 2019, 132, 82–87. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian, Version 09, Expanding the Limits of Computational Chemistry; Gaussian, Inc.: Wallingford, CT, USA, 2013. [Google Scholar]
- Xiang, Y.F.; Pei, Y.; Qu, C.; Lai, Z.C.; Ren, Z.; Yang, K.; Xiong, S.; Zhang, Y.J.; Yang, C.R.; Wang, D.; et al. In vitro anti-herpes simplex virus activity of 1,2,4,6-tetra-O-galloyl-D-glucose from Phyllanthus emblica L. (Euphorbiaceae). Phytother. Res. 2011, 25, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Jakhar, R.; Sharma, C.; Paul, S.; Kang, S.C. Immunosuppressive potential of astemizole against LPS activated T cell proliferation and cytokine secretion in RAW macrophages, zebrafish larvae and mouse splenocytes by modulating MAPK signaling pathway. Int. Immunopharmacol. 2018, 65, 268–278. [Google Scholar] [CrossRef]
- Chen, F.Y.; Ni, Y.; Ye, Y.P.; Sun, H.X.; Li, X.Y.; Xu, S.F. Stephanthraniline A inhibits the proliferation and activation of T cells in vitro and in vivo. Eur. J. Pharmacol. 2012, 685, 186–197. [Google Scholar] [CrossRef]
- Wang, W.; Wang, C.Y. Integrated miRNA and mRNA omics reveal the anti-cancerous mechanism of Licochalcone B on Human Hepatoma Cell HepG2. Food Chem. Toxicil. 2021, 150, 112096. [Google Scholar] [CrossRef]
- Hasanpourghadia, M.; Majidb, N.A.; Mustafaa, M.R. Activation of autophagy by stress-activated signals as a cellular self-defense mechanism against the cytotoxic effects of MBIC in human breast cancer cells in vitro. Biochem. Pharmacol. 2018, 1552, 174–186. [Google Scholar] [CrossRef] [PubMed]
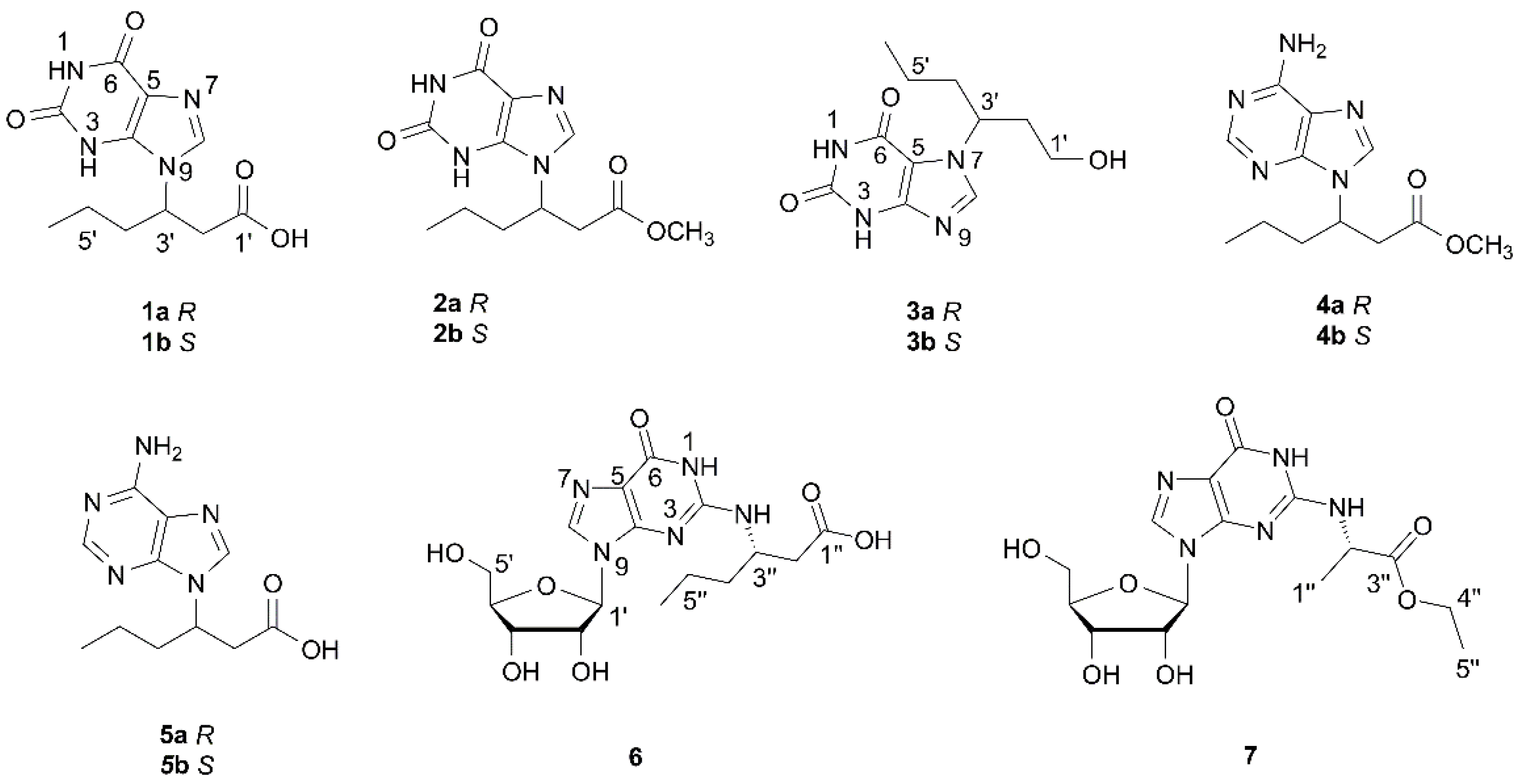
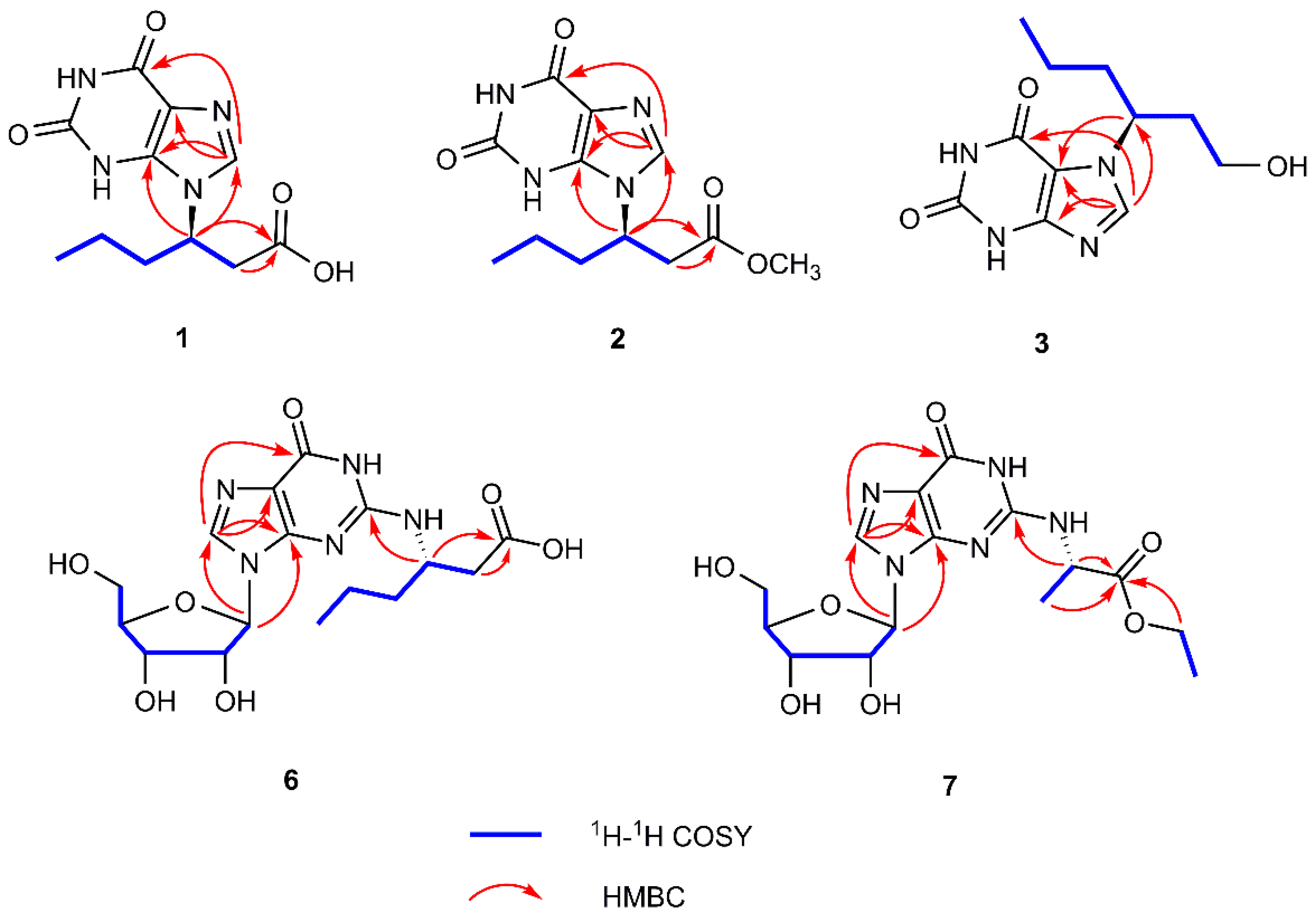

| Position | 1 a | 2 b | 3 b | |||
|---|---|---|---|---|---|---|
| ΔH | ΔC | ΔH | ΔC | ΔH | ΔC | |
| 2 | 152.8, s | 150.9, s | 150.2, s | |||
| 4 | 142.6, s | 140.6, s | 151.1, s | |||
| 5 | 115.9, s | 114.7, s | 106.2, s | |||
| 6 | 159.9, s | 157.9, s | 155.4, s | |||
| 8 | 7.99 (s) | 136.4, d | 7.89 (s) | 134.6, d | 8.03 (s) | 142.5, d |
| 1′a | 173.4, s | 170.5, s | 3.31 (m) | 57.3, t | ||
| 1′b | 3.15 (m) | |||||
| 2′a | 2.99 (dd, 17.0, 9.3) | 40.2, t | 3.05 (dd, 17.0, 9.4) | 38.4, t | 2.12 (m) | 37.2, t |
| 2′b | 2.97 (dd, 17.0, 5.4) | 3.00 (dd, 17.0, 5.2) | 1.98 (m) | |||
| 3′ | 4.70 (m) | 54.5, d | 4.71 (m) | 51.6, d | 4.63 (m) | 55.1, d |
| 4′ | 1.91 (m) | 38.0, t | 1.76 (m) | 36.3, t | Ha: 1.90 (m)Hb: 1.70 (m) | 36.2, t |
| 5′a | 1.29 (m) | 20.2, t | 1.18 (m) | 18.4, t | 1.10 (m) | 18.7, t |
| 5′b | 1.18 (m) | 1.00 (m) | 1.02 (m) | |||
| 6′ | 0.93 (t-like, 7.3) | 13.9, q | 0.81 (t-like, 7.3) | 13.4, q | 0.82 (t-like, 7.3) | 13.5, q |
| OCH3 | 3.51 (s) | 51.5, q |
| Position | 6 | 7 | ||
|---|---|---|---|---|
| ΔH | ΔC | ΔH | ΔC | |
| 2 | 154.2, s | 153.7, s | ||
| 4 | 152.2, s | 152.1, s | ||
| 5 | 115.9, s | 117.2, s | ||
| 6 | 158.3, s | 158.7, s | ||
| 8 | 8.45 (s) | 138.1, d | 8.38 (s) | 138.2, d |
| 1ʹ | 5.93 (brd, 4.4) | 90.5, d | 5.91 (brd, 4.6) | 89.8, d |
| 2ʹ | 4.64 (t-like, 5.1) | 75.6, d | 4.54 (t-like,5.0) | 75.9, d |
| 3ʹ | 4.33 (t-like, 5.1) | 71.6, d | 4.30 (t-like, 5.0) | 71.8, d |
| 4ʹ | 4.10 (m) | 86.7, d | 4.08 (m) | 86.7, d |
| 5ʹ | Ha: 3.88 (dd, 12.1, 3.2); Hb: 3.77 (dd, 12.1, 3.9) | 62.7, t | Ha: 3.85 (dd, 12.1, 3.1); Hb: 3.74 (dd, 12.1, 3.7) | 62.8, t |
| 1″ | 175.2, s | 1.51 (d, 7.2) | 18.1, q | |
| 2″ | Ha: 2.62 (dd, 15.8, 5.8); Hb: 2.59 (dd, 15.8, 6.1) | 39.8, t | 4.62 (q, 7.2) | 51.2, d |
| 3″ | 4.42 (m) | 49.0, d | 174.8, s | |
| 4″ | 1.65 (m) | 37.6, t | 4.23 (m) | 62.8, t |
| 5″ | 1.45 (m) | 20.4, t | 1.29 (t, 1.7) | 14.6, q |
| 6″ | 0.97 (t-like, 7.4) | 14.3, q |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Yan, Y.-M.; Wang, D.-W.; Cheng, Y.-X. Parvaxanthines D–F and Asponguanosines C and D, Racemic Natural Hybrids from the Insect Cyclopelta parva. Molecules 2021, 26, 3531. https://doi.org/10.3390/molecules26123531
Chen H, Yan Y-M, Wang D-W, Cheng Y-X. Parvaxanthines D–F and Asponguanosines C and D, Racemic Natural Hybrids from the Insect Cyclopelta parva. Molecules. 2021; 26(12):3531. https://doi.org/10.3390/molecules26123531
Chicago/Turabian StyleChen, Heng, Yong-Ming Yan, Dai-Wei Wang, and Yong-Xian Cheng. 2021. "Parvaxanthines D–F and Asponguanosines C and D, Racemic Natural Hybrids from the Insect Cyclopelta parva" Molecules 26, no. 12: 3531. https://doi.org/10.3390/molecules26123531







