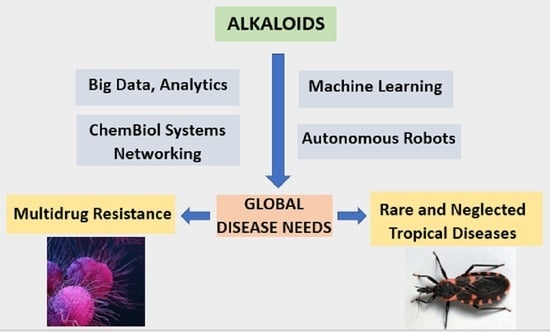
Alkaloids in Contemporary Drug Discovery to Meet Global Disease Needs
Abstract
:1. Introduction
1.1. Alkaloids, Drug Discovery, and the Fourth Industrial Revolution
1.2. Global Disease Burden and the Need for New Drugs
2. Introduction to Alkaloids
2.1. Background and Origins
2.2. The Sourcing of Alkaloids
2.3. Alkaloids in Drug Discovery for Tropical and Neglected Diseases
2.4. Antibiotic Drug Discovery
2.5. Constraints for Alkaloids in Drug Discovery
3. Discovery Strategies
3.1. Improved Collaborative Approaches
3.2. Targeted Discovery Based on In Silico Binding
3.3. Alkaloids to Overcome Drug Resistance
3.4. Transformation of Alkaloids
3.4.1. Chemical Transformations
3.4.2. Microbial Transformations
3.4.3. Biocatalysis
3.4.4. Application of Nanotechnology
3.4.5. Repurposing and the Follow-Up of Known Alkaloids
3.5. Genomics-Based Discovery
3.6. Applications of Metagenomics
4. Alkaloids and the Fourth Industrial Revolution
4.1. Industry 4.0 (4IR) and the Quintuple Helix
4.2. Artificial Intelligence in Drug Discovery
4.3. Machine Learning
5. A Way Forward
The Need for a Third Class of Medicinal Agents
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Debnath, B.; Singh, W.S.; Das, M.; Goswami, S.; Singh, M.K.; Maiti, D.; Manna, K. Role of plant alkaloids on human health: A review of biological activities. Mater. Today Chem. 2018, 9, 56–72. [Google Scholar] [CrossRef]
- Funayama, S.; Cordell, G.A. Alkaloids—A Treasury of Poisons and Medicines; Academic Press: New York, NY, USA, 2015; p. 284. [Google Scholar]
- Butler, M.S. The role of natural product chemistry in drug discovery. J. Nat. Prod. 2004, 67, 2141–2153. [Google Scholar] [CrossRef] [PubMed]
- Carter, G.T. Natural products and Pharma 2011: Strategic changes spur new opportunities. Nat. Prod. Rep. 2011, 28, 1783–1789. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Sun, H.; Zhang, A.-H.; Xu, H.-Y.; Yan, G.-L.; Han, Y.; Wang, X.-J. Natural alkaloids: Basic aspects, biological roles, and future perspectives. Chin. J. Nat. Med. 2014, 12, 401–406. [Google Scholar] [CrossRef]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef] [Green Version]
- Shen, B. A new golden age of natural products drug discovery. Cell 2015, 163, 1297–1300. [Google Scholar] [CrossRef] [Green Version]
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 35, 8–53. [Google Scholar] [CrossRef] [Green Version]
- Thomford, N.E.; Senthebane, D.A.; Rowe, A.; Munro, D.; Seele, P.; Maroyi, A.; Dzobo, K. Natural products for drug discovery in the 21st century: Innovations for novel drug discovery. Int. J. Mol. Sci. 2018, 19, 1578. [Google Scholar] [CrossRef] [Green Version]
- Bernardini, S.; Tiezzi, A.; Masci, V.L.; Ovidi, E. Natural products for human health: An historical overview of the drug discovery approaches. Nat. Prod. Res. 2018, 32, 1926–1950. [Google Scholar] [CrossRef]
- Li, F.; Wang, Y.; Li, D.; Chen, Y.; Dou, Q.P. Are we seeing a resurgence in the use of natural products for new drug discovery? Expert Opin. Drug Discov. 2019, 14, 417–420. [Google Scholar] [CrossRef]
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar] [CrossRef]
- Xu, X. New concepts and approaches for drug discovery based on traditional Chinese medicine. Drug Discov. Today Technol. 2006, 3, 247–253. [Google Scholar] [CrossRef]
- Singh, R.K.; Tiwari, S.P.; Rai, A.K.; Mohapatra, T.M. Cyanobacteria: An emerging source for drug discovery. J. Antibiot. 2011, 64, 401–412. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.-Y.; Hou, J.-J.; Long, H.-L.; Yang, W.-Z.; Liang, J.; Guo, D.-a. TCM-based new drug discovery and development in China. Chin. J. Nat. Med. 2014, 12, 241–250. [Google Scholar] [CrossRef]
- Schwab, K. The Fourth Industrial Revolution; World Economic Forum: Geneva, Switzerland, 2016; p. 192. [Google Scholar]
- Kim, S.Y.; The Fourth Industrial Revolution and the Triple Helix. Daegu: Triple Helix Association Triple Helix International Conference. 2017. Available online: https://www.triplehelixassociation.org/wp-content/uploads/2017/07/Theme-paper-THC2017.pdf (accessed on 18 March 2021).
- Carayannis, G.; Campbell, D.F.J. Triple helix, Quadruple Helix and Quintuple Helix and how do knowledge, innovation and the environment relate to each other? A proposed framework for a trans-disciplinary analysis of sustainable development and social ecology. Int. J. Soc. Ecol. Sustain. Dev. 2010, 1, 41–69. [Google Scholar] [CrossRef]
- Barth, T.D. The idea of a green new deal in a Quintuple Helix model of knowledge, know-how and innovation. Int. J. Soc. Ecol. Sustain. Dev. 2013, 1, 1–15. [Google Scholar] [CrossRef]
- Daley, S.-K.; Cordell, G.A. Natural Products, the Fourth Industrial Revolution, and the Quintuple Helix. Nat. Prod. Commun. 2021, 16, 31. [Google Scholar] [CrossRef]
- Cordell, G.A.; Colvard, M.D. Natural products and traditional medicine: Turning on a paradigm. J. Nat. Prod. 2012, 75, 514–525. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Lopez, A.D. (Eds.) The Global Burden of Disease. A Comprehensive Assessment of Mortality and Disability from Diseases, Injuries and Risk Factors in 1990 and Projected to 2020; GBD Series; Harvard School of Public Health, World Health Organization, World Bank: Cambridge, MA, USA, 1996; Volume I. [Google Scholar]
- Editorial. Global health: Time for radical change? Lancet 2020, 396, 1129. [Google Scholar] [CrossRef]
- Vos, T.; Lim, S.S.; Abbafati, C.; Abbas, K.M.; Abbasi, M.; Abbasifard, M.; Abbasi-Kangevari, M.; Abbastabar, H.; Abd-Allah, F.; Abdelalim, A.; et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef]
- Karimkhani, C.; Wanga, V.; Coffeng, L.E.; Naghavi, P.; Dellavalle, R.P.; Naghavi, M. Global burden of cutaneous leishmaniasis: A cross-sectional analysis from the Global Burden of Disease Study. Lancet Infect. Dis. 2016, 16, 584–591. [Google Scholar] [CrossRef]
- Stanaway, J.D.; Shepard, D.S.; Undurraga, E.A.; Halasa, Y.A.; Coffeng, L.E.; Brady, O.; Hay, S.; Bedi, N.; Bensenor, I.M.; Castañeda-Orjuela, C.; et al. The global burden of dengue: An analysis from the Global Burden of Disease Study. Lancet Infect. Dis. 2016, 16, 712–723. [Google Scholar] [CrossRef] [Green Version]
- Herricks, J.R.; Hotez, P.J.; Wanga, V.; Coffeng, L.E.; Haagsma, J.A.; Basáñez, M.-G.; Buckle, G.; Budke, C.M.; Carabin, H.; Fèvre, E.M.; et al. The global burden of disease study 2013: What does it mean for the NTDs? PLoS Neglected Trop. Dis. 2017, 11, e0005424. [Google Scholar] [CrossRef] [Green Version]
- Kyu, H.H.; Maddison, E.R.; Henry, N.J.; Mumford, J.E.; Barber, R.; Shields, C.; Brown, J.C.; Nguyen, G.; Carter, A.; Wolock, T.M.; et al. The global burden of tuberculosis: Results from the Global Burden of Disease Study. Lancet Infect. Dis. 2018, 18, 261–284. [Google Scholar] [CrossRef] [Green Version]
- Pisarski, K. The global burden of disease of zoonotic parasitic diseases: Top 5 contenders for priority consideration. Trop. Med. Infect. Dis. 2019, 4, 44. [Google Scholar] [CrossRef] [Green Version]
- Devereaux, A.L.; Mercer, S.L.; Cunningham, C.W. DARK classics in chemical neuroscience: Morphine. ACS Chem. Neurosci. 2018, 9, 2395–2407. [Google Scholar] [CrossRef]
- Kaufman, T.S.; Rúveda, E.A. The quest for quinine: Those who won the battles and those who won the war. Angew. Chem. Int. Ed. 2005, 44, 854–885. [Google Scholar] [CrossRef]
- Cushnie, T.T.; Cushnie, B.; Lamb, A.J. Alkaloids: An overview of their antibacterial, antibiotic-enhancing and antivirulence activities. Int. J. Antimicrob. Agents 2014, 44, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Cordell, G.A.; Quinn-Beattie, M.L.; Farnsworth, N.R. The potential of alkaloids in drug discovery. Phytother. Res. 2001, 15, 183–205. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Gavia, D.J.; Tang, Y. Biosynthesis of fungal indole alkaloids. Nat. Prod. Rep. 2014, 31, 1474–1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Q.; Mustafa, N.R.; Tang, K.; Choi, Y.H.; Verpoorte, R. Monoterpenoid indole alkaloids biosynthesis and its regulation in Catharanthus roseus: A literature review from genes to metabolites. Phytochem. Rev. 2016, 15, 221–250. [Google Scholar] [CrossRef]
- Thamm, A.M.K.; Qu, Y.; De Luca, V. Discovery and metabolic engineering of iridoid/secoiridoid and monoterpenoid indole alkaloid biosynthesis. Phytochem. Rev. 2016, 15, 339–361. [Google Scholar] [CrossRef]
- Singh, A.; Menéndez-Perdomo, I.M.; Facchini, P.J. Benzylisoquinoline alkaloid biosynthesis in opium poppy: An update. Phytochem. Rev. 2019, 18, 1457–1482. [Google Scholar] [CrossRef]
- Roddan, R.; Ward, J.M.; Keep, N.H.; Hailes, H.C. Pictet-Spenglerases in alkaloid biosynthesis: Future applications in bio-catalysis. Curr. Opin. Chem. Biol. 2020, 55, 69–76. [Google Scholar] [CrossRef]
- Desgagné-Penix, I. Biosynthesis of alkaloids in Amaryllidaceae plants: A review. Phytochem. Rev. 2021, 20, 409–431. [Google Scholar] [CrossRef]
- Lichman, B.R. The scaffold-forming steps of plant alkaloid biosynthesis. Nat. Prod. Rep. 2021, 38, 103–129. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Lipinski, C. Rule of five in 2015 and beyond: Target and ligand structural limitations, ligand chemistry structure and drug discovery project decisions. Adv. Drug Deliv. Rev. 2016, 101, 34–41. [Google Scholar] [CrossRef]
- Martens, E.; Demain, A.L. The antibiotic resistance crisis, with a focus on the United States. J. Antibiot. 2017, 70, 520–526. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.; Handelsman, J. Status of the microbial census. Microbiol. Mol. Biol. Rev. 2004, 68, 686–691. [Google Scholar] [CrossRef] [Green Version]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef]
- Hawksworth, J. Mushrooms: The extent of the unexplored potential. Int. J. Med. Mushrooms 2001, 3, 333–337. [Google Scholar] [CrossRef]
- Hugenholtz, P.; Hooper, S.D.; Kyrpides, N.C. Focus: Synergistetes. Environ. Microbiol. 2009, 11, 1327–1329. [Google Scholar] [CrossRef]
- Vasas, G.; Borbely, G.; Nánási, P.; Nanasi, P. Alkaloids from cyanobacteria with diverse powerful bioactivities. Mini Rev. Med. Chem. 2010, 10, 946–955. [Google Scholar] [CrossRef]
- Shah, S.A.A.; Akhter, N.; Auckloo, B.N.; Khan, I.; Lu, Y.; Wang, K.; Wu, B.; Guo, Y.W. Structural diversity, biological properties and applications of natural products from cyanobacteria. A review. Mar. Drugs 2017, 15, 354. [Google Scholar] [CrossRef] [Green Version]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.-M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [Green Version]
- Gouda, S.; Das, G.; Sen, S.K.; Shin, H.S.; Patra, J.K. Endophytes: A treasure house of bioactive compounds of medicinal im-portance. Front. Microbiol. 2016, 7, 1538. [Google Scholar] [CrossRef] [Green Version]
- Daley, S.-K.; Cordell, G.A. Biologically significant and recently isolated alkaloids from endophytic fungi. J. Nat. Prod. 2021, 84, 871–897. [Google Scholar] [CrossRef]
- World Health Organization. Investing to Overcome the Global Impact of Neglected Tropical Diseases; Third WHO Report on Neglected Tropical Diseases; World Health Organization: Geneva, Switzerland, 2015; p. 211. [Google Scholar]
- United Nations Sustainable Development Goals. Available online: https://www.un.org/sustainabledevelopment/sustainable-development-goals/ (accessed on 5 April 2021).
- Access to Medicine Foundation. Access to Medicine Index; Access to Medicine Foundation: Amsterdam, The Netherlands, 2021; p. 237. [Google Scholar]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 80, 770–803. [Google Scholar] [CrossRef]
- WHO. The Twenty Conditions Identified by WHO as NTDs Are: Buruli Ulcer, Chagas Disease, Dengue and Chikungunya, Dracunculiasis, Echinococcosis, Foodborne Trematodiases, Human African Trypanosomiasis, Leishmaniasis, Leprosy, Lymphatic Filariasis, Myce-Toma, Chromoblastomycosis and Other Deep Mycoses, Onchocerciasis, Rabies, Scabies and Other Ectoparasitoses, Schistosomiasis, Soil-Transmitted Helminthiases, Snakebite Envenoming, Taeniasis and Cysticercosis, Trachoma and Yaws; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- World Health Organization. Ending the Neglect to Attain Sustainable Development Goals. A Road Map for Neglected Tropical Diseases 2021–2030; WHO: Geneva, Switzerland, 2020; p. 55. [Google Scholar]
- Capela, R.; Moreira, R.; Lopes, F. An overview of drug resistance in protozoal diseases. Int. J. Mol. Sci. 2019, 20, 5748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keiser, J. Antimalarials in the treatment of schistosomiasis. Curr. Pharm. Des. 2012, 18, 3531–3538. [Google Scholar] [CrossRef] [PubMed]
- Utzinger, J.; Raso, G.; Brooker, S.; De Savigny, D.; Tanner, M.; Ørnbjerg, N.; Singer, B.H.; N’Goran, E.K. Schistosomiasis and neglected tropical diseases: Towards integrated and sustainable control and a word of caution. Parasitology 2009, 136, 1859–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schafer, T.W.; Hale, B.R. Gastrointestinal complications of schistosomiasis. Curr. Gastroenterol. Rep. 2001, 3, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Cioli, D.; Pica-Mattoccia, L.; Archer, S. Antischistosomal drugs: Past, present … and future? Pharmacol. Ther. 1995, 68, 35–85. [Google Scholar] [CrossRef]
- Utzinger, J.; Shuhua, X.; N’Goran, E.K.; Bergquist, R.; Tanner, M. The potential of artemether for the control of schistosomiasis. Int. J. Parasitol. 2001, 31, 1549–1562. [Google Scholar] [CrossRef]
- Coles, G.; Bruce, J.; Kinoti, G.; Mutahi, W.; Dias, L.; Rocha, R.; Katz, N. The potential for drug resistance in schistosomiasis. Parasitol. Today 1987, 3, 349–350. [Google Scholar] [CrossRef]
- Danso-Appiah, A.; De Vlas, S.J. Interpreting low praziquantel cure rates of Schistosoma mansoni infections in Senegal. Trends Parasitol. 2002, 18, 125–129. [Google Scholar] [CrossRef]
- Mostafa, O.M.S.; Eid, R.A.; Adly, M.A. Antischistosomal activity of ginger (Zingiber officinale) against Schistosoma mansoni harbored in C57 mice. Parasitol. Res. 2011, 109, 395–403. [Google Scholar] [CrossRef]
- Guimarães, M.A.; De Oliveira, R.N.; Véras, L.M.C.; Lima, D.F.; Campelo, Y.D.M.; Campos, S.A.; Kuckelhaus, S.A.S.; Pinto, P.L.S.; Eaton, P.; Mafud, A.C.; et al. Anthelmintic activity in vivo of epiisopiloturine against juvenile and adult worms of Schistosoma mansoni. PLoS Negl. Trop. Dis. 2015, 9, e0003656. [Google Scholar] [CrossRef]
- Chatterjee, A.; Dutta, C.P. Alkaloids of Piper longum Linn. I. Structure and synthesis of piperlongumine and piperlonguminine. Tetrahedron 1967, 23, 1769–1781. [Google Scholar] [CrossRef]
- Bezerra, D.P.; Pessoa, C.; de Moraes, M.O.; Saker-Neto, N.; Silveira, E.R.; Costa-Lotufo, L.V. Overview of the therapeutic potential of piplartine (piperlongumine). Eur. J. Pharm. Sci. 2013, 48, 453–463. [Google Scholar] [CrossRef]
- De Moraes, J.; Nascimento, C.; Lopes, P.O.M.V.; Nakano, E.; Yamaguchi, L.F.; Kato, M.; Kawano, T. Schistosoma mansoni: In vitro schistosomicidal activity of piplartine. Exp. Parasitol. 2011, 127, 357–364. [Google Scholar] [CrossRef]
- Bezerra, D.P.; Militão, G.C.G.; de Castro, F.O.; Pessoa, C.; de Moraes, M.O.; Silveira, E.R.; Lima, M.A.S.; Elmiro, F.J.M.; Costa-Lotufo, L.V. Piplartine induces inhibition of leukemia cell proliferation triggering both apoptosis and necrosis pathways. Toxicol. Vitr. 2007, 21, 1–8. [Google Scholar] [CrossRef]
- De Moraes, J.; Keiser, J.; Ingram, K.; Nascimento, C.; Yamaguchi, L.F.; Bittencourt, C.R.; Bemquerer, M.P.; Leite, J.R.; Kato, M.J.; Nakano, E. In vitro synergistic interaction between amide piplartine and antimicrobial peptide dermaseptin against Schistosoma mansoni schistosomula and adult worms. Curr. Med. Chem. 2013, 20, 301–309. [Google Scholar] [CrossRef]
- Bezerra, D.P.; Moura, D.J.; Rosa, R.M.; de Vasconcellos, M.C.; Silva, A.C.R.; de Moraes, M.O.; Silveira, E.R.; Lima, M.A.; Henriques, J.A.; Costa-Lotufo, L.V.; et al. Evaluation of the genotoxicity of piplartine, an alkamide of Piper tuberculatum, in yeast and mammalian V79 cells. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2008, 652, 164–174. [Google Scholar] [CrossRef]
- Schaab, E.H.; Crotti, A.E.M.; Iamamoto, Y.; Kato, M.; Lotufo, L.V.C.; Lopes, N.P. Biomimetic oxidation of piperine and piplartine catalyzed by iron(III) and manganese(III) porphyrins. Biol. Pharm. Bull. 2010, 33, 912–916. [Google Scholar] [CrossRef] [Green Version]
- Raj, L.; Ide, T.; Gurkar, A.U.; Foley, M.A.; Schenone, M.; Li, X.; Tolliday, N.J.; Golub, T.R.; Carr, S.A.; Shamji, A.F.; et al. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nat. Cell Biol. 2011, 475, 231–234. [Google Scholar] [CrossRef]
- Prasad, K.B.; Reddy, G.A.K.; Joy, J.M.; Rasheed, A.; Dalith, D. Natural antifilarial drugs: A review. Int. J. Pharmacol. Toxicol. 2011, 1, 1–10. [Google Scholar]
- Bulman, C.A.; Bidlow, C.M.; Lustigman, S.; Cho-Ngwa, F.; Williams, D.; Rascón, J.A.A.; Tricoche, N.; Samje, M.; Bell, A.; Suzuki, B.; et al. Repurposing auranofin as a lead candidate for treatment of lymphatic filariasis and onchocerciasis. PLoS Negl. Trop. Dis. 2015, 9, e0003534. [Google Scholar] [CrossRef]
- Wei, G.; Wei, D.; Du, Y. An alternative total synthesis of solamargine. Sci. China Ser. B Chem. 2012, 55, 1247–1251. [Google Scholar] [CrossRef] [Green Version]
- Murray, H.W.; Berman, J.D.; Davies, C.R.; Saravia, N.G. Advances in leishmaniasis. Lancet 2005, 366, 1561–1577. [Google Scholar] [CrossRef]
- Amato, V.S.; Tuon, F.F.; Bacha, H.A.; Neto, V.A.; Nicodemo, A.C. Mucosal leishmaniasis. Acta Trop. 2008, 105, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Pandey, H.P.; Sundar, S. Visceral leishmaniasis (kala-azar): Challenges ahead. Indian J. Med Res. 2006, 123, 331–344. [Google Scholar] [PubMed]
- Cheuka, P.M.; Mayoka, G.; Mutai, P.; Chibale, K. The role of natural products in drug discovery and development against neglected tropical diseases. Molecules 2016, 22, 58. [Google Scholar] [CrossRef] [Green Version]
- Hughes, D.; Andersson, D.I. Evolutionary consequences of drug resistance: Shared principles across diverse targets and or-ganisms. Nat. Rev. Genet. 2015, 16, 459–471. [Google Scholar] [CrossRef]
- Dadgostar, P. Antimicrobial resistance: Implications and costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [Green Version]
- Nathan, C.; Cars, O. Antibiotic resistance—problems, progress, and prospects. N. Engl. J. Med. 2014, 371, 1761–1763. [Google Scholar] [CrossRef]
- Wohlleben, W.; Mast, Y.; Stegmann, E.; Ziemert, N. Antibiotic drug discovery. Microb. Biotechnol. 2016, 9, 541–548. [Google Scholar] [CrossRef] [Green Version]
- Brown, E.D.; Wright, G.D. Antibacterial drug discovery in the resistance era. Nature 2016, 529, 336–343. [Google Scholar] [CrossRef]
- Wright, G.D. Opportunities for natural products in 21st century antibiotic discovery. Nat. Prod. Rep. 2017, 34, 694–701. [Google Scholar] [CrossRef]
- Bhattarai, K.; Bastola, R.; Baral, B. Antibiotic drug discovery: Challenges and perspectives in the light of emerging antibiotic resistance. Adv. Genet. 2020, 105, 229–292. [Google Scholar] [CrossRef]
- Kraus, C.N. Low hanging fruit in infectious disease drug development. Curr. Opin. Microbiol. 2008, 11, 434–438. [Google Scholar] [CrossRef]
- Butler, M.S.; Buss, A.D. Natural products—The future scaffolds for novel antibiotics? Biochem. Pharmacol. 2006, 71, 919–929. [Google Scholar] [CrossRef]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States, Atlanta, GA, CDC. 2019. Available online: https://www.cdc.gov/drugresistance/index.html (accessed on 5 April 2021).
- Butaye, P.; Devriese, L.A.; Haesebrouck, F. Antimicrobial growth promoters used in animal feed: Effects of less well-known antibiotics on Gram-positive bacteria. Clin. Microbiol. Rev. 2003, 16, 175–188. [Google Scholar] [CrossRef] [Green Version]
- Casewell, M.; Friis, C.; Marco, E.; McMullin, P.; Phillips, I. The European ban on growth-promoting antibiotics and emerging consequences for human and animal health. J. Antimicrob. Chemother. 2003, 52, 159–161. [Google Scholar] [CrossRef] [Green Version]
- Drawz, S.M.; Papp-Wallace, K.M.; Bonomo, R.A. New β-lactamase inhibitors: A therapeutic renaissance in an MDR world. Antimicrob. Agents Chemother. 2014, 58, 1835–1846. [Google Scholar] [CrossRef] [Green Version]
- Bueno, J. Antimicrobial adjuvants drug discovery, the challenge of avoid the resistance and recover the susceptibility of mul-tidrug-resistant strains. J. Microb. Biochem. Technol. 2016, 8, 169–176. [Google Scholar] [CrossRef]
- Viveiros, M.; Amaral, L. Enhancement of antibiotic activity against polydrug resistant Mycobacterium tuberculosis by pheno-thiazines. Int. J. Antimicrob. Agents 2001, 17, 225–228. [Google Scholar] [CrossRef]
- Dastidar, S.G.; Kristiansen, J.E.; Molnar, J.; Amaral, L. Role of phenothiazines and structurally similar compounds of plant origin in the fight against infections by drug resistant bacteria. Antibiotics 2013, 2, 58–72. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.; Feng, Y.; Tsao, S.; Wang, N.; Curtain, R.; Wang, Y. Berberine and Coptidis Rhizoma as novel antineoplastic agents: A review of traditional use and biomedical investigations. J. Ethnopharmacol. 2009, 126, 5–17. [Google Scholar] [CrossRef] [Green Version]
- Tillhon, M.; Ortiz, L.M.G.; Lombardi, P.; Scovassi, A.I. Berberine: New perspectives for old remedies. Biochem. Pharmacol. 2012, 84, 1260–1267. [Google Scholar] [CrossRef]
- Imenshahidi, M.; Hosseinzadeh, H. Berberis vulgaris and berberine: An update review. Phytother. Res. 2016, 30, 1745–1764. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, F.; Song, Y.; Liu, H. The status of and trends in the pharmacology of berberine: A bibliometric review [1985–2018]. Chin. Med. 2020, 15, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Sun, T.; Li, X.-D.; Hong, J.; Liu, C.; Zhang, X.-L.; Zheng, J.-P.; Xu, Y.-J.; Ou, Z.-Y.; Zheng, J.-L.; Yu, D.-J. Inhibitory effect of two traditional Chinese medicine monomers, berberine and matrine, on the quorum sensing system of antimicrobial-resistant Escherichia coli. Front. Microbiol. 2019, 10, 2584. [Google Scholar] [CrossRef]
- Yu, H.-H.; Kim, K.-J.; Cha, J.-D.; Kim, H.-K.; Lee, Y.-E.; Choi, N.-Y.; You, Y.O. Antimicrobial activity of berberine alone and in combination with ampicillin or oxacillin against methicillin-resistant Staphylococcus aureus. J. Med. Food 2005, 8, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Habtemariam, S. Berberine and inflammatory bowel disease: A concise review. Pharmacol. Res. 2016, 113, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Jin, X.; Liang, C.; Bu, F.; Pan, D.; He, Q.; Ming, Y.; Little, P.; Du, H.; Liang, S.; et al. Berberine for diarrhea in children and adults: A systematic review and meta-analysis. Ther. Adv. Gastroenterol. 2020, 13, 1756284820961299. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Yang, L.X.; Jamshaid, F. New development of novel berberine derivatives against bacteria. Mini Rev. Med. Chem. 2020, 20, 716–724. [Google Scholar] [CrossRef]
- Kim, J.H.; Ryu, Y.B.; Lee, W.S.; Kim, Y.H. Neuraminidase inhibitory activities of quaternary isoquinoline alkaloids from Co-rydalis turtschaninovii rhizome. Bioorg. Med. Chem. 2014, 22, 604–6052. [Google Scholar] [CrossRef] [Green Version]
- Cascioferro, S.; Totsika, M.; Schillaci, D. Sortase A: An ideal target for anti-virulence drug development. Microb. Pathog. 2014, 77, 105–112. [Google Scholar] [CrossRef] [Green Version]
- Guan, G.; Wang, H. The anticancer effect of sanguinarine: A review. Curr. Pharm. Des. 2018, 24, 2760–2764. [Google Scholar] [CrossRef]
- Godowski, K.C. Antimicrobial action of sanguinarine. J. Clin. Dent. 1989, 1, 96–101. [Google Scholar]
- Wennström, J.; Lindhe, J. Some effects of a sanguinarine-containing mouth rinse on developing plaque and gingivitis. J. Clin. Periodont. 1985, 12, 867–872. [Google Scholar] [CrossRef]
- Zhong, H.; Hu, D.-D.; Hu, G.-H.; Su, J.; Bi, S.; Zhang, Z.-E.; Wang, Z.; Zhang, R.-L.; Xu, Z.; Jiang, Y.-Y.; et al. Activity of sanguinarine against Candida albicans biofilms. Antimicrob. Agents Chemother. 2017, 61, e02259–e02316. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.-Y.; Shen, J.-Y.; Li, X.-L.; Xu, Y.; Hao, G.-J.; Pan, X.-Y.; Wang, G.-X.; Yin, W.-L. Effect of sanguinarine from the leaves of Macleaya cordata against Ichthyophthirius multifiliis in grass carp (Ctenopharyngodon idella). Parasitol. Res. 2010, 107, 1035–1042. [Google Scholar] [CrossRef]
- Yang, X.-J.; Miao, F.; Yao, Y.; Cao, F.-J.; Yang, R.; Ma, Y.-N.; Qin, B.-F.; Zhou, L. In vitro antifungal activity of sanguinarine and chelerythrine derivatives against phytopathogenic fungi. Molecules 2012, 17, 13026–13035. [Google Scholar] [CrossRef]
- Zhang, S.-M.; Coultas, K.A. Identification of plumbagin and sanguinarine as effective chemotherapeutic agents for treatment of schistosomiasis. Int. J. Parasitol. Drugs Drug Resist. 2013, 3, 28–34. [Google Scholar] [CrossRef] [Green Version]
- Meng, L.-H.; Li, X.-M.; Liu, Y.; Wang, B.-G. Polyoxygenated dihydropyrano[2,3-c]pyrrole-4,5-dione derivatives from the marine mangrove-derived endophytic fungus Penicillium brocae MA-231 and their antimicrobial activity. Chin. Chem. Lett. 2015, 26, 610–612. [Google Scholar] [CrossRef]
- Bontemps, N.; Bry, D.; López-Legentil, S.; Simon-Levert, A.; Long, C.; Banaigs, B. Structures and antimicrobial activities of pyridoacridine alkaloids isolated from different chromotypes of the ascidian Cystodytes dellechiajei. J. Nat. Prod. 2010, 73, 1044–1048. [Google Scholar] [CrossRef]
- Won, T.H.; Jeon, J.-E.; Lee, S.-H.; Rho, B.J.; Oh, K.-B.; Shin, J. Beta-carboline alkaloids derived from the ascidian Synoicum sp. Bioorg. Med. Chem. 2012, 20, 4082–4087. [Google Scholar] [CrossRef]
- Moore, K.S.; Wehrli, S.; Roder, H.; Rogers, M.; Forrest, J.N.; McCrimmon, D.; Zasloff, M. Squalamine: An aminosterol antibiotic from the shark. Proc. Natl. Acad. Sci. USA 1993, 90, 1354–1358. [Google Scholar] [CrossRef] [Green Version]
- Alhanout, K.; Malesinki, S.; Vidal, N.; Peyrot, V.; Rolain, J.M.; Brunel, J.M. New insights into the antibacterial mechanism of action of squalamine. J. Antimicrob. Chemother. 2010, 65, 1688–1693. [Google Scholar] [CrossRef]
- Alves, R.R.N.; Rosa, I.M.L. Biodiversity, traditional medicine and public health: Where do they meet? J. Ethnobiol. Ethnomed. 2007, 3, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Alkemade, R.; Reid, R.S.; van den Berg, M.; de Leeuw, J.; Jeuken, M. Assessing the impacts of livestock production on biodi-versity in rangeland ecosystems. Proc. Natl. Acad. Sci. USA 2013, 110, 20900–20905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Cuervo, A.M.; de Lima, L.S.; Dallmeier, F.; Garate, P.; Bravo, A.; Vanthomme, H. Twenty years of land cover change in the southeastern Peruvian Amazon: Implications for biodiversity conservation. Reg. Environ. Chang. 2020, 20, 1–14. [Google Scholar] [CrossRef]
- Cordell, G.A. Biodiversity and drug discovery—A symbiotic relationship. Phytochemistry 2000, 55, 463–480. [Google Scholar] [CrossRef]
- Cordell, G.A. Ecopharmacognosy and the responsibilities of natural product research to sustainability. Phytochem. Lett. 2015, 11, 332–346. [Google Scholar] [CrossRef]
- Cordell, G.A. Cognate and cognitive ecopharmacognosy—In an anthropogenic era. Phytochem. Lett. 2017, 20, 540–549. [Google Scholar] [CrossRef]
- Winnikoff, J.R.; Glukhov, E.; Watrous, J.; Dorrestein, P.C.; Gerwick, W.H. Quantitative molecular networking to profile marine cyanobacterial metabolomes. J. Antibiot. 2014, 67, 105–112. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.-Y.; Ke, J.-P.; Wang, W.; Kong, Y.-S.; Zhang, P.; Ling, T.-J.; Bao, G.-H. Discovery of neolignan glycosides with acetylcholin-esterase inhibitory activity from Huangjinya green tea guided by ultra-performance liquid chromatography-tandem mass spectrometry data and Global Natural Product Social molecular networking. J. Agric. Food Chem. 2019, 67, 11986–11993. [Google Scholar] [CrossRef]
- Corley, D.G.; Durley, R.C. Strategies for database dereplication of natural products. J. Nat. Prod. 1994, 57, 1484–1490. [Google Scholar] [CrossRef]
- Hubert, J.; Nuzillard, J.-M.; Renault, J.-H. Dereplication strategies in natural product research: How many tools and method-ologies behind the same concept? Phytochem. Rev. 2017, 16, 55–95. [Google Scholar] [CrossRef]
- Wolfender, J.-L.; Marti, G.; Queiroz, E.F. Advances in techniques for profiling crude extracts and for the rapid identification of natural products: Dereplication, quality control and metabolomics. Curr. Org. Chem. 2010, 14, 1808–1832. [Google Scholar] [CrossRef]
- Yuliana, N.D.; Jahangir, M.; Verpoorte, R.; Choi, Y.H. Metabolomics for the rapid dereplication of bioactive compounds from natural sources. Phytochem. Rev. 2013, 12, 293–304. [Google Scholar] [CrossRef]
- Pérez-Victoria, I.; Martín, J.; Reyes, F. Combined LC/UV/MS and NMR strategies for the dereplication of marine natural products. Planta Med. 2016, 82, 857–871. [Google Scholar] [CrossRef] [Green Version]
- Allard, P.-M.; Péresse, T.; Bisson, J.; Gindro, K.; Marcourt, L.; Pham, V.C.; Roussi, F.; Litaudon, M.; Wolfender, J.-L. Integration of molecular networking and in-silico MS/MS fragmentation for natural products dereplication. Anal. Chem. 2016, 88, 3317–3323. [Google Scholar] [CrossRef]
- Cordell, G.A.; Beecher, C.W.W.; Kinghorn, A.D.; Pezzuto, J.M.; Constant, H.L.; Fang, L.; Seo, E.-K.; Long, L.; Cui, B.-L.; Barrilos, K.S. The dereplication of natural products. In Studies in Natural Products Chemistry; Atta-ur-Rahman, Ed.; Elsevier Science Publishers: Amsterdam, The Netherlands, 1997; Volume 19, pp. 749–791. [Google Scholar]
- Cordell, G.A.; Shin, Y.G. Finding the needle in the haystack. The dereplication of natural product extracts. Pure Appl. Chem. 1999, 71, 1089–1094. [Google Scholar] [CrossRef] [Green Version]
- Baell, J.B. Feeling nature’s PAINS: Natural products, natural product drugs, and pan assay interference compounds (PAINS). J. Nat. Prod. 2016, 79, 616–628. [Google Scholar] [CrossRef]
- Baell, J.B.; Holloway, G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bisson, J.; McAlpine, J.B.; Friesen, J.B.; Chen, S.-N.; Graham, J.; Pauli, G.F. Can invalid bioactives undermine natural product-based drug discovery? J. Med. Chem. 2016, 59, 1671–1690. [Google Scholar] [CrossRef] [PubMed]
- Powell, R.G.; Smith, C.R., Jr.; Weisleder, D.; Muthard, D.A.; Clardy, J. Sesbanine, a novel cytotoxic alkaloid from Sesbania drummondii. J. Am. Chem. Soc. 1979, 101, 2784–2785. [Google Scholar] [CrossRef]
- Powell, R.G.; Smith, C.R.; Weisleder, D.; Matsumoto, G.; Clardy, J.; Kozlowski, J. Sesbanimide, a potent antitumor substance from Sesbania drummondii seed. J. Am. Chem. Soc. 1983, 105, 3739–3741. [Google Scholar] [CrossRef]
- Choules, M.P.; Klein, L.L.; Lankin, D.C.; McAlpine, J.B.; Cho, S.-H.; Cheng, J.; Lee, H.; Suh, J.-W.; Jaki, B.U.; Franzblau, S.G.; et al. Residual complexity does impact organic chemistry and drug discovery: The case of rufomyazine and rufomycin. J. Org. Chem. 2018, 83, 6664–6672. [Google Scholar] [CrossRef]
- Jaki, B.U.; Franzblau, S.; Chadwick, L.R.; Lankin, D.C.; Zhang, F.; Wang, Y.; Pauli, G. Purity−activity relationships of natural products: The case of anti-TB active ursolic acid. J. Nat. Prod. 2008, 71, 1742–1748. [Google Scholar] [CrossRef]
- Nwaka, S.; Ridley, R.G. Virtual drug discovery and development for neglected diseases through public–private partnerships. Nat. Rev. Drug Discov. 2003, 2, 919–928. [Google Scholar] [CrossRef]
- Ioset, J.-R.; Chatelain, E. Drug discovery and development for neglected diseases: The DNDi model. Drug Des. Dev. Ther. 2011, 5, 175–181. [Google Scholar] [CrossRef] [Green Version]
- Johnston, K.L.; Ford, L.; Taylor, M.J. Overcoming the challenges of drug discovery for neglected tropical diseases: The A·WOL experience. J. Biomol. Screen. 2014, 19, 335–343. [Google Scholar] [CrossRef] [Green Version]
- Sunyoto, T. Partnerships for better neglected disease drug discovery and development: How have we fared? Expert Opin. Drug Discov. 2020, 15, 531–537. [Google Scholar] [CrossRef]
- Simpkin, V.L.; Renwick, M.J.; Kelly, R.; Mossialos, E. Incentivising innovation in antibiotic drug discovery and development: Progress, challenges and next steps. J. Antibiot. 2017, 70, 1087–1096. [Google Scholar] [CrossRef] [Green Version]
- Reed, J.C.; White, E.L.; Aube, J.; Lindsley, C.; Li, M.; Sklar, L.; Schreiber, S. The NIH’s role in accelerating translational sciences. Nat. Biotechnol. 2012, 30, 16–19. [Google Scholar] [CrossRef]
- Jarvis, L.M. NIH initiative aims to partner academics with pharmaceutical companies to revive failed drug candidates. Chem. Eng. News 2012, 90, 41–43. [Google Scholar]
- Roberts, J.P. Incentives aim to boost antibiotic development. Nat. Biotechnol. 2012, 30, 735. [Google Scholar] [CrossRef]
- Tralau-Stewart, C.J.; Wyatt, C.A.; Kleyn, D.E.; Ayad, A. Drug discovery: New models for industry–academic partnerships. Drug Discov. Today 2009, 14, 95–101. [Google Scholar] [CrossRef]
- Cragg, G.M.; Newman, A.D.J.; Snader, K.M. Natural products in drug discovery and development. J. Nat. Prod. 1997, 60, 52–60. [Google Scholar] [CrossRef]
- Light, D.W. Addressing health care disparities: A radical perspective and proposal. Front. Sociol. 2020, 5, 29. [Google Scholar] [CrossRef]
- Zicker, F.; Faid, M.; Reeder, J.; Aslanyan, G. Building coherence and synergy among global health initiatives. Health Res. Policy Syst. 2015, 13, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Young, R.; Bekele, T.; Gunn, A.; Chapman, N.; Chowdhary, V.; Corrigan, K.; Dahora, L.; Martinez, S.; Permar, S.; Persson, J.; et al. Developing new health technologies for neglected diseases: A pipeline portfolio review and cost model. Gates Open Res. 2018, 2, 23. [Google Scholar] [CrossRef]
- Lagunin, A.A.; Goel, R.; Gawande, D.; Pahwa, P.; Gloriozova, T.A.; Dmitriev, A.; Ivanov, S.; Rudik, A.V.; Konova, V.I.; Pogodin, P.V.; et al. Chemo- and bioinformatics resources for in silico drug discovery from medicinal plants beyond their traditional use: A critical review. Nat. Prod. Rep. 2014, 31, 1585–1611. [Google Scholar] [CrossRef]
- Olğaç, A.; Orhan, I.E.; Banoglu, E. The potential role of in silico approaches to identify novel bioactive molecules from natural resources. Future Med. Chem. 2017, 9, 1665–1686. [Google Scholar] [CrossRef]
- Park, K. A review of computational drug repurposing. Transl. Clin. Pharmacol. 2019, 27, 59–63. [Google Scholar] [CrossRef] [Green Version]
- Scotti, L.; Ishiki, H.; Mendonça, F.; Silva, M.; Scotti, M. In-silico analyses of natural products on leishmania enzyme targets. Mini Rev. Med. Chem. 2015, 15, 253–269. [Google Scholar] [CrossRef]
- Argüelles, A.J.; Cordell, G.A.; Maruenda, H. Molecular docking and binding mode analysis of plant alkaloids as in vitro and in silico inhibitors of trypanothione reductase from Trypanosoma cruzi. Nat. Prod. Commun. 2016, 11, 57–62. [Google Scholar] [CrossRef] [Green Version]
- Sobarzo-Sánchez, E.; Bilbao-Ramos, P.; Dea-Ayuela, M.; González-Díaz, H.; Yañez, M.; Uriarte, E.; Santana, L.; Martínez-Sernández, V.; Bolás-Fernández, F.; Ubeira, F.M. Synthetic oxoisoaporphine alkaloids: In vitro, in vivo and in silico assessment of antileishmanial activities. PLoS ONE 2013, 8, e77560. [Google Scholar] [CrossRef]
- Powers, C.N.; Setzer, W.N. An in-silico investigation of phytochemicals as antiviral agents against dengue fever. Comb. Chem. High Throughput Screen. 2016, 19, 516–536. [Google Scholar] [CrossRef] [Green Version]
- Behera, D.R.; Bhatnagar, S. In vitro and in silico efficacy of isolated alkaloid compounds from Rauvolfia tetraphylla L. against bovine filarial parasite Setaria cervi: A drug discovery approach. J. Parasit. Dis. 2019, 43, 103–112. [Google Scholar] [CrossRef]
- Casciaro, B.; Calcaterra, A.; Cappiello, F.; Mori, M.; Loffredo, M.R.; Ghirga, F.; Mangoni, M.L.; Botta, B.; Quaglio, D. Nigritanine as a new potential antimicrobial alkaloid for the treatment of Staphylococcus aureus-induced infections. Toxins 2019, 11, 511. [Google Scholar] [CrossRef] [Green Version]
- Laudadio, E.; Cedraro, N.; Mangiaterra, G.; Citterio, B.; Mobbili, G.; Minnelli, C.; Bizzaro, D.; Biavasco, F.; Galeazzi, R. Natural alkaloid berberine activity against Pseudomonas aeruginosa MexXY-mediated aminoglycoside resistance: In silico and in vitro studies. J. Nat. Prod. 2019, 82, 1935–1944. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, G.R.; Maurya, A.; Yadav, D.K.; Singh, V.; Khan, F.; Gupta, M.K.; Singh, M.; Darokar, M.P.; Srivastava, S.K. Synergy of clavine alkaloid ‘chanoclavine’ with tetracycline against multi-drug-resistant E. coli. J. Biomol. Struct. Dyn. 2019, 37, 1307–1325. [Google Scholar] [CrossRef] [PubMed]
- Annunziato, G. Strategies to overcome antimicrobial resistance (AMR) making use of non-essential target inhibitors: A review. Int. J. Mol. Sci. 2019, 20, 5844. [Google Scholar] [CrossRef] [Green Version]
- Shiraishi, N.; Akiyama, S.-i.; Nakagawa, M.; Kobayashi, M.; Kuwano, M. Effect of bisbenzylisoquinoline (bis-coclaurine) alkaloids on multidrug resistance in KB human cancer cells. Cancer Res. 1987, 47, 2413–2416. [Google Scholar]
- Nakajima, A.; Yamamoto, Y.; Taura, K.; Hata, K.; Fukumoto, M.; Uchinami, H.; Yonezawa, K.; Yamaoka, Y. Beneficial effect of cepharanthine on overcoming drug-resistance of hepatocellular carcinoma. Int. J. Oncol. 2004, 24, 635–645. [Google Scholar] [CrossRef]
- Xu, W.; Chen, S.; Wang, X.; Wu, H.; Yamada, H.; Hirano, T. Bisbenzylisoquinoline alkaloids and P-glycoprotein function: A structure activity relationship study. Bioorg. Med. Chem. 2020, 28, 115553. [Google Scholar] [CrossRef]
- Saifah, E.; Puripattanavong, J.; Likhitwitayawuid, K.; Cordell, G.A.; Chai, H.; Pezzuto, J.M. Bisamides from Aglaia species: Structure analysis and potential to reverse drug resistance with cultured cells. J. Nat. Prod. 1993, 56, 473–477. [Google Scholar] [CrossRef]
- You, M.; Ma, X.-J.; Mukherjee, R.; Farnsworth, N.R.; Cordell, G.A.; Kinghorn, A.D.; Pezzuto, J.M. Indole alkaloids from Peschiera laeta that enhance vinblastine-mediated cytotoxicity with multidrug-resistant cells. J. Nat. Prod. 1994, 57, 1517–1522. [Google Scholar] [CrossRef]
- You, M.; Wickramaratne, D.B.M.; Silva, G.L.; Chai, H.; Chagwedera, T.E.; Farnsworth, N.R.; Cordell, G.A.; Kinghorn, A.D.; Pezzuto, J.M. (-)-Roemerine, an aporphine alkaloid from Annona senegalensis that reverses the multidrug-resistance phenotype with cultured cells. J. Nat. Prod. 1995, 58, 598–604. [Google Scholar] [CrossRef]
- Silva, G.L.; Cui, B.; Chávez, D.; You, M.; Chai, H.B.; Rasoanaivo, P.; Lynn, S.M.; O’Neill, M.J.; Lewis, J.A.; Besterman, J.M.; et al. Modulation of the multidrug-resistance phenotype by new tropane alkaloid aromatic esters from Erythroxylum pervillei. J. Nat. Prod. 2001, 64, 1514–1520. [Google Scholar] [CrossRef]
- Baumert, C.; Hilgeroth, A. Recent advances in the development of P-gp inhibitors. Anticancer Agents Med. Chem. 2009, 9, 415–436. [Google Scholar] [CrossRef]
- Huang, X.-C.; Xiao, X.; Zhang, Y.-K.; Talele, T.T.; Salim, A.A.; Chen, Z.-S.; Capon, R.J. Lamellarin O, a pyrrole alkaloid from an Australian marine sponge, Ianthella sp., reverses BCRP mediated drug resistance in cancer cells. Mar. Drugs 2014, 12, 3818–3837. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-P.; Zhao, W.; Xue, R.; Zhou, Z.-X.; Liu, F.; Han, Y.-X.; Ren, G.; Peng, Z.-G.; Cen, S.; Chen, H.-S.; et al. Oxymatrine inhibits hepatitis B infection with an advantage of overcoming drug-resistance. Antivir. Res. 2011, 89, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Frisoni, P.; Bacchio, E.; Bilel, S.; Talarico, A.; Gaudio, R.M.; Barbieri, M.; Neri, M.; Marti, M. Novel synthetic opioids: The pathologist’s point of view. Brain Sci. 2018, 8, 170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salle, S.; Bodeau, S.; Dhersin, A.; Ferdonnet, M.; Goncalves, R.; Lenski, M.; Lima, B.; Martin, M.; Outreville, J.; Vaucel, J.; et al. Novel synthetic opioids: A review of the literature. Toxicol. Anal. Clin. 2019, 31, 298–316. [Google Scholar] [CrossRef]
- Kyzer, J.L.; Wenthur, C.J. Classics in chemical neuroscience: Buprenorphine. ACS Chem. Neurosci. 2020, 11, 1385–1399. [Google Scholar] [CrossRef] [PubMed]
- Hamilton-Miller, J. Development of the semi-synthetic penicillins and cephalosporins. Int. J. Antimicrob. Agents 2008, 31, 189–192. [Google Scholar] [CrossRef]
- Etebu, E.; Arikekpar, I. Antibiotics: Classification and mechanisms of action with emphasis on molecular perspectives. Int. J. Appl. Microbiol. Biotechnol. Res. 2016, 4, 90–101. [Google Scholar]
- Kennedy, J.P.; Williams, L.; Bridges, T.M.; Daniels, R.N.; Weaver, D.; Lindsley, C.W. Application of combinatorial chemistry science on modern drug discovery. J. Comb. Chem. 2008, 10, 345–354. [Google Scholar] [CrossRef]
- Kolb, H.C.; Sharpless, K.B. The growing impact of click chemistry on drug discovery. Drug Discov. Today 2003, 8, 1128–1137. [Google Scholar] [CrossRef]
- Lombardino, J.G.; Lowe, J.A. The role of the medicinal chemist in drug discovery—Then and now. Nat. Rev. Drug Discov. 2004, 3, 853–862. [Google Scholar] [CrossRef]
- Hou, J.; Liu, X.; Shen, J.; Zhao, G.; Wang, P.G. The impact of click chemistry in medicinal chemistry. Expert Opin. Drug Discov. 2012, 7, 489–501. [Google Scholar] [CrossRef]
- Jiang, X.; Hao, X.; Jing, L.; Wu, G.; Kang, D.; Liu, X.; Zhan, P. Recent applications of click chemistry in drug discovery. Expert Opin. Drug Discov. 2019, 14, 779–789. [Google Scholar] [CrossRef]
- Cordell, G.A. Natural products in drug discovery—Creating a new vision. Phytochem. Rev. 2002, 1, 261–273. [Google Scholar] [CrossRef]
- Ramallo, I.A.; Salazar, M.O.; Méndez, L.; Furlán, R.L.E. Chemically engineered extracts: Source of bioactive compounds. Accounts Chem. Res. 2011, 44, 241–250. [Google Scholar] [CrossRef]
- Ramallo, I.A.; Alonso, V.L.; Rua, F.; Serra, E.; Furlán, R.L.E. A bioactive Trypanosoma cruzi bromodomain inhibitor from chemically engineered extracts. ACS Comb. Sci. 2018, 20, 220–228. [Google Scholar] [CrossRef]
- Du, Y.; Sun, J.; Gong, Q.; Wang, Y.; Fu, P.; Zhu, W. New α-pyridones with quorum-sensing inhibitory activity from diversity-enhanced extracts of a Streptomyces sp. derived from marine algae. J. Agric. Food Chem. 2018, 66, 1807–1812. [Google Scholar] [CrossRef]
- Salazar, M.O.; Osella, M.I.; Arcusin, D.E.J.; Lescano, L.E.; Furlan, R.L.E. New α-glucosidase inhibitors from a chemically en-gineered essential oil of Origanum vulgare L. Ind. Crop. Prod. 2020, 156, 112855. [Google Scholar] [CrossRef]
- Tomohara, K.; Ito, T.; Furusawa, K.; Hasegawa, N.; Tsuge, K.; Kato, A.; Adachi, I. Multiple production of α,α-disubstituted amino acid derivatives through direct chemical derivatization of natural plant extracts: An apparently difficult but successful route. Tetrahedron Lett. 2017, 58, 3143–3147. [Google Scholar] [CrossRef]
- Kamauchi, H.; Noji, M.; Kinoshita, K.; Takanami, T.; Koyama, K. Coumarins with an unprecedented tetracyclic skeleton and coumarin dimers from chemically engineered extracts of a marine-derived fungus. Tetrahedron 2018, 74, 2846–2856. [Google Scholar] [CrossRef]
- Zhang, J.-L.; Xu, W.; Zhou, Z.-R.; Li, J.; Jiang, L.-L.; Zhang, X.-X.; Jiang, R.-W. Antineoplastic constituents from the chemical diversified extract of Radix puerariae. Chem. Biodivers. 2019, 16, e1800408. [Google Scholar] [CrossRef] [Green Version]
- Hogg, J.A. Steroids, the steroid community, and Upjohn in perspective: A profile of innovation. Steroids 1992, 57, 593–616. [Google Scholar] [CrossRef]
- Rathbone, D.A.; Lister, D.L.; Bruce, N.C. Biotransformation of alkaloids. In The Alkaloids: Chemistry and Biology; Elsevier BV: Amsterdam, The Netherlands, 2001; Volume 57, pp. 1–74. [Google Scholar]
- Rathbone, D.A.; Bruce, N.C. Microbial transformation of alkaloids. Curr. Opin. Microbiol. 2002, 5, 274–281. [Google Scholar] [CrossRef]
- Boonstra, B.; Rathbone, D.A.; French, C.E.; Walker, E.H.; Bruce, N.C. cofactor regeneration by a soluble pyridine nucleotide transhydrogenase for biological production of hydromorphone. Appl. Environ. Microbiol. 2000, 66, 5161–5166. [Google Scholar] [CrossRef] [Green Version]
- Boonstra, B.; Rathbone, D.A.; Bruce, N.C. Engineering novel biocatalytic routes for production of semisynthetic opiate drugs. Biomol. Eng. 2001, 18, 41–47. [Google Scholar] [CrossRef]
- Kiener, A.; Roduit, J.-P.; Wellig, A. Renewable functionalized pyridines derived from microbial metabolites of the alkaloid (S)-nicotine. Heterocycles 1997, 45, 1687. [Google Scholar] [CrossRef]
- Baitsch, D.; Sandu, C.; Brandsch, R.; Igloi, G.L. Gene cluster on pAO1 of Arthrobacter nicotinovorans involved in degradation of the plant alkaloid nicotine: Cloning, purification, and characterization of 2,6-dihydroxypyridine 3-hydroxylase. J. Bacteriol. 2001, 183, 5262–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, S.; Clastre, M.; Courdavault, V.; O’Connor, S.E. De novo production of the plant-derived alkaloid strictosidine in yeast. Proc. Natl. Acad. Sci. USA 2015, 112, 3205–3210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fossati, E.; Ekins, A.; Narcross, L.; Zhu, Y.; Falgueyret, J.-P.; Beaudoin, G.A.W.; Facchini, P.J.; Martin, V.J.J. Reconstitution of a 10-gene pathway for synthesis of the plant alkaloid dihydrosanguinarine in Saccharomyces cerevisiae. Nat. Commun. 2014, 5, 3283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Courdavault, V.; O’Connor, S.E.; Oudin, A.; Besseau, S.; Papon, N. Towards the microbial production of plant-derived anticancer drugs. Trends Cancer 2020, 6, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T. Biocatalysis in organic chemistry and biotechnology: Past, present, and future. J. Am. Chem. Soc. 2013, 135, 12480–12496. [Google Scholar] [CrossRef]
- Rosenthaler, L. Durch Enzyme bewirkte asymmetrische Synthesen. Biochem. Z. 1908, 14, 238–253. [Google Scholar]
- Sheldon, R.A.; Woodley, J.M. Role of biocatalysis in sustainable chemistry. Chem. Rev. 2018, 118, 801–838. [Google Scholar] [CrossRef]
- Kiss, G.; Celebi-Olçüm, N.; Moretti, R.; Baker, D.; Houk, K.N. Computational enzyme design. Angew. Chem. Int. Ed. 2013, 52, 5700–5725. [Google Scholar] [CrossRef]
- Wallace, S.; Balskus, E.P. Opportunities for merging chemical and biological synthesis. Curr. Opin. Biotechnol. 2014, 30, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Galanie, S.; Entwistle, D.; Lalonde, J. Engineering biosynthetic enzymes for industrial natural product synthesis. Nat. Prod. Rep. 2020, 37, 1122–1143. [Google Scholar] [CrossRef]
- Hyster, T.K.; Knörr, L.; Ward, T.R.; Rovis, T. Biotinylated Rh(III) complexes in engineered streptavidin for accelerated asym-metric C-H activation. Science 2012, 338, 500–503. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Zheng, J.; Zhang, X.; Wang, Z. Interfacing a phosphate catalytic reaction with a microbial metabolism for the production of azaphilone alkaloids. React. Chem. Eng. 2020, 5, 2048–2052. [Google Scholar] [CrossRef]
- Turner, N.J.; O’Reilly, E. Biocatalytic retrosynthesis. Nat. Chem. Biol. 2013, 9, 285–288. [Google Scholar] [CrossRef]
- Cordell, G.A.; Lemos, T.L.G.; Monte, F.J.Q.; De Mattos, M.C. Vegetables as chemical reagents. J. Nat. Prod. 2007, 70, 478–492. [Google Scholar] [CrossRef]
- Xu, B.; Watkins, R.; Wu, L.; Zhang, C.; Davis, R. Natural product-based nanomedicine: Recent advances and issues. Int. J. Nanomed. 2015, 10, 6055–6074. [Google Scholar] [CrossRef] [Green Version]
- Kralova, K.; Jampilek, J. Responses of medicinal and aromatic plants to engineered nanoparticles. Appl. Sci. 2021, 11, 1813. [Google Scholar] [CrossRef]
- Karakaş, Ö. Effect of silver nanoparticles on production of indole alkaloids in Isatis constricta. Iran. J. Sci. Technol. Trans. A Sci. 2020, 44, 621–627. [Google Scholar] [CrossRef]
- Sahibzada, M.U.K.; Sadiq, A.; Faidah, H.S.; Khurram, M.; Amin, M.U.; Haseeb, A.; Kakar, M. Berberine nanoparticles with enhanced in vitro bioavailability: Characterization and antimicrobial activity. Drug Des. Dev. Ther. 2018, 12, 303–312. [Google Scholar] [CrossRef] [Green Version]
- Baldim, I.; Oliveira, W.P.; Kadian, V.; Rao, R.; Yadav, N.; Mahant, S.; Lucarini, M.; Durazzo, A.; Da Ana, R.; Capasso, R.; et al. Natural ergot alkaloids in ocular pharmacotherapy: Known molecules for novel nanoparticle-based delivery systems. Biomolecules 2020, 10, 980. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, Y.; Chen, L. Network-based drug repositioning. Mol. BioSyst. 2013, 9, 1268–1281. [Google Scholar] [CrossRef]
- Baker, N.C.; Ekins, S.; Williams, A.J.; Tropsha, A. A bibliometric review of drug repurposing. Drug Discov. Today 2018, 23, 661–672. [Google Scholar] [CrossRef]
- Portincasa, P. Colchicine, biologic agents and more for the treatment of Familial Mediterranean Fever. The Old, the New, and the Rare. Curr. Med. Chem. 2015, 23, 60–86. [Google Scholar] [CrossRef] [PubMed]
- Panic, G.; Duthaler, U.; Speich, B.; Keiser, J. Repurposing drugs for the treatment and control of helminth infections. Int. J. Parasitol. Drugs Drug Resist. 2014, 4, 185–200. [Google Scholar] [CrossRef] [Green Version]
- Turner, N.; Zeng, X.-Y.; Osborne, B.; Rogers, S.; Ye, J.-M. Repurposing drugs to target the diabetes epidemic. Trends Pharmacol. Sci. 2016, 37, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Bhakta, S. The prospect of repurposing immunomodulatory drugs for adjunctive chemotherapy against tuberculosis: A critical review. Antibiotics 2021, 10, 91. [Google Scholar] [CrossRef] [PubMed]
- Abdelaleem, M.; Ezzat, H.; Osama, M.; Megahed, A.; Alaa, W.; Gaber, A.; Shafei, A.; Refaat, A. Prospects for repurposing CNS drugs for cancer treatment. Oncol. Rev. 2019, 13, 411. [Google Scholar] [CrossRef] [PubMed]
- Bayazeid, O.; Yalçın, F.N. Biological targets of 92 alkaloids isolated from Papaver genus: A perspective based on in silico pre-dictions. Med. Chem. Res. 2021, 30, 574–585. [Google Scholar] [CrossRef]
- Gu, J.; Gui, Y.; Chen, L.; Yuan, G.; Lu, H.-Z.; Xu, X. Use of natural products as chemical library for drug discovery and network pharmacology. PLoS ONE 2013, 8, e62839. [Google Scholar] [CrossRef] [Green Version]
- Schlesinger, N.; Firestein, B.L.; Brunetti, L. Colchicine in COVID-19: An old drug, new use. Curr. Pharmacol. Rep. 2020, 6, 137–145. [Google Scholar] [CrossRef]
- Cusinato, J.; Cau, Y.; Calvani, A.M.; Mori, M. Repurposing drugs for the management of COVID-19. Expert Opin. Ther. Patents 2021, 31, 295–307. [Google Scholar] [CrossRef]
- Levin, J.M.; Oprea, T.I.; Davidovich, S.; Clozel, T.; Overington, J.P.; Vanhaelen, Q.; Cantor, C.R.; Bischof, E.; Zhavoronkov, A. Artificial intelligence, drug repurposing and peer review. Nat. Biotechnol. 2020, 38, 1127–1131. [Google Scholar] [CrossRef] [PubMed]
- Borquaye, L.S.; Gasu, E.N.; Ampomah, G.B.; Kyei, L.K.; Amarh, M.A.; Mensah, N.K.; Nartey, D. Alkaloids from Cryptolepis sanguinolenta as potential inhibitors of SARS-CoV-2 viral proteins: An in silico study. BioMed Res. Int. 2020, 2020, 5324560. [Google Scholar] [CrossRef]
- Ghosh, R.; Chakraborty, A.; Biswas, A.; Chowdhuri, S. Identification of alkaloids from Justicia adhatoda as potent SARS CoV-2 main protease inhibitors: An in silico perspective. J. Mol. Struct. 2021, 1229, 129489. [Google Scholar] [CrossRef]
- Garg, S.; Roy, A. In silico analysis of selected alkaloids against main protease (Mpro) of SARS-CoV-2. Chem. Biol. Interact. 2020, 332, 109309. [Google Scholar] [CrossRef]
- Fournet, A.; Gantier, J.-C.; Gautheret, A.; Leysalles, L.; Munos, M.H.; Mayrargue, J.; Moskowitz, H.; Cavé, A.; Hocquemiller, R. The activity of 2-substituted quinoline alkaloids in BALB/c mice infected with Leishmania donovani. J. Antimicrob. Chemother. 1994, 33, 537–544. [Google Scholar] [CrossRef] [Green Version]
- Fournet, A.; Ferreira, M.E.; de Arias, A.R.; Ortiz, S.T.; Fuentes, S.; Nakayama, H.; Schinini, A.; Hocquemiller, R. In vivo efficacy of oral and intralesional administration of 2-substituted quinolines in experimental treatment of new world cutaneous leishmaniasis caused by Leishmania amazonensis. Antimicrob. Agents Chemother. 1996, 40, 2447–2451. [Google Scholar] [CrossRef] [Green Version]
- Muhammad, I.; Dunbar, D.C.; Khan, S.I.; Tekwani, B.L.; Bedir, E.; Takamatsu, S.; Ferreira, D.; Walker, L.A. Antiparasitic alkaloids from Psychotria klugii. J. Nat. Prod. 2003, 66, 962–967. [Google Scholar] [CrossRef] [PubMed]
- Nakao, Y.; Shiroiwa, T.; Murayama, S.; Matsunaga, S.; Goto, Y.; Matsumoto, Y.; Fusetani, N. Identification of renieramycin a as an antileishmanial substance in a marine sponge Neopetrosia sp. Mar. Drugs 2004, 2, 55–62. [Google Scholar] [CrossRef] [Green Version]
- Cordell, G.A.; Angerhofer, C.K.; Pezzuto, J.M. Recent studies on cytotoxic, anti-HIV and antimalarial agents from plants. Pure Appl. Chem. 1994, 66, 2283–2286. [Google Scholar] [CrossRef]
- Hopkins, A.L. Network pharmacology. Nat. Biotechnol. 2007, 25, 1110–1111. [Google Scholar] [CrossRef]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef]
- Kibble, M.; Saarinen, N.; Tang, J.; Wennerberg, K.; Mäkelä, S.; Aittokallio, T. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Nat. Prod. Rep. 2015, 32, 1249–1266. [Google Scholar] [CrossRef]
- Doroghazi, J.R.; Albright, J.C.; Goering, A.W.; Ju, K.-S.; Haines, R.R.; Tchalukov, K.A.; Labeda, D.P.; Kelleher, N.L.; Metcalf, W.W. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 2014, 10, 963–968. [Google Scholar] [CrossRef]
- Challis, G.L. Exploitation of the Streptomyces coelicolor A3(2) genome sequence for discovery of new natural products and biosynthetic pathways. J. Ind. Microbiol. Biotechnol. 2014, 41, 219–232. [Google Scholar] [CrossRef]
- Nett, M.; Ikeda, H.; Moore, B.S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009, 26, 1362–1384. [Google Scholar] [CrossRef]
- Donadio, S.; Monciardini, P.; Sosio, M. Polyketide synthases and nonribosomal peptide synthetases: The emerging view from bacterial genomics. Nat. Prod. Rep. 2007, 24, 1073–1109. [Google Scholar] [CrossRef]
- Letzel, A.-C.; Pidot, S.J.; Hertweck, C. A genomic approach to the cryptic secondary metabolome of the anaerobic world. Nat. Prod. Rep. 2013, 30, 392–428. [Google Scholar] [CrossRef]
- Calteau, A.; Fewer, D.P.; Latifi, A.; Coursin, T.; Laurent, T.; Jokela, J.; Kerfeld, C.A.; Sivonen, K.; Piel, J.; Gugger, M. Phylum-wide comparative genomics unravel the diversity of secondary metabolism in cyanobacteria. BMC Genom. 2014, 15, 977. [Google Scholar] [CrossRef] [Green Version]
- Wilson, M.C.; Mori, T.; Rückert, C.; Uria, A.R.; Helf, M.J.; Takada, K.; Gernert, C.; Steffens, U.A.E.; Heycke, N.; Schmitt, S.; et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nat. Cell Biol. 2014, 506, 58–62. [Google Scholar] [CrossRef] [Green Version]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism from biochemistry to genomics. Nat. Rev. Microbiol. 2005, 3, 937–947. [Google Scholar] [CrossRef]
- Fischbach, M.A.; Walsh, C.T. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem. Rev. 2006, 106, 3468–3496. [Google Scholar] [CrossRef]
- Singh, M.; Chaudhary, S.; Sareen, D. Non-ribosomal peptide synthetases: Identifying the cryptic gene clusters and decoding the natural product. J. Biosci. 2017, 42, 175–187. [Google Scholar] [CrossRef]
- Winter, J.M.; Behnken, S.; Hertweck, C. Genomics-inspired discovery of natural products. Curr. Opin. Chem. Biol. 2011, 15, 22–31. [Google Scholar] [CrossRef]
- Rutledge, P.J.; Challis, G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. [Google Scholar] [CrossRef]
- Blin, K.; Andreu, V.P.; de los Santos, E.L.C.; Del Carratore, F.; Lee, S.Y.; Medema, M.H.; Weber, T. The antiSMASH database version 2: A comprehensive resource on secondary metabolite biosynthetic gene clusters. Nucl. Acids Res. 2019, 47, D625–D630. [Google Scholar]
- Flissi, A.; Ricart, E.; Campart, C.; Chevalier, M.; Dufresne, Y.; Michalik, J.; Flahaut, C.; Lisacek, F.; Leclère, V.; Pupin, M. Norine: Update of the nonribosomal peptide resource. Nucl. Acids Res. 2020, 48, D465–D469. [Google Scholar]
- Zierep, P.F.; Ceci, A.T.; Dobrusin, I.; Rockwell-Kollmann, S.C.; Günther, S. SeMPI 2.0—A web server for PKS and NRPS predictions combined with metabolite screening in natural product databases. Metabolites 2021, 11, 13. [Google Scholar]
- Ugai, T.; Minami, A.; Gomi, K.; Oikawa, H. Genome mining approach for harnessing the cryptic gene cluster in Alternaria solani: Production of PKS-NRPS hybrid metabolite, didymellamide B. Tetrahedron Lett. 2016, 57, 2793–2796. [Google Scholar]
- Chen, R.; Zhang, Q.; Tan, B.; Zheng, L.; Li, H.; Zhu, Y.; Zhang, C. Genome mining and activation of a silent PKS/NRPS gene cluster direct the production of totopotensamides. Org. Lett. 2017, 19, 5697–5700. [Google Scholar]
- Tang, S.; Zhang, W.; Li, Z.; Li, H.; Geng, C.; Huang, X.; Lu, X. Discovery and characterization of a PKS-NRPS hybrid in Aspergillus terreus by genome mining. J. Nat. Prod. 2020, 83, 473–480. [Google Scholar]
- Suroto, D.A.; Kitani, S.; Arai, M.; Ikeda, H.; Nihira, T. Characterization of the biosynthetic gene cluster for cryptic phthoxazolin A in Streptomyces avermitilis. PLoS ONE 2018, 13, e0190973. [Google Scholar]
- Niehs, S.P.; Dose, B.; Scherlach, K.; Pidot, S.J.; Stinear, T.P.; Hertweck, C. Genome mining reveals endopyrroles from a nonribosomal peptide assembly line triggered in fungal–bacterial symbiosis. ACS Chem. Biol. 2019, 14, 1811–1818. [Google Scholar] [CrossRef]
- Chiang, Y.-M.; Lee, K.-H.; Sanchez, J.F.; Keller, N.P.; Wang, C.C.C. Unlocking fungal cryptic natural products. Nat. Prod. Commun. 2009, 4, 1505–1510. [Google Scholar]
- Zerikly, M.; Challis, G.L. Strategies for the discovery of new natural products by genome mining. ChemBioChem 2009, 10, 625–633. [Google Scholar] [CrossRef]
- Weber, T.; Charusanti, P.; Musiol-Kroll, E.M.; Jiang, X.; Tong, Y.; Kim, H.U.; Lee, S.Y. Metabolic engineering of antibiotic factories: New tools for antibiotic production in actinomycetes. Trends Biotechnol. 2015, 33, 15–26. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, H.; Chen, H.; Jing, X.; Zheng, W.; Li, R.; Sun, T.; Liu, J.; Fu, J.; Huo, L. Discovery of recombinases enables genome mining of cryptic biosynthetic gene clusters in Burkholderiales species. Proc. Natl. Acad. Sci. USA 2018, 115, E4255–E4263. [Google Scholar]
- Zheng, W.; Wang, X.; Zhou, H.; Zhang, Y.; Li, A.; Bian, X. Establishment of recombineering genome editing system in Paraburkholderia megapolitana empowers activation of silent biosynthetic gene clusters. Microb. Biotechnol. 2020, 13, 397–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prihoda, D.; Maritz, J.M.; Klempir, O.; Dzamba, D.; Woelk, C.H.; Hazuda, D.J.; Bitton, D.A.; Hannigan, G.D. The application potential of machine learning and genomics for understanding natural product diversity, chemistry, and therapeutic translatability. Nat. Prod. Rep. 2021. [Google Scholar] [CrossRef] [PubMed]
- Clardy, J.; Walsh, C.J. Lessons from natural molecules. Nature 2004, 432, 829–837. [Google Scholar] [CrossRef]
- Lorenz, P.; Eck, J. Metagenomics and industrial applications. Nat. Rev. Microbiol. 2005, 3, 510–516. [Google Scholar] [CrossRef]
- Lefevre, F.; Robe, P.; Jarrin, C.; Ginolhac, A.; Zago, C.; Auriol, D.; Vogel, T.M.; Simonet, P.; Nalin, R. Drugs from hidden bugs: Their discovery via untapped resources. Res. Microbiol. 2008, 159, 153–161. [Google Scholar] [CrossRef]
- Simon, C.; Daniel, R. Achievements and new knowledge unraveled by metagenomic approaches. Appl. Microbiol. Biotechnol. 2009, 85, 265–276. [Google Scholar] [CrossRef] [Green Version]
- Riesenfeld, C.S.; Schloss, P.D.; Handelsman, J. Metagenomics: Genomic analysis of microbial communities. Annu. Rev. Genet. 2004, 38, 525–552. [Google Scholar] [CrossRef] [Green Version]
- Frias-Lopez, J.; Shi, Y.; Tyson, G.W.; Coleman, M.L.; Schuster, S.C.; Chisholm, S.W.; DeLong, E.F. Microbial community gene expression in ocean surface waters. Proc. Natl. Acad. Sci. USA 2008, 105, 3805–3810. [Google Scholar] [CrossRef] [Green Version]
- Banik, J.J.; Brady, S.F. Cloning and characterization of new glycopeptide gene clusters found in an environmental DNA megalibrary. Proc. Natl. Acad. Sci. USA 2008, 105, 17273–17277. [Google Scholar] [CrossRef] [Green Version]
- Donia, M.S.; Ruffner, D.E.; Cao, S.; Schmidt, E.W. Accessing the hidden majority of marine natural products through metagenomics. ChemBioChem 2011, 12, 1230–1236. [Google Scholar] [CrossRef]
- Trindade, M.; van Zyl, L.J.; Navarro-Fernández, J.; Elrazak, A.A. Targeted metagenomics as a tool to tap into marine natural product diversity for the discovery and production of drug candidates. Front. Microbiol. 2015, 6, 890. [Google Scholar]
- Mahapatra, G.P.; Raman, S.; Nayak, S.; Gouda, S.; Das, G.; Patra, J.K. Metagenomics approaches in discovery and development of new bioactive compounds from marine actinomycetes. Curr. Microbiol. 2019, 77, 645–656. [Google Scholar]
- Carayannis, E.G.; Campbell, D.F.J. “Mode 3” and “Quadruple Helix”: Toward a 21st century fractal innovation ecosystem. Int. J. Technol. Manag. 2009, 46, 201–234. [Google Scholar] [CrossRef] [Green Version]
- Carayannis, E.G.; Barth, T.D.; Campbell, D.F.J. The Quintuple Helix innovation model: Global warming as a challenge and a driver for innovation. J. Innov. Entrep. 2012, 1, 2. [Google Scholar] [CrossRef] [Green Version]
- Martins, V.W.B.; Rampasso, I.S.; Anholon, R.; Quelhas, O.L.G.; Filho, W. Knowledge management in the context of sustainability: Literature review and opportunities for future research. J. Clean. Prod. 2019, 229, 489–500. [Google Scholar] [CrossRef]
- Cordell, G.A. Cyberecoethnopharmacolomics. J. Ethnopharmacol. 2019, 244, 112134. [Google Scholar] [CrossRef]
- Ou-Yang, S.-S.; Lu, J.-Y.; Kong, X.-Q.; Liang, Z.-J.; Luo, C.; Jiang, H. Computational drug discovery. Acta Pharmacol. Sin. 2012, 33, 1131–1140. [Google Scholar]
- Smith, J.S.; Roitberg, A.E.; Isayev, O. Transforming computational drug discovery with machine learning and AI. ACS Med. Chem. Lett. 2018, 9, 1065–1069. [Google Scholar] [CrossRef]
- Schneider, G. Automating drug discovery. Nat. Rev. Drug Discov. 2018, 17, 97. [Google Scholar] [CrossRef]
- Chen, H.; Kogej, T.; Engkvist, O. Cheminformatics in drug discovery, an industrial perspective. Mol. Inform. 2018, 37, 1800041. [Google Scholar]
- Chan, H.C.S.; Shan, H.; Dahoun, T.; Vogel, H.; Yuan, S. Advancing drug discovery via artificial intelligence. Trends Pharmacol. Sci. 2019, 40, 592–604. [Google Scholar]
- Schneider, P.; Walters, W.P.; Plowright, A.T.; Sieroka, N.; Listgarten, J.; Goodnow, R.A.; Fisher, J.; Jansen, J.M.; Duca, J.S.; Rush, T.S.; et al. Rethinking drug design in the artificial intelligence era. Nat. Rev. Drug Discov. 2020, 19, 353–364. [Google Scholar]
- Zhao, L.; Ciallella, H.L.; Aleksunes, L.M.; Zhu, H. Advancing computer-aided drug discovery (CADD) by big data and data-driven machine learning modeling. Drug Discov. Today 2020, 25, 1624–1638. [Google Scholar]
- MacConnell, A.B.; Price, A.K.; Paegel, B.M. An integrated microfluidic processor for DNA-encoded combinatorial library functional screening. ACS Comb. Sci. 2017, 19, 181–192. [Google Scholar] [CrossRef]
- Baranczak, A.; Tu, N.P.; Marjanovic, J.; Searle, P.A.; Vasudevan, A.; Djuric, S.W. Integrated platform for expedited synthesis–purification–testing of small molecule libraries. ACS Med. Chem. Lett. 2017, 8, 461–465. [Google Scholar] [CrossRef] [Green Version]
- Clark, A.J.; Negron, C.; Hauser, K.; Sun, M.; Wang, L.; Abel, R.; Friesner, R.A. Relative binding affinity prediction of charge-changing sequence mutations with FEP in protein-protein interfaces. J. Mol. Biol. 2019, 431, 1481–1493. [Google Scholar] [CrossRef]
- Capel, A.J.; Rimington, R.P.; Lewis, M.P.; Christie, S.D.R. 3D Printing for chemical, pharmaceutical and biological applications. Nat. Rev. Chem. 2018, 2, 422–436. [Google Scholar] [CrossRef]
- Hartings, M.R.; Ahmed, Z. Chemistry from 3D printed objects. Nat. Rev. Chem. 2019, 3, 305–314. [Google Scholar] [CrossRef]
- Sparkes, A.; Aubrey, W.; Byrne, E.; Clare, A.; Khan, M.N.; Liakata, M.; Markham, M.; Rowland, J.; Soldatova, L.N.; Whelan, K.E.; et al. Towards robot scientists for autonomous scientific discovery. Autom. Exp. 2010, 2, 1–11. [Google Scholar]
- Lehár, J.; Krueger, A.S.; Avery, W.; Heilbut, A.M.; Johansen, L.M.; Price, E.R.; Rickles, R.J.; Short, G.F., III; Staunton, J.E.; Jin, X.; et al. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat. Biotechnol. 2009, 27, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Kola, I.; Landis, J. Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov. 2004, 3, 711–716. [Google Scholar]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar]
- Wu, M.; Ma, C.; Wu, Y.; Li, S. Simultaneous LC analysis of five bioactive alkaloids in an anti-angiogenesis herbal formula, Qing-Luo-Yin. Chromatographia 2008, 68, 579–585. [Google Scholar] [CrossRef]
- Li, S.; Zhang, B.; Zhang, N. Network target for screening synergistic drug combinations with application to traditional Chinese medicine. BMC Syst. Biol. 2011, 5, S10. [Google Scholar] [CrossRef] [Green Version]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef] [Green Version]
- Tarca, A.L.; Carey, V.J.; Chen, X.-w.; Romero, R.; Drăghici, S. Machine learning and its applications to biology. PLoS Comput. Biol. 2007, 3, e116. [Google Scholar] [CrossRef]
- Chen, H.; Engkvist, O.; Wang, Y.; Olivecrona, M.; Blaschke, T. The rise of deep learning in drug discovery. Drug Discov. Today 2018, 23, 1241–1250. [Google Scholar]
- Lo, Y.-C.; Rensi, S.E.; Torng, W.; Altman, R.B. Machine learning in chemoinformatics and drug discovery. Drug Discov. Today 2018, 23, 1538–1546. [Google Scholar]
- Stephenson, N.; Shane, E.; Chase, J.; Rowland, J.; Ries, D.; Justice, N.; Zhang, J.; Chan, L.; Cao, R. Survey of machine learning techniques in drug discovery. Curr. Drug Metab. 2019, 20, 185–193. [Google Scholar] [CrossRef]
- Gini, G.; Zanoli, F.; Gamba, A.; Raitano, G.; Benfenati, E. Could deep learning in neural networks improve the QSAR models? SAR QSAR Environ. Res. 2019, 30, 617–642. [Google Scholar] [CrossRef]
- Hu, S.; Chen, P.; Gu, P.; Wang, B. A deep learning-based chemical system for QSAR prediction. IEEE J. Biomed. Health Inform. 2020, 24, 3020–3028. [Google Scholar]
- Aliper, A.; Plis, S.; Artemov, A.; Ulloa, A.; Mamoshina, P.; Zhavoronkov, A. Deep learning applications for predicting pharmacological properties of drugs and drug repurposing using transcriptomic data. Mol. Pharm. 2016, 13, 2524–2530. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.; Zhao, Z.; Wang, R.; Wei, G.-W. TopP-S: Persistent homology-based multi-task deep neural networks for simultaneous predictions of partition coefficient and aqueous solubility. J. Comput. Chem. 2018, 39, 1444–1454. [Google Scholar] [CrossRef] [Green Version]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.N.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A deep learning approach to antibiotic discovery. Cell 2020, 180, 688–702. [Google Scholar] [CrossRef] [Green Version]
- Ekins, S.; Siqueira-Neto, J.L.; McCall, L.-I.; Sarker, M.; Yadav, M.; Ponder, E.L.; Kallel, E.A.; Kellar, D.; Chen, S.; Arkin, M.; et al. Machine learning models and pathway genome data base for Trypanosoma cruzi drug discovery. PLoS Negl. Trop. Dis. 2015, 9, e0003878. [Google Scholar]
- Egieyeh, S.; Syce, J.; Malan, S.F.; Christoffels, A. Predictive classifier models built from natural products with antimalarial bioactivity using machine learning approach. PLoS ONE 2018, 13, e0204644. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Stork, C.; Hirte, S.; Kirchmair, J. NP-Scout: Machine learning approach for the quantification and visualization of the natural product-likeness of small molecules. Biomolecules 2019, 9, 43. [Google Scholar]
- Grisoni, F.; Merk, D.; Friedrich, L.; Schneider, G. Design of natural-product-inspired multitarget ligands by machine learning. ChemMedChem 2019, 14, 1129–1134. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.; Bernardes, G.J.L. Machine learning for target discovery in drug development. Curr. Opin. Chem. Biol. 2020, 56, 16–22. [Google Scholar] [CrossRef]
- Capecchi, A.; Reymond, J.-L. Assigning the origin of microbial natural products by chemical space map and machine learning. Biomolecules 2020, 10, 1385. [Google Scholar] [CrossRef] [PubMed]
- Ekins, S.; Puhl, A.C.; Zorn, K.M.; Lane, T.R.; Russo, D.P.; Klein, J.J.; Hickey, A.J.; Clark, A.M. Exploiting machine learning for end-to-end drug discovery and development. Nat. Mat. 2019, 18, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Zhavoronkov, A. Artificial intelligence for drug discovery, biomarker development, and generation of novel chemistry. Mol. Pharm. 2018, 15, 4311–4313. [Google Scholar] [CrossRef] [Green Version]
- Vamathevan, J.; Clark, D.; Czodrowski, P.; Dunham, I.; Ferran, E.; Lee, G.; Li, B.; Madabhushi, A.; Shah, P.; Spitzer, M.; et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 2019, 18, 463–477. [Google Scholar] [CrossRef]
- Davies, D.W.; Butler, K.T.; Isayev, O.; Walsh, A. Materials discovery by chemical analogy: Role of oxidation states in structure prediction. Faraday Discuss. 2018, 211, 553–568. [Google Scholar] [CrossRef] [Green Version]
- Mayr, A.; Klambauer, G.; Unterthiner, T.; Steijaert, M.; Wegner, J.K.; Ceulemans, H.; Clevert, D.A.; Hochreiter, S. Large-scale comparison of machine learning methods for drug target prediction on ChEMBL. Chem. Sci. 2018, 9, 5441–5451. [Google Scholar]
- Sorokina, M.; Steinbeck, C. Review on natural products databases: Where to find data in 2020. J. Cheminform. 2020, 12, 1–51. [Google Scholar] [CrossRef] [Green Version]
- Allard, P.-M.; Bisson, J.; Azzollini, A.; Pauli, G.F.; Cordell, G.A.; Wolfender, J.-L. Pharmacognosy in the digital era: Shifting to contextualized metabolomics. Curr. Opin. Biotechnol. 2018, 54, 57–64. [Google Scholar] [CrossRef]
- Zampieri, G.; Vijayakumar, S.; Yaneske, E.; Angione, C. Machine and deep learning meet genome-scale metabolic modeling. PLoS Comput. Biol. 2019, 15, e1007084. [Google Scholar] [CrossRef]
- Wouters, O.J.; McKee, M.; Luyten, J. Estimated research and development investment needed to bring a new medicine to market, 2009–2018. J. Am. Med. Assoc. 2020, 323, 844–853. [Google Scholar] [CrossRef]
- Henry, D.; Lexchin, J. The pharmaceutical industry as a medicines provider. Lancet 2002, 360, 1590–1595. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daley, S.-k.; Cordell, G.A. Alkaloids in Contemporary Drug Discovery to Meet Global Disease Needs. Molecules 2021, 26, 3800. https://doi.org/10.3390/molecules26133800
Daley S-k, Cordell GA. Alkaloids in Contemporary Drug Discovery to Meet Global Disease Needs. Molecules. 2021; 26(13):3800. https://doi.org/10.3390/molecules26133800
Chicago/Turabian StyleDaley, Sharna-kay, and Geoffrey A. Cordell. 2021. "Alkaloids in Contemporary Drug Discovery to Meet Global Disease Needs" Molecules 26, no. 13: 3800. https://doi.org/10.3390/molecules26133800