Biotransformation of Timosaponin BII into Seven Characteristic Metabolites by the Gut Microbiota
Abstract
:1. Introduction
2. Results
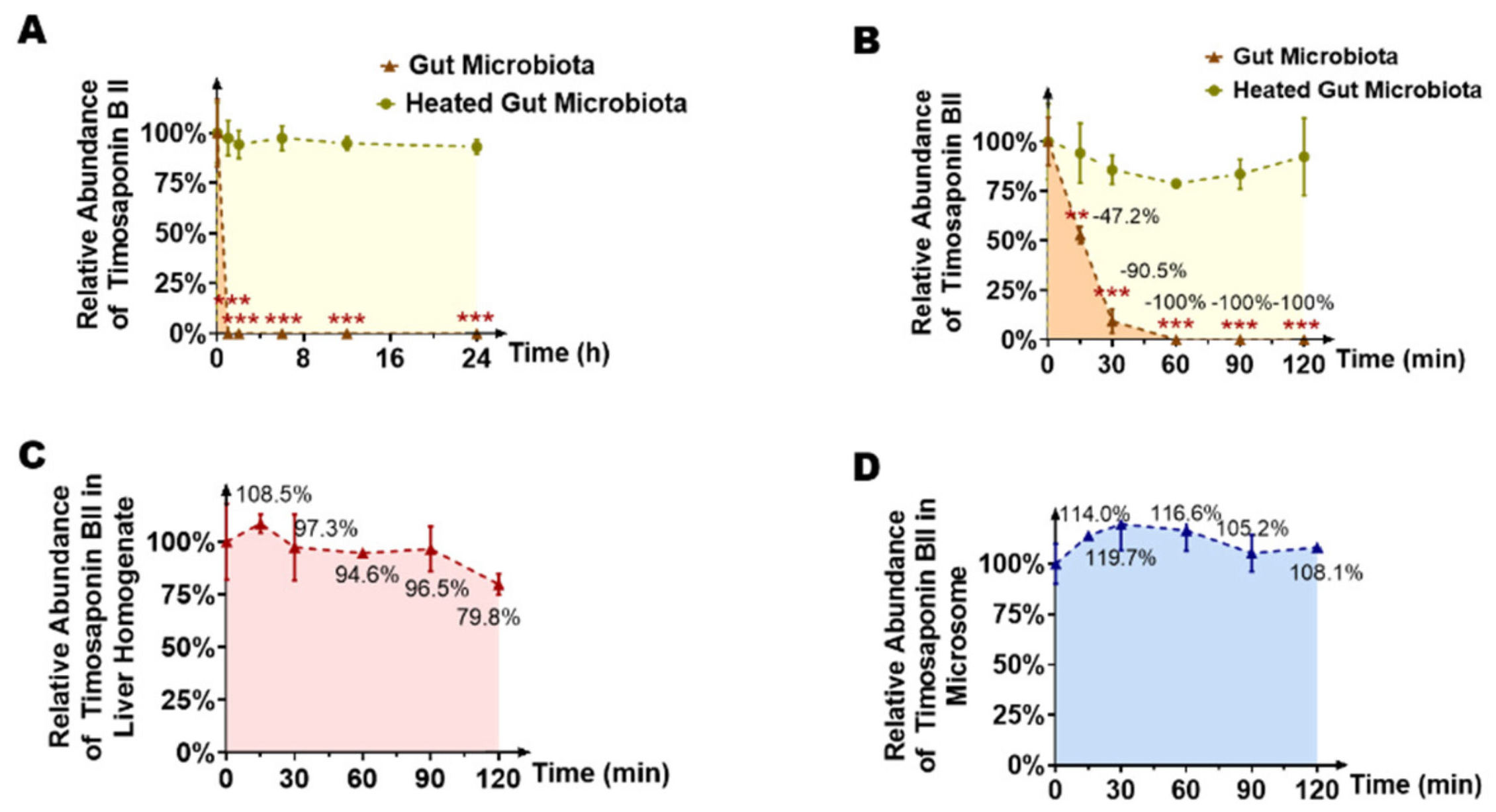
2.1. Timosaponin BII Was Significantly Metabolized by the Gut Microbiota
2.2. Timosaponin BII Was Transformed to Seven Metabolites by the Gut Microbiota
2.3. Identification of Seven Gut Microbiota-Metabolites by HPLC/MS-Q-TOF
2.4. Temporal Characteristics of Main Timosaponin BII Metabolites (Timosaponin AIII, Timosaponin AI and Sarsasapogenin) by Gut Microbiota
3. Discussion
4. Materials and Methods
4.1. Instruments and Reagents
4.2. Animals
4.3. Determination of Timosaponin BII by LC-MS/MS
4.4. Identification of the Metabolites of Timosaponin BII by LC/MS-Q-TOF
4.5. In Vitro Incubation of Timosaponin BII with Gut Microbiota
4.6. In Vitro Incubation of Timosaponin BII with Liver Microsomes and Liver Homogenate
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Ni, Y.; Gong, X.G.; Lu, M.; Chen, H.M.; Wang, Y. Mitochondrial ROS burst as an early sign in sarsasapogenin-induced apoptosis in HepG2 cells. Cell Biol. Int. 2008, 32, 337–343. [Google Scholar] [CrossRef]
- Kang, Y.J.; Chung, H.J.; Nam, J.W.; Park, H.J.; Seo, E.K.; Kim, Y.S.; Lee, D.; Lee, S.K. Cytotoxic and antineoplastic activity of timosaponin A-III for human colon cancer cells. J. Nat. Prod. 2011, 74, 701–706. [Google Scholar] [CrossRef]
- Cong, Y.; Wang, L.; Peng, R.; Zhao, Y.; Bai, F.; Yang, C.; Liu, X.; Wang, D.; Ma, B.; Cong, Y. Timosaponin AIII induces antiplatelet and antithrombotic activity via Gq-mediated signaling by the thromboxane A2 receptor. Sci. Rep. 2016, 6, 38757. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.L.; Guo, C.R.; Cui, L.L.; Ruan, S.X.; Zhang, C.F.; Ji, D.; Yang, Z.L.; Li, F. Timosaponin B-II ameliorates diabetic nephropathy via TXNIP, mTOR, and NF-κB signaling pathways in alloxan-induced mice. Drug Des. Devel. Ther. 2015, 9, 6247–6258. [Google Scholar]
- Ren, L.X.; Luo, Y.F.; Li, X.; Wu, Y.L. Antidepressant activity of sarsasapogenin from Anemarrhena asphodeloides Bunge (Liliaceae). Pharmazie 2007, 62, 78–79. [Google Scholar] [PubMed]
- Li, T.J.; Qiu, Y.; Yang, P.Y.; Rui, Y.C.; Chen, W.S. Timosaponin B-II improves memory and learning dysfunction induced by cerebral ischemia in rats. Neurosci. Lett. 2007, 421, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Jung, K.; Kim, D.H. Timosaponin AIII, a saponin isolated from Anemarrhena asphodeloides, ameliorates learning and memory deficits in mice. Pharmacol. Biochem. Behav. 2009, 93, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dan, Y.; Yang, D.; Hu, Y.; Zhang, L.; Zhang, C.; Zhu, H.; Cui, Z.; Li, M.; Liu, Y. The genus Anemarrhena Bunge: A review on ethnopharmacology, phytochemistry and pharmacology. J. Ethnopharmacol. 2014, 153, 42–60. [Google Scholar] [CrossRef]
- Nakashima, N.; Kimura, I.; Kimura, M.; Matsuura, H. Isolation of pseudoprototimosaponin AIII from rhizomes of Anemarrhena asphodeloides and its hypoglycemic activity in streptozotocin-induced diabetic mice. J. Nat. Prod. 1993, 56, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Ou, G.; Chen, W.; Yang, M.; Li, Q.; Zhang, Y.; Wan, D.; Du, L.; Ma, B. Preventive effect of nasal Timosaponin BII-loaded temperature-/ion-sensitive in situ hydrogels on Alzheimer’s disease. J. Tradit. Chin. Med Sci. 2021, 8, 59–64. [Google Scholar] [CrossRef]
- Lu, W.-Q.; Qiu, Y.; Li, T.-J.; Tao, X.; Sun, L.-N.; Chen, W.-S. Antiplatelet and antithrombotic activities of timosaponin B-II, an extract of Anemarrhena asphodeloides. Clin. Exp. Pharmacol. Physiol. 2011, 38, 430–434. [Google Scholar] [CrossRef]
- Lu, W.-Q.; Qiu, Y.; Li, T.-J.; Tao, X.; Sun, L.-N.; Chen, W.-S. Timosaponin B-II inhibits pro-inflammatory cytokine induction by lipopolysaccharide in BV2 cells. Arch. Pharmacal Res. 2009, 32, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Cai, F.; Sun, L.; Gao, S.; Yang, Y.; Yang, Q.; Chen, W. A rapid and sensitive liquid chromatography-tandem mass spectrometric method for the determination of timosaponin B-II in blood plasma and a study of the pharmacokinetics of saponin in the rat. J. Pharm. Biomed. Anal. 2008, 48, 1411–1416. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Fu, Z.; Li, Z.; Hu, P.; Xue, R.; Chen, M.; Xiang, T.; Huang, C. In-vivo and In-vitro Metabolism Study of Timosaponin B-II Using HPLC-ESI-MSn. Chromatographia 2015, 78, 1175–1184. [Google Scholar] [CrossRef]
- Illiano, P.; Brambilla, R.; Parolini, C. The mutual interplay of gut microbiota, diet and human disease. FEBS J. 2019, 287, 833–855. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Saha, S.; Khanna, S. Therapies to modulate gut microbiota: Past, present and future. World J. Gastroenterol. 2020, 26, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-T.; Huang, H.-L.; Li, Y.-Q.; Xu, H.-M.; Zhou, Y.-J. Therapeutic advances in non-alcoholic fatty liver disease: A microbiota-centered view. World J. Gastroenterol. 2020, 26, 1901–1911. [Google Scholar] [CrossRef]
- Al Bander, Z.; Nitert, M.D.; Mousa, A.; Naderpoor, N. The Gut Microbiota and Inflammation: An Overview. Int. J. Environ. Res. Public Health 2020, 17, 7618. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Song, X.; Khan, S.; Li, Y.; Guo, Z.; Li, C.; Wang, S.; Dong, W.; Liu, W.; Wang, B.; et al. The gut microbiota at the intersection of bile acids and intestinal carcinogenesis: An old story, yet mesmerizing. Int. J. Cancer 2020, 146, 1780–1790. [Google Scholar] [CrossRef]
- Wang, Y.; Tong, Q.; Ma, S.-R.; Zhao, Z.-X.; Pan, L.-B.; Cong, L.; Han, P.; Peng, R.; Yu, H.; Lin, Y.; et al. Oral berberine improves brain dopa/dopamine levels to ameliorate Parkinson’s disease by regulating gut microbiota. Signal Transduct. Target. Ther. 2021, 6, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Han, P.; Ma, S.; Peng, R.; Wang, C.; Kong, W.; Cong, L.; Fu, J.; Zhang, Z.; Yu, H.; et al. Abnormal metabolism of gut microbiota reveals the possible molecular mechanism of nephropathy induced by hyperuricemia. Acta Pharm. Sin. B 2020, 10, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; He, J.; Jiaojiao, H. The influence of gut microbiota on drug metabolism and toxicity. Expert Opin. Drug Metab. Toxicol. 2016, 12, 31–40. [Google Scholar] [CrossRef] [Green Version]
- Peng, R.; Ma, S.-R.; Fu, J.; Han, P.; Pan, L.-B.; Zhang, Z.-W.; Yu, H.; Wang, Y. Transforming of Triptolide into Characteristic Metabolites by the Gut Microbiota. Molecules 2020, 25, 606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, I.D.; Nicholson, J. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl. Res. 2017, 179, 204–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.B.; Zhao, Z.X.; Peng, R.; Pan, L.B.; Fu, J.; Ma, S.R.; Han, P.; Cong, L.; Zhang, Z.W.; Sun, L.X.; et al. Gut Microbiota-Based Pharmacokinetics and the Antidepressant Mechanism of Paeoniflorin. Front Pharmacol. 2019, 10, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.X.; Fu, J.; Ma, S.R.; Peng, R.; Yu, J.B.; Cong, L.; Pan, L.B.; Zhang, Z.G.; Tian, H.; Che, C.T.; et al. Gut-brain axis metabolic pathway regulates antidepressant efficacy of albiflorin. Theranostics 2018, 8, 5945–5959. [Google Scholar] [CrossRef]
- Zhao, Y.; Jiang, T.; Han, B.; Lu, L.; Feng, B.; Kang, L.; Zhang, J.; Fang, Y.; Xiong, C.; Jia, D. Preparation of some metabolites of timosaponin BII by biotransformation in vitro. Process Biochem. 2015, 50, 2182–2187. [Google Scholar] [CrossRef]
- Hu, Y.-M.; Yu, Z.-L.; Fong, W.-F. Stereoselective biotransformation of timosaponin A-III by Saccharomyces cerevisiae. J. Microbiol. Biotechnol. 2011, 21, 582–589. [Google Scholar] [CrossRef]
- Zhang, M.; Qu, J.; Gao, Z.; Qi, Q.; Yin, H.; Zhu, L.; Wu, Y.; Liu, W.; Yang, J.; Huang, X. Timosaponin AIII Induces G2/M Arrest and Apoptosis in Breast Cancer by Activating the ATM/Chk2 and p38 MAPK Signaling Pathways. Front Pharmacol. 2020, 11, 601468. [Google Scholar] [CrossRef]
- Sy, L.K.; Yan, S.C.; Lok, C.N.; Man, R.Y.; Che, C.M. Timosaponin A-III induces autophagy preceding mitochondria-mediated apoptosis in HeLa cancer cells. Cancer Res. 2008, 68, 10229–10237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, B.P.; Feng, B.; Huang, H.Z.; Cong, Y.W. Biotransformation of Chinese Herbs and Their Ingredients. World Sci. Technol. 2010, 12, 150–154. [Google Scholar]
- Wang, S.; Huai, J.; Shang, Y.; Xie, L.; Cao, X.; Liao, J.; Zhang, T.; Dai, R. Screening for natural inhibitors of 5-lipoxygenase from Zi-shen pill extract by affinity ultrafiltration coupled with ultra performance liquid chromatography-mass spectrometry. J. Ethnopharmacol. 2020, 254, 112733. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.L.; Wang, Y.; Zhao, X.P. Screening and identification of DPP-4 inhibitors from Xiaokean formula by a fluorescent probe. Zhongguo Zhong Yao Za Zhi 2016, 41, 1241–1245. [Google Scholar]
- Ren, L.X.; Luo, Y.F.; Li, X.; Zuo, D.Y.; Wu, Y.L. Antidepressant-like effects of sarsasapogenin from Anemarrhena asphodeloides BUNGE (Liliaceae). Biol. Pharm. Bull 2006, 29, 2304–2306. [Google Scholar] [CrossRef] [Green Version]
- Hidalgo, M.; Oruna-Concha, M.J.; Kolida, S.; Walton, G.E.; Kallithraka, S.; Spencer, J.P.; de Pascual-Teresa, S. Metabolism of anthocyanins by human gut microflora and their influence on gut bacterial growth. J. Agric. Food Chem. 2012, 60, 3882–3890. [Google Scholar] [CrossRef]
- Michlmayr, H.; Kneifel, W. β-Glucosidase activities of lactic acid bacteria: Mechanisms, impact on fermented food and human health. FEMS Microbiol. Lett. 2014, 352, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Bang, S.H.; Hyun, Y.J.; Shim, J.; Hong, S.W.; Kim, D.H. Metabolism of rutin and poncirin by human intestinal microbiota and cloning of their metabolizing α-L-rhamnosidase from Bifidobacterium dentium. J. Microbiol. Biotechnol. 2015, 25, 18–25. [Google Scholar] [CrossRef] [Green Version]
- Riva, A.; Kolimár, D.; Spittler, A.; Wisgrill, L.; Herbold, C.W.; Abrankó, L.; Berry, D. Conversion of Rutin, a Prevalent Dietary Flavonol, by the Human Gut Microbiota. Front Microbiol. 2020, 11, 585428. [Google Scholar] [CrossRef] [PubMed]
- Sellick, C.A.; Campbell, R.N.; Reece, R.J. Chapter 3 Galactose Metabolism in Yeast—Structure and Regulation of the Leloir Pathway Enzymes and the Genes Encoding Them. In International Review of Cell and Molecular Biology; Academic Press: Cambridge, MA, USA, 2008; Volume 269, pp. 111–150. [Google Scholar]
- Liu, Z.; Zhu, D.; Lv, L.; Li, Y.; Dong, X.; Zhu, Z.; Chai, Y. Metabolism profile of timosaponin B-II in urine after oral administration to rats by ultrahigh-performance liquid chromatography/quadrupole-time-of-flight mass spectrometry. Rapid. Commun. Mass Spectrom. 2012, 26, 1955–1964. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Hong, S.; Yang, P.; Sun, Y.; Wang, Y.; Zhang, P.; Jiang, W.; Gu, Y. Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria. Nat. Commun. 2021, 12, 790. [Google Scholar] [CrossRef]
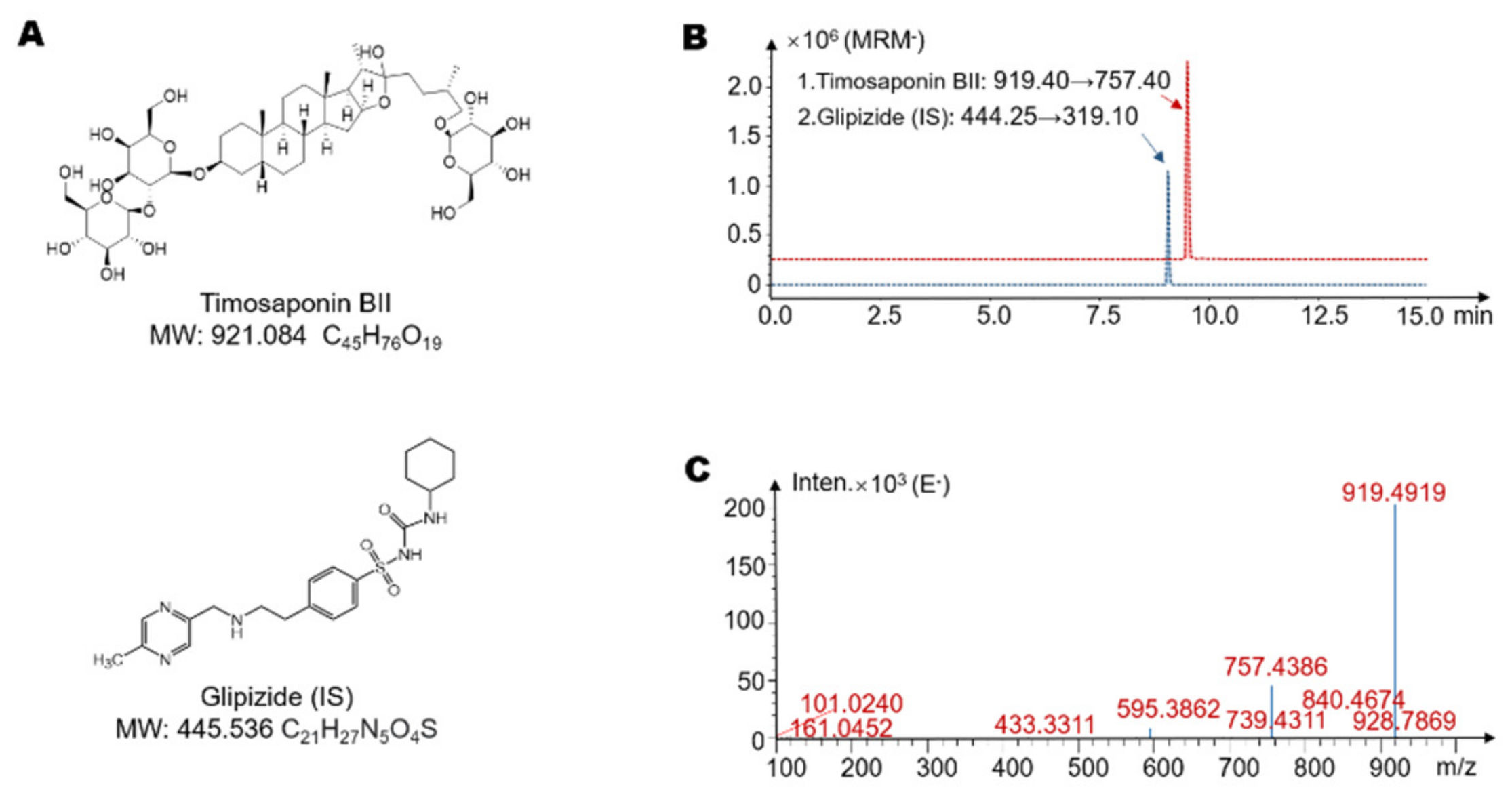
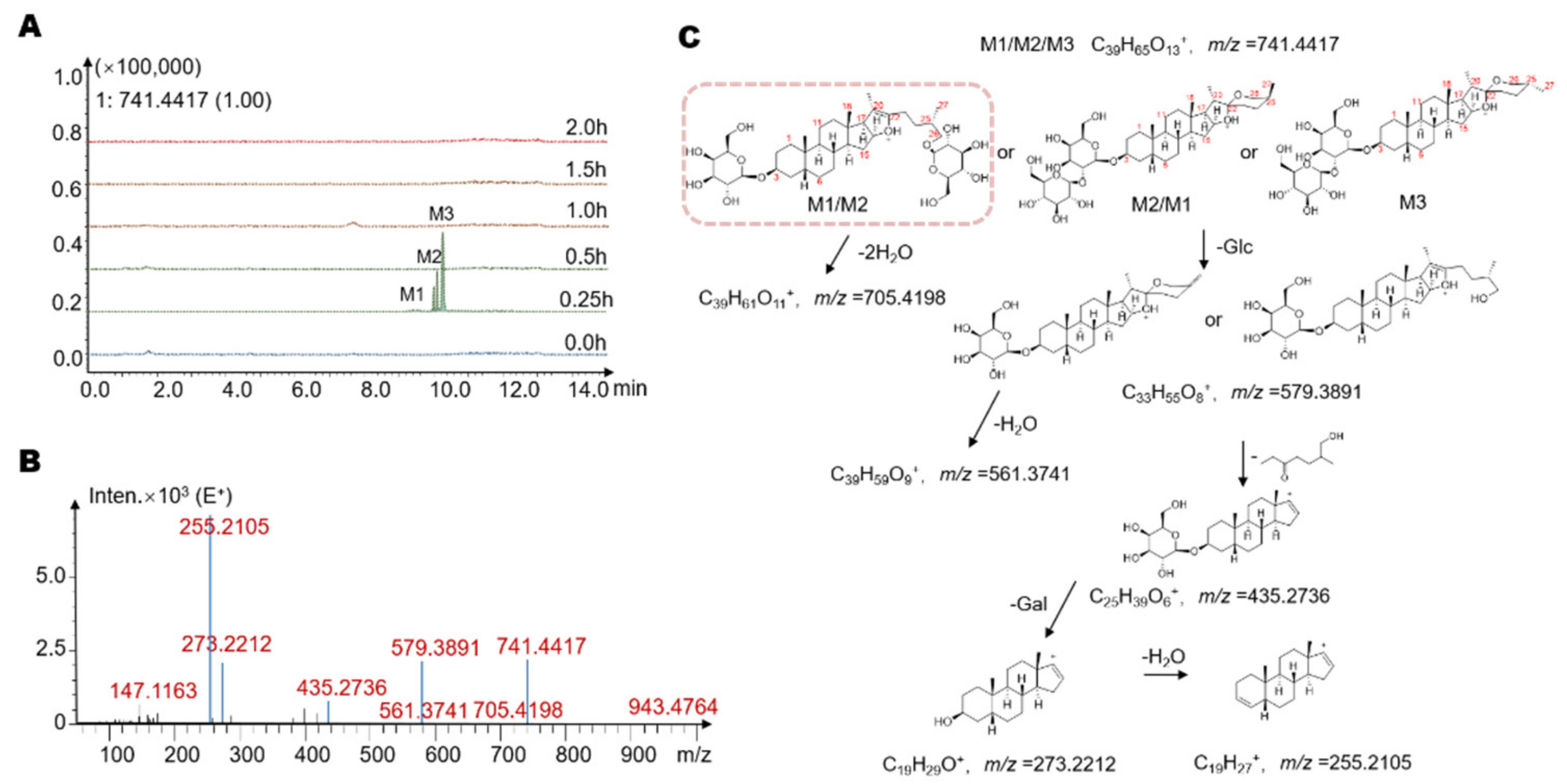
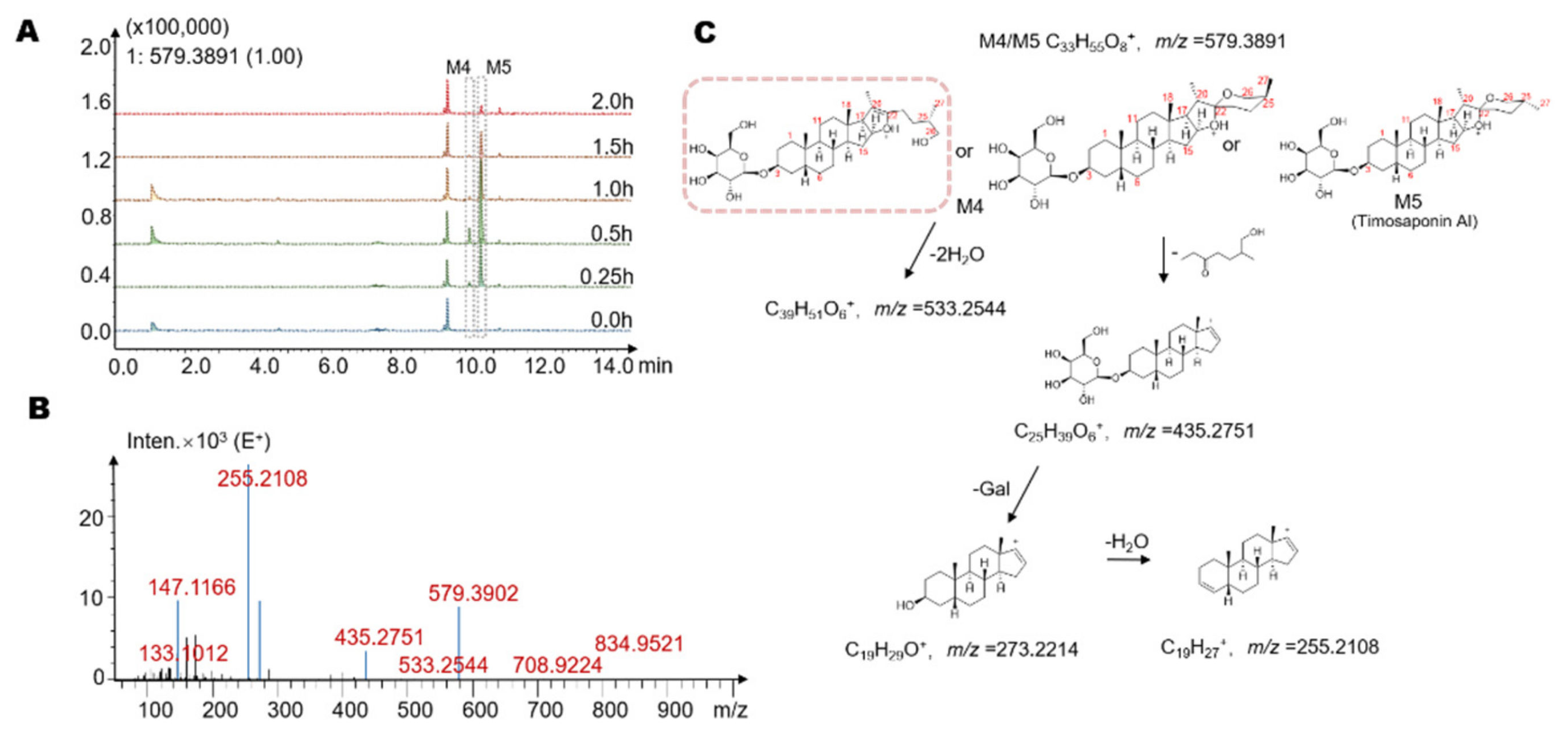
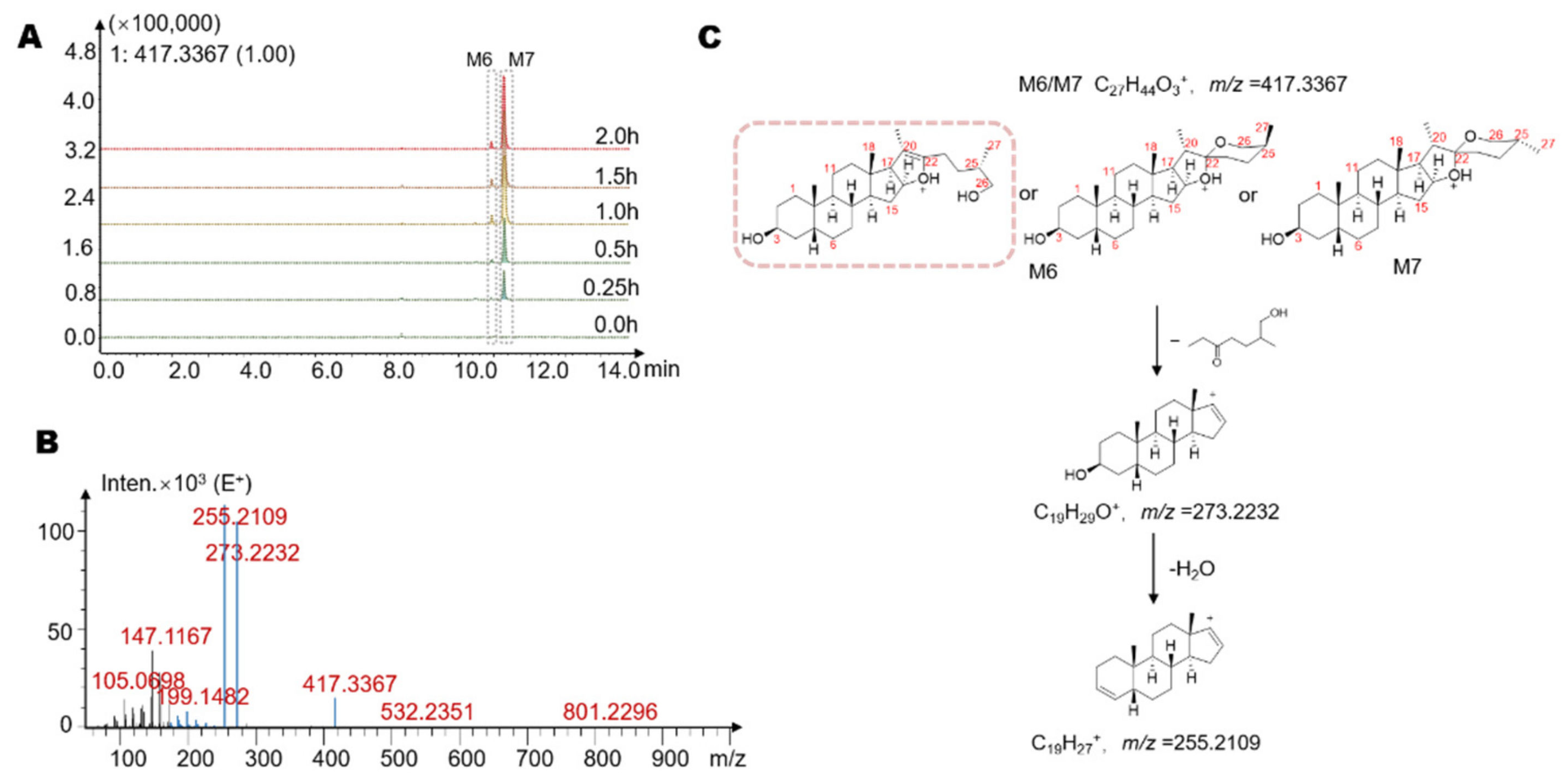
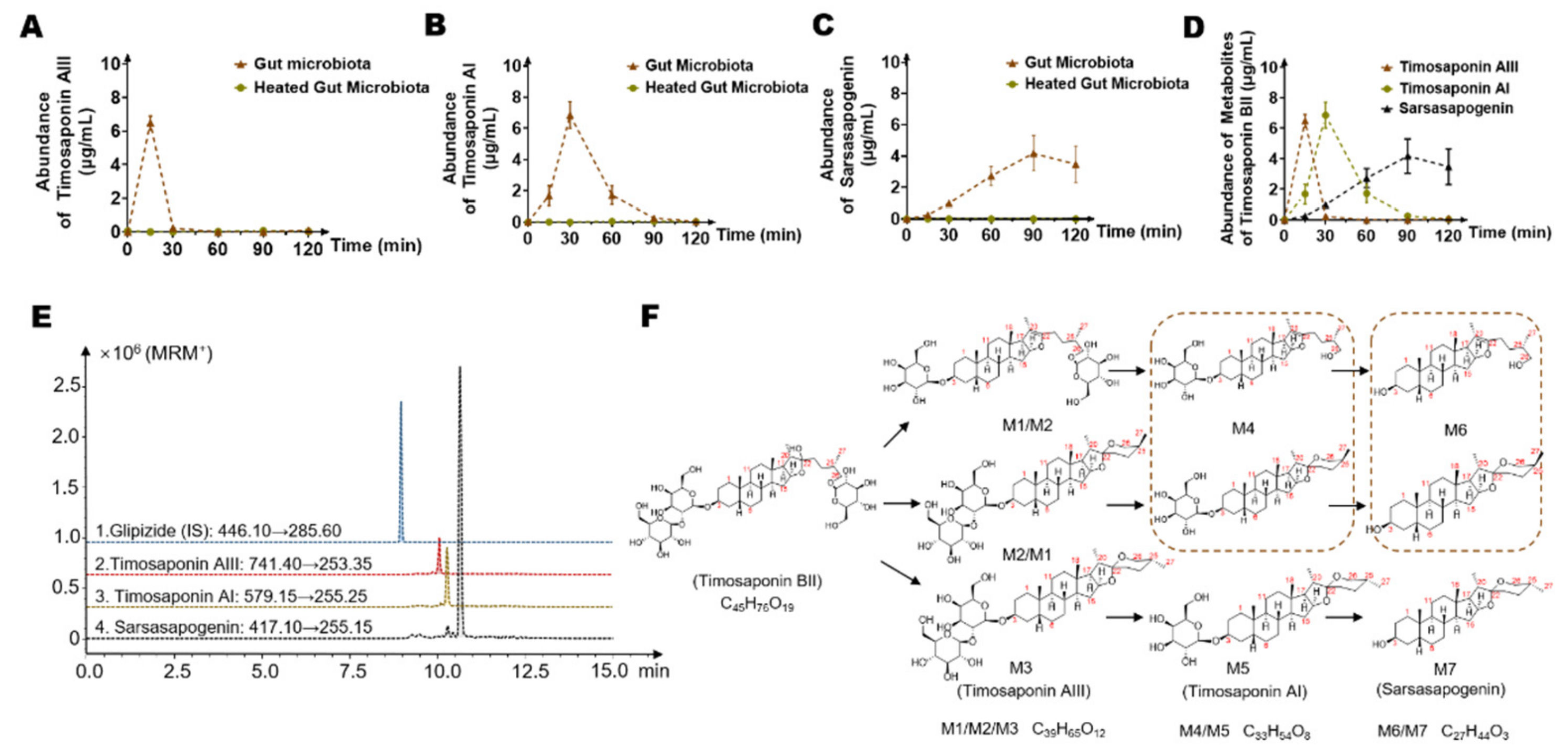
| Metabolites | Retention Time (min) | Reaction * | Predicted Molecular Weight | Molecular Formula | Fragment Characteristics | |
|---|---|---|---|---|---|---|
| MS1/[M + H]+ | MS/MS | |||||
| M1 | 9.9 | −Glc ** | 740.93 | C39H65O12 | 741 | 705, 579, 561, 435, 273, 255 |
| M2 | 10.1 | −Glc | 740.93 | C39H65O12 | 741 | 705, 579, 561, 435, 273, 255 |
| M3 | 10.2 | −Glc | 740.93 | C39H65O12 | 741 | 705, 579, 561, 435, 273, 255 |
| M4 | 10.3 | −2Glc | 578.38 | C33H54O8 | 579 | 533, 435, 273, 255 |
| M5 | 10.6 | −2Glc | 578.38 | C33H54O8 | 579 | 533, 435, 273, 255 |
| M6 | 10.9 | −2Glc-Gal *** | 416.65 | C27H44O3 | 417 | 273, 255 |
| M7 | 11.3 | −2Glc-Gal | 416.65 | C27H44O3 | 417 | 273, 255 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, G.-M.; Yu, H.; Pan, L.-B.; Ma, S.-R.; Xu, H.; Zhang, Z.-W.; Han, P.; Fu, J.; Yang, X.-Y.; Keranmu, A.; et al. Biotransformation of Timosaponin BII into Seven Characteristic Metabolites by the Gut Microbiota. Molecules 2021, 26, 3861. https://doi.org/10.3390/molecules26133861
Dong G-M, Yu H, Pan L-B, Ma S-R, Xu H, Zhang Z-W, Han P, Fu J, Yang X-Y, Keranmu A, et al. Biotransformation of Timosaponin BII into Seven Characteristic Metabolites by the Gut Microbiota. Molecules. 2021; 26(13):3861. https://doi.org/10.3390/molecules26133861
Chicago/Turabian StyleDong, Guo-Ming, Hang Yu, Li-Bin Pan, Shu-Rong Ma, Hui Xu, Zheng-Wei Zhang, Pei Han, Jie Fu, Xin-Yu Yang, Adili Keranmu, and et al. 2021. "Biotransformation of Timosaponin BII into Seven Characteristic Metabolites by the Gut Microbiota" Molecules 26, no. 13: 3861. https://doi.org/10.3390/molecules26133861
APA StyleDong, G.-M., Yu, H., Pan, L.-B., Ma, S.-R., Xu, H., Zhang, Z.-W., Han, P., Fu, J., Yang, X.-Y., Keranmu, A., Niu, H.-T., Jiang, J.-D., & Wang, Y. (2021). Biotransformation of Timosaponin BII into Seven Characteristic Metabolites by the Gut Microbiota. Molecules, 26(13), 3861. https://doi.org/10.3390/molecules26133861





