Analysis of Volatile Molecules Present in the Secretome of the Fungal Pathogen Candida glabrata
Abstract
:1. Introduction
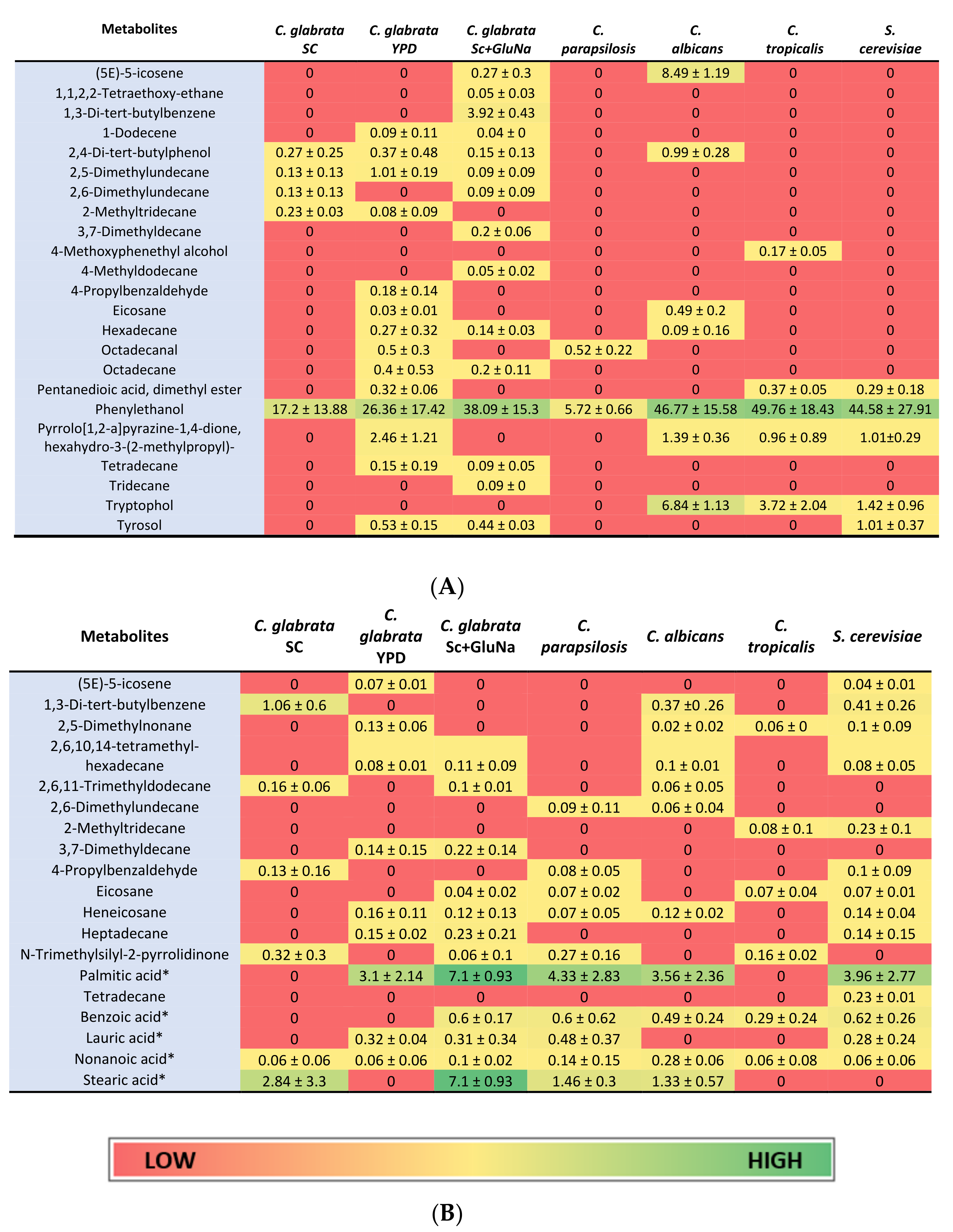
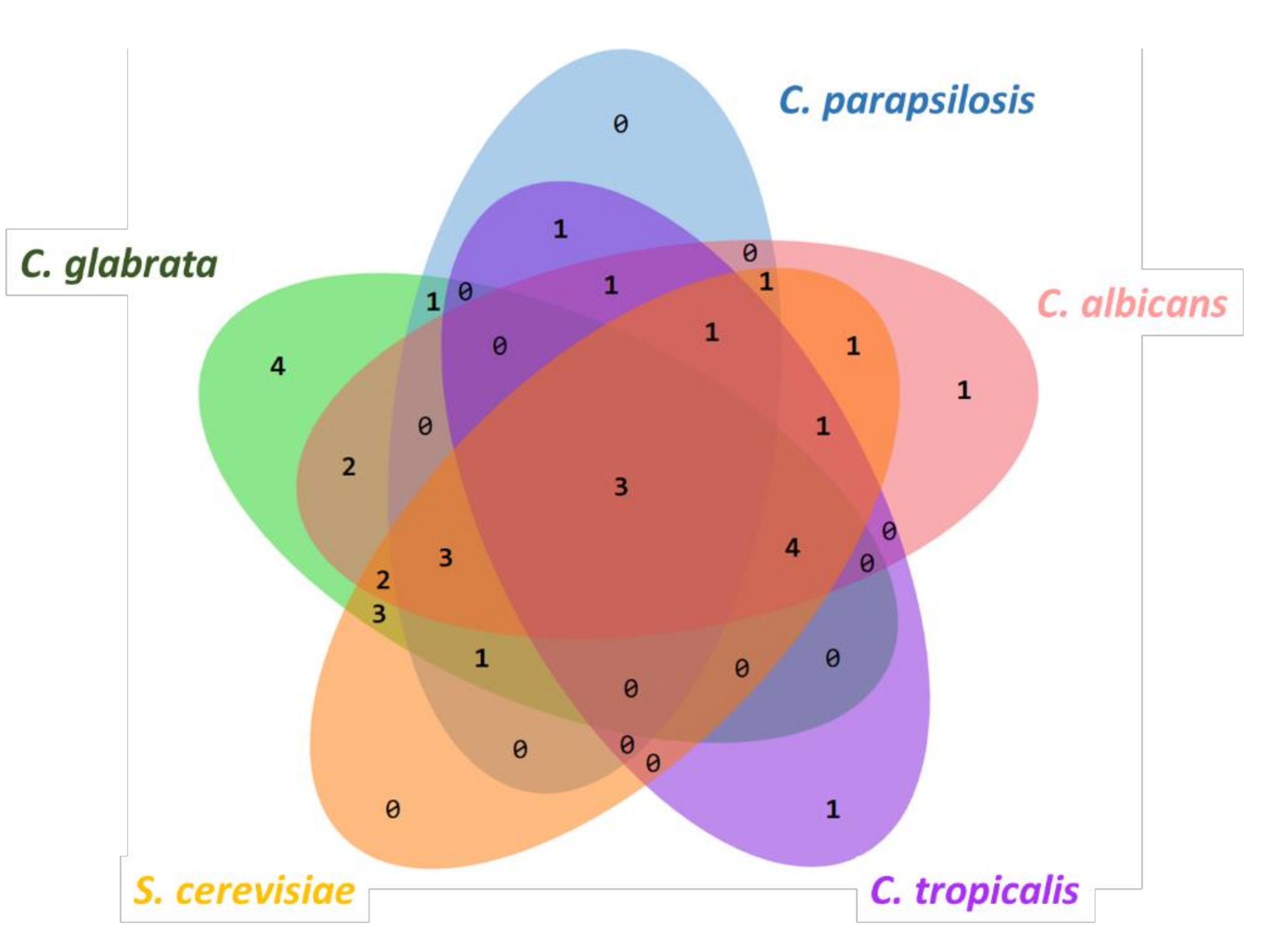
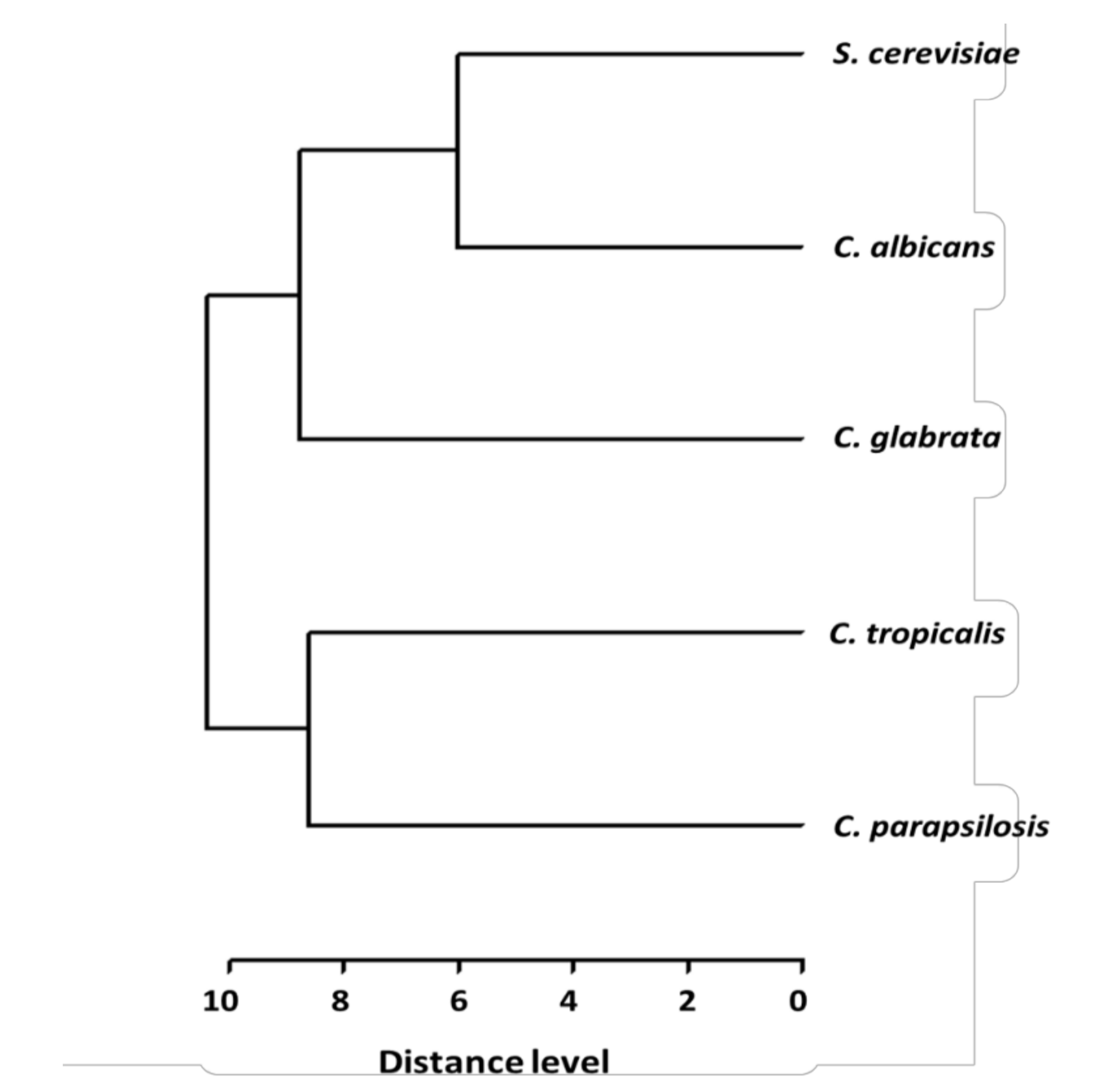
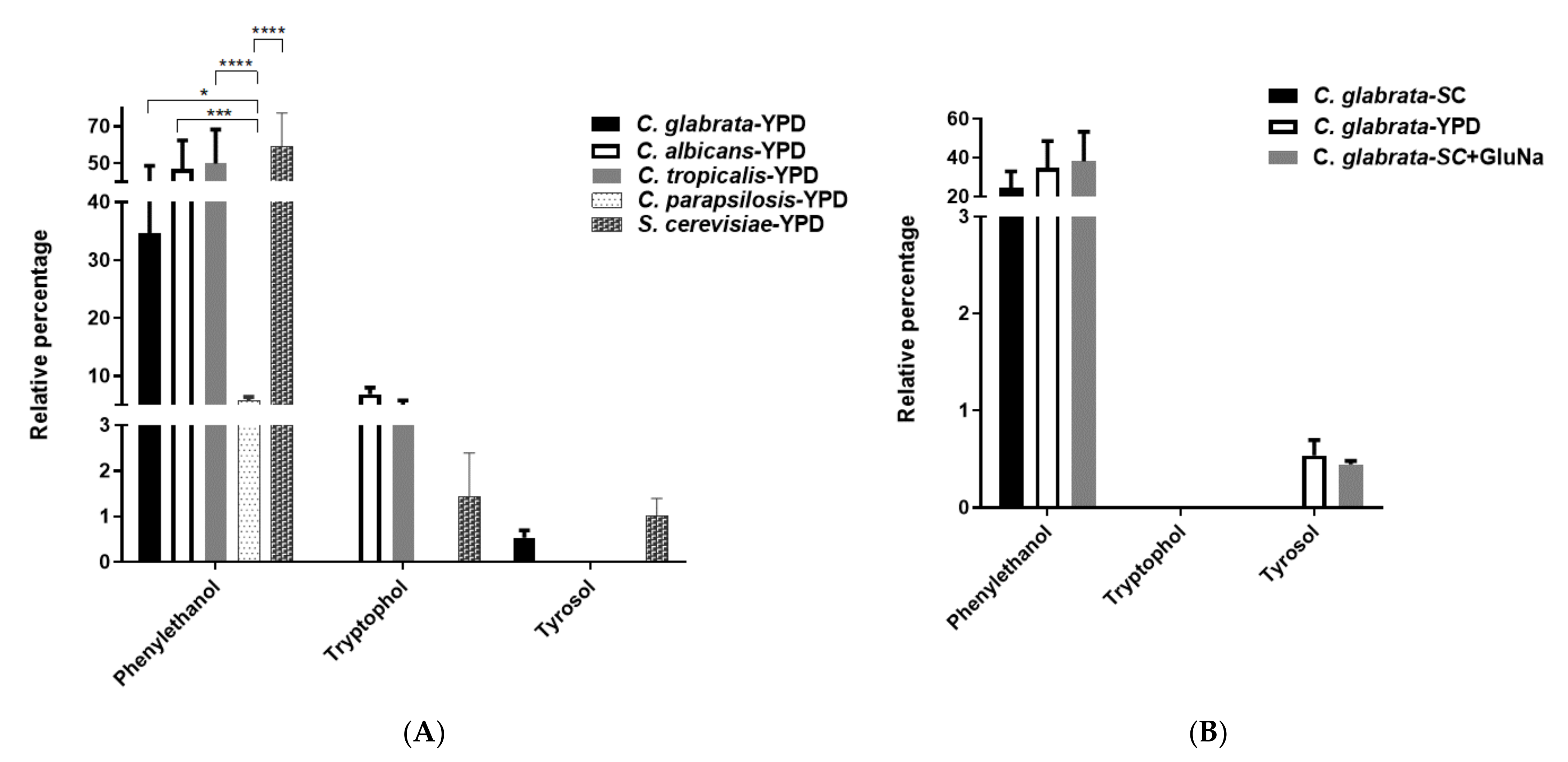
2. Results
3. Discussion
4. Materials and Methods
4.1. Strains
| Strains | Source of Reference | ||
|---|---|---|---|
| C. glabrata | BG14 | ura3Δ:Tn903 Neo R Ura- | [39] |
| C. parapsilosis | ATCC-22019 | Clinical Isolate | Lab. collection |
| C. tropicalis | ATCC-750 | Clinical Isolate | Lab. collection |
| C. albicans Sc5314 | ATCC MYA2876 | Clinical Isolate | Lab. collection |
| S. cerevisiae S288c | ATCC 504208 | MATα SUC2 mal mel gal2 CUP1 | Lab. collection |
4.2. Media and Growth Conditions
4.3. Sample Preparation for GC-MS Analysis
4.4. GC-MS Spectrometry Analysis
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sardi, J.C.O.; Scorzoni, L.; Bernardi, T.; Fusco-Almeida, A.M.; Mendes Giannini, M.J.S. Candida Species: Current Epidemiology, Pathogenicity, Biofilm Formation, Natural Antifungal Products and New Therapeutic Options. J. Med. Microbiol. 2013, 62, 10–24. [Google Scholar] [CrossRef]
- Toda, M.; Williams, S.R.; Berkow, E.L.; Farley, M.M.; Harrison, L.H.; Bonner, L.; Marceaux, K.M.; Hollick, R.; Zhang, A.Y.; Schaffner, W.; et al. Population-Based Active Surveillance for Culture-Confirmed Candidemia—Four Sites, United States, 2012–2016. MMWR. Surveill. Summ. 2019, 68, 1–15. [Google Scholar] [CrossRef]
- Honigberg, S.M. Cell Signals, Cell Contacts, and the Organization of Yeast Communities. Eukaryot. Cell 2011, 10, 466–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Fink, G.R. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 2006, 20, 1150–1161. [Google Scholar] [CrossRef] [Green Version]
- Yashiroda, Y.; Yoshida, M. Intraspecies Cell-Cell Communication in Yeast. FEMS Yeast Res. 2019, 19. [Google Scholar] [CrossRef]
- Deveau, A.; Piispanen, A.E.; Jackson, A.A.; Hogan, D.A. Farnesol Induces Hydrogen Peroxide Resistance in Candida Albicans Yeast by Inhibiting the Ras-Cyclic AMP Signaling Pathway. Eukaryot. Cell 2010, 9, 569–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pappas, P.G.; Lionakis, M.S.; Arendrup, M.C.; Ostrosky-Zeichner, L.; Kullberg, B.J. Invasive Candidiasis. Nat. Rev. Dis. Prim. 2018, 4, 18026. [Google Scholar] [CrossRef]
- Baddley, J.W.; Patel, M.; Bhavnani, S.M.; Moser, S.A.; Andes, D.R. Association of Fluconazole Pharmacodynamics With Mortality in Patients With Candidemia. Antimicrob. Agents Chemother. 2008, 52, 3022–3028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castaño, I.; Cormack, B.; De Las Peñas, A. Virulence of the Opportunistic Pathogen Mushroom Candida glabrata. Rev. Latinoam. Microbiol. 2007, 48, 66–69. [Google Scholar]
- Rodrigues, C.F.; Rodrigues, M.E.; Silva, S.C.; Henriques, M. Candida Glabrata Biofilms: How Far Have We Come? J. Fungi 2017, 3, 11. [Google Scholar] [CrossRef] [Green Version]
- Riera, M.; Mogensen, E.; D’Enfert, C.; Janbon, G. New Regulators of Biofilm Development in Candida Glabrata. Res. Microbiol. 2012, 163, 297–307. [Google Scholar] [CrossRef]
- Kołaczkowska, A.; Kołaczkowski, M. Drug Resistance Mechanisms and Their Regulation in Non-Albicans Candidaspecies. J. Antimicrob. Chemother. 2016, 71, 1438–1450. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, S.; Sae-Tia, S.; Fries, B.C. Candidiasis and Mechanisms of Antifungal Resistance. Antibiotics 2020, 9, 312. [Google Scholar] [CrossRef]
- Briones-Martin-Del-Campo, M.; Orta-Zavalza, E.; Juarez-Cepeda, J.; Gutierrez-Escobedo, G.; Cañas-Villamar, I.; Castaño, I.; Peñas, A.D.L. The Oxidative Stress Response of the Opportunistic Fungal Pathogen Candida Glabrata. Rev. Iberoam. Micol. 2014, 31, 67–71. [Google Scholar] [CrossRef]
- Gutiérrez-Escobedo, G.; Orta-Zavalza, E.; Castaño, I.; Peñas, A.D.L. Role of Glutathione in the Oxidative Stress Response in the Fungal Pathogen Candida Glabrata. Curr. Genet. 2013, 59, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, M.; Kumar, N.; Kaur, R. Global Secretome Characterization of the Pathogenic Yeast Candida Glabrata. J. Proteome Res. 2019, 19, 49–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliver, J.C.; Laghi, L.; Parolin, C.; Foschi, C.; Marangoni, A.; Liberatore, A.; Dias, A.L.T.; Cricca, M.; Vitali, B. Metabolic Profiling of Candida Clinical Isolates of Different Species and Infection Sources. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Semreen, M.H.; Soliman, S.S.M.; Saeed, B.Q.; Alqarihi, A.; Uppuluri, P.; Ibrahim, A.S. Metabolic Profiling of Candida Auris, a Newly-Emerging Multi-Drug Resistant Candida Species, by GC-MS. Molecules 2019, 24, 399. [Google Scholar] [CrossRef] [Green Version]
- Costa, C.P.; Bezerra, A.R.; Almeida, A.; Rocha, S.M. Candida Species (Volatile) Metabotyping through Advanced Comprehensive Two-Dimensional Gas Chromatography. Microorganisms 2020, 8, 1911. [Google Scholar] [CrossRef]
- Škrbić, B.; Durisic-Mladenovic, N.; Cvejanov, J. Principal Component Analysis of Trace Elements in Serbian Wheat. J. Agric. Food Chem. 2005, 53, 2171–2175. [Google Scholar] [CrossRef]
- Škrbić, B.D.; Buljovčić, M.; Jovanović, G.; Antić, I. Seasonal, Spatial Variations and Risk Assessment of Heavy Elements in Street Dust from Novi Sad, Serbia. Chemosphere 2018, 205, 452–462. [Google Scholar] [CrossRef] [PubMed]
- Deorukhkar, S.C.; Saini, S.; Mathew, S. Non-Albicans CandidaInfection: An Emerging Threat. Interdiscip. Perspect. Infect. Dis. 2014, 2014, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Vazquez, J.A.; Dembry, L.M.; Sanchez, V.; Vazquez, M.A.; Sobel, J.D.; Dmuchowski, C.; Zervos, M.J. Nosocomial Candida Glabrata Colonization: An Epidemiologic Study. J. Clin. Microbiol. 1998, 36, 421–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, S.-Y.; Lee, L.-N.; Jerng, J.-S.; Yu, C.-J.; Hsueh, P.-R. Candida Glabrata Fungaemia in Intensive Care Units. Clin. Microbiol. Infect. 2008, 14, 136–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mashaly, G.; Shrief, R. Candida Glabrata Complex from Patients With Healthcare-Associated Infections in Mansoura University Hospitals, Egypt: Distribution, Antifungal Susceptibility and Effect of Fluconazole and Polymyxin B Combination. Germs 2019, 9, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Marcet-Houben, M.; Gabaldón, T. The Tree Versus the Forest: The Fungal Tree of Life and the Topological Diversity Within the Yeast Phylome. PLoS ONE 2009, 4, e4357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, S.; Kebaara, B.W.; Atkin, A.L.; Nickerson, K.W. Regulation of Aromatic Alcohol Production in Candida Albicans. Appl. Environ. Microbiol. 2008, 74, 7211–7218. [Google Scholar] [CrossRef] [Green Version]
- Hazelwood, L.A.; Daran, J.-M.; van Maris, A.J.A.; Pronk, J.T.; Dickinson, J.R. The Ehrlich Pathway for Fusel Alcohol Production: A Century of Research on Saccharomyces Cerevisiae Metabolism. Appl. Environ. Microbiol. 2008, 74, 2259–2266. [Google Scholar] [CrossRef] [Green Version]
- Etschmann, M.M.W.; Bluemke, W.; Sell, D.; Schrader, J. Biotechnological production of 2-phenylethanol. Appl. Microbiol. Biotechnol. 2002, 59, 1–8. [Google Scholar] [CrossRef]
- Lingappa, B.T.; Prasad, M.; Lingappa, Y.; Hunt, D.F.; Biemann, K. Phenethyl Alcohol and Tryptophol: Autoantibiotics Produced by the Fungus Candida Albicans. Science 1969, 163, 192–194. [Google Scholar] [CrossRef]
- Martins, M.; Henriques, M.; Azeredo, J.; Rocha, S.M.; Coimbra, M.A.; Oliveira, R. Morphogenesis Control in Candida Albicans and Candida Dubliniensis through Signaling Molecules Produced by Planktonic and Biofilm Cells. Eukaryot. Cell 2007, 6, 2429–2436. [Google Scholar] [CrossRef] [Green Version]
- Los Santos, F.J.P.-D.; García-Ortega, L.F.; Robledo-Márquez, K.; Guzmán-Moreno, J.; Riego-Ruiz, J.G.-M.A.L. Transcriptome Analysis Unveils Gln3 Role in Amino Acids Assimilation and Fluconazole Resistance in Candida Glabrata. J. Microbiol. Biotechnol. 2021, 31, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Aneja, M.; Gianfagna, T.J.; Hebbar, P.K. Trichoderma Harzianum Produces Nonanoic Acid, an Inhibitor of Spore Germination and Mycelial Growth of Two Cacao Pathogens. Physiol. Mol. Plant Pathol. 2005, 67, 304–307. [Google Scholar] [CrossRef]
- Jang, Y.-W.; Jung, J.-Y.; Lee, I.-K.; Kang, S.-Y.; Yun, B.-S. Nonanoic Acid, an Antifungal Compound from Hibiscus Syriacus Ggoma. Mycobiology 2012, 40, 145–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavergne, F.D.; Broeckling, C.D.; Cockrell, D.M.; Haley, S.D.; Peairs, F.B.; Jahn, C.E.; Heuberger, A.L. GC-MS Metabolomics to Evaluate the Composition of Plant Cuticular Waxes for Four Triticum Aestivum Cultivars. Int. J. Mol. Sci. 2018, 19, 249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nandhini, U.S.; Sangareshwari, S.; Lata, K. Gas chromatography-mass spectrometry analysis of bioactive constituents from the marine streptomyces. Asian J. Pharm. Clin. Res. 2015, 8, 244–246. [Google Scholar]
- Cain, C.E.; Bell, O.E.; White, H.B.; Sulya, L.L.; Smith, R.R. Hydrocarbons from Human Meninges and Meningiomas. Biochim. Biophys. Acta (BBA) Lipids Lipid Metab. 1967, 144, 493–500. [Google Scholar] [CrossRef]
- Lintas, C.; Balduzzi, A.M.; Bernardini, M.P.; Di Muccio, A. Distribution of Hydrocarbons in Bovine Tissues. Lipids 1979, 14, 298–303. [Google Scholar] [CrossRef]
- Cormack, B.P.; Falkow, S. Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics 1999, 151, 979–987. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Ramos, J.E.; Bautista, E.; Gutiérrez-Escobedo, G.; Mancilla-Montelongo, G.; Castaño, I.; González-Chávez, M.M.; De Las Peñas, A. Analysis of Volatile Molecules Present in the Secretome of the Fungal Pathogen Candida glabrata. Molecules 2021, 26, 3881. https://doi.org/10.3390/molecules26133881
López-Ramos JE, Bautista E, Gutiérrez-Escobedo G, Mancilla-Montelongo G, Castaño I, González-Chávez MM, De Las Peñas A. Analysis of Volatile Molecules Present in the Secretome of the Fungal Pathogen Candida glabrata. Molecules. 2021; 26(13):3881. https://doi.org/10.3390/molecules26133881
Chicago/Turabian StyleLópez-Ramos, Juan Ernesto, Elihú Bautista, Guadalupe Gutiérrez-Escobedo, Gabriela Mancilla-Montelongo, Irene Castaño, Marco Martín González-Chávez, and Alejandro De Las Peñas. 2021. "Analysis of Volatile Molecules Present in the Secretome of the Fungal Pathogen Candida glabrata" Molecules 26, no. 13: 3881. https://doi.org/10.3390/molecules26133881
APA StyleLópez-Ramos, J. E., Bautista, E., Gutiérrez-Escobedo, G., Mancilla-Montelongo, G., Castaño, I., González-Chávez, M. M., & De Las Peñas, A. (2021). Analysis of Volatile Molecules Present in the Secretome of the Fungal Pathogen Candida glabrata. Molecules, 26(13), 3881. https://doi.org/10.3390/molecules26133881





