Heavy Vacuum Gas Oil Upregulates the Rhamnosyltransferases and Quorum Sensing Cascades of Rhamnolipids Biosynthesis in Pseudomonas sp. AK6U
Abstract
:1. Introduction
2. Results
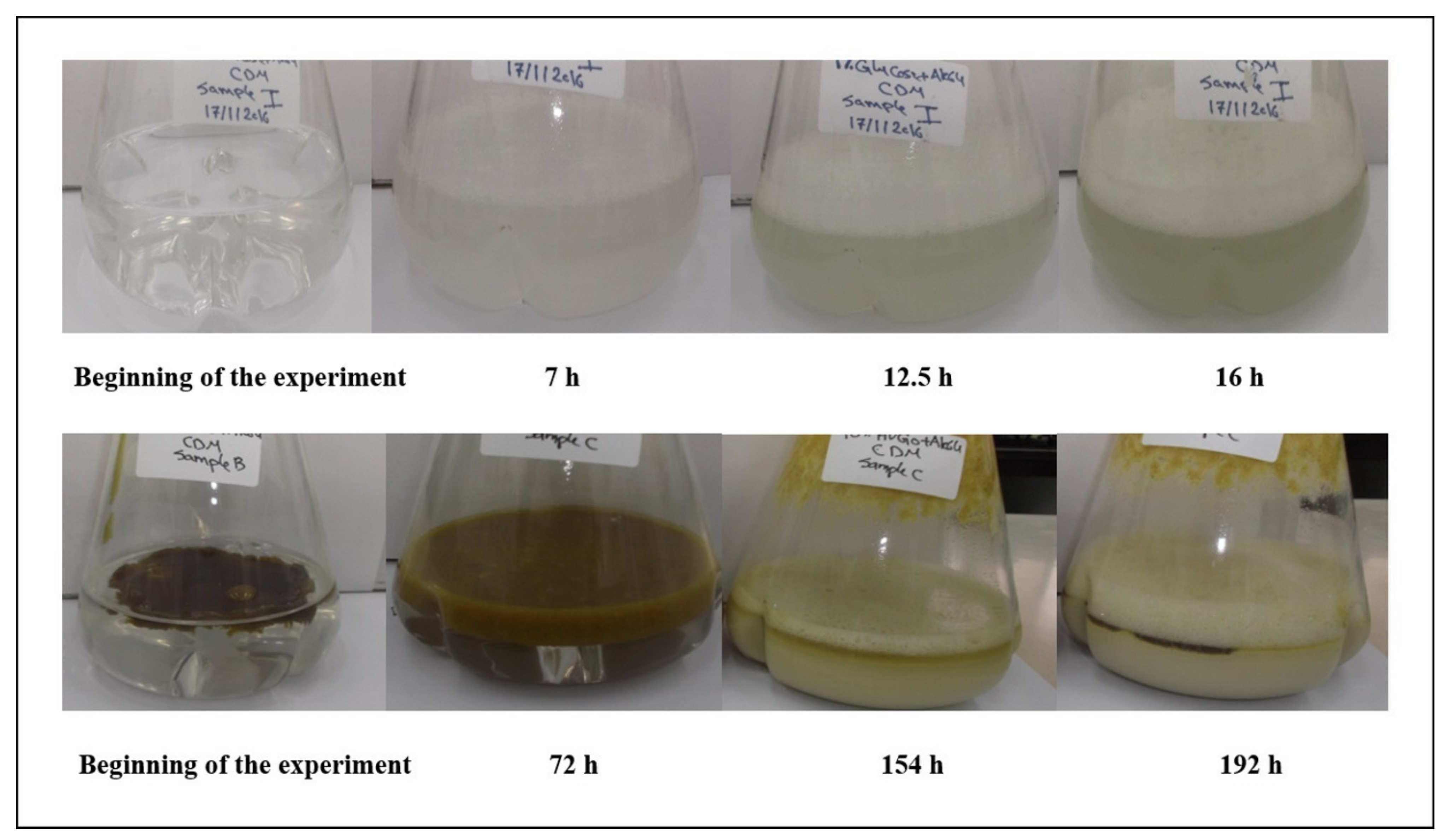
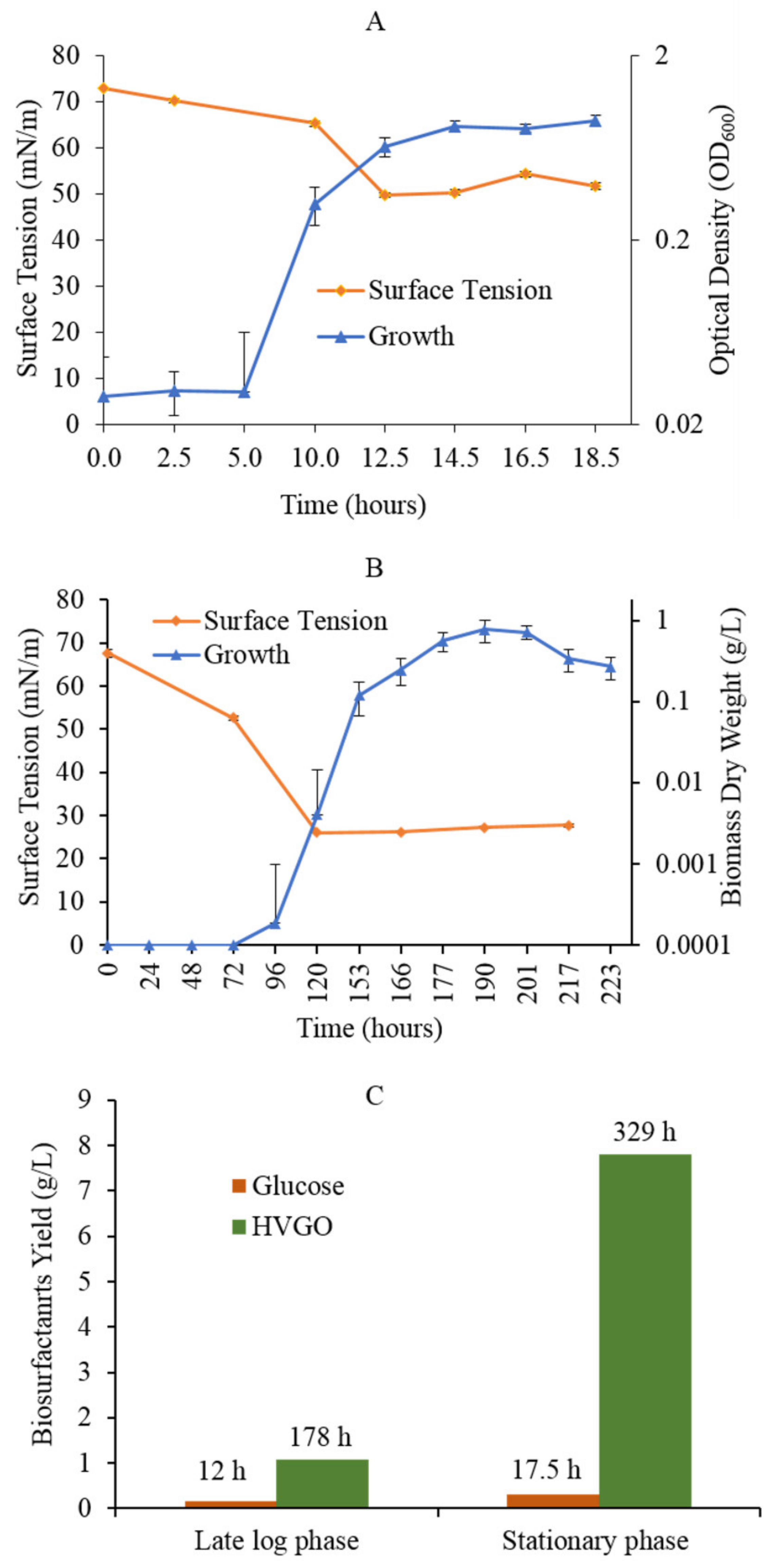
2.1. The AK6U Strain Utilizes HVGO as a Sole Carbon Source for Growth and Biosurfactants Production
2.2. HVGO Stimulates Biosurfactants Production
2.3. The Rhamnolipids Composition in the HVGO Culture Is Different from That of the Glucose Culture
2.4. Isolation of RNA and cDNA Synthesis
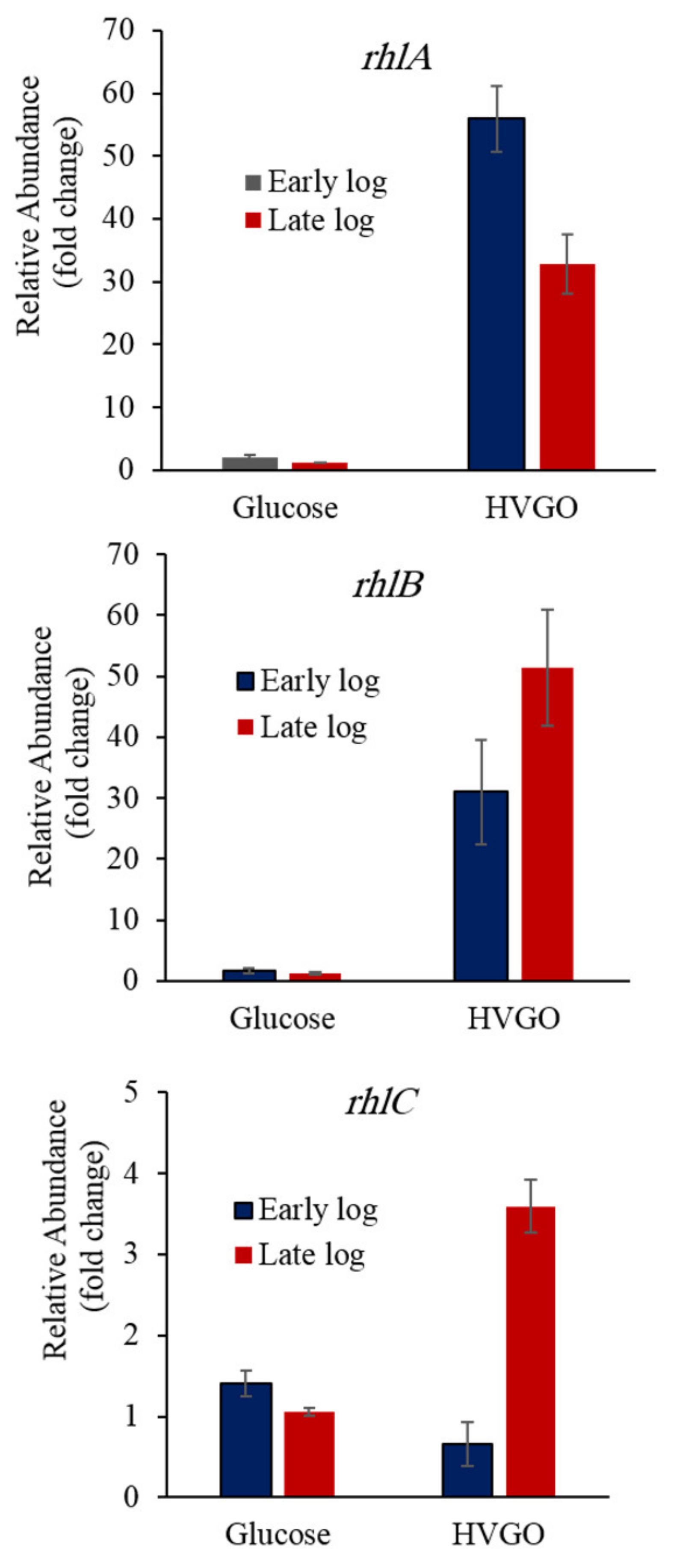
2.5. HVGO Induces the Expression of the Rhamnolipids Biosynthesis Genes
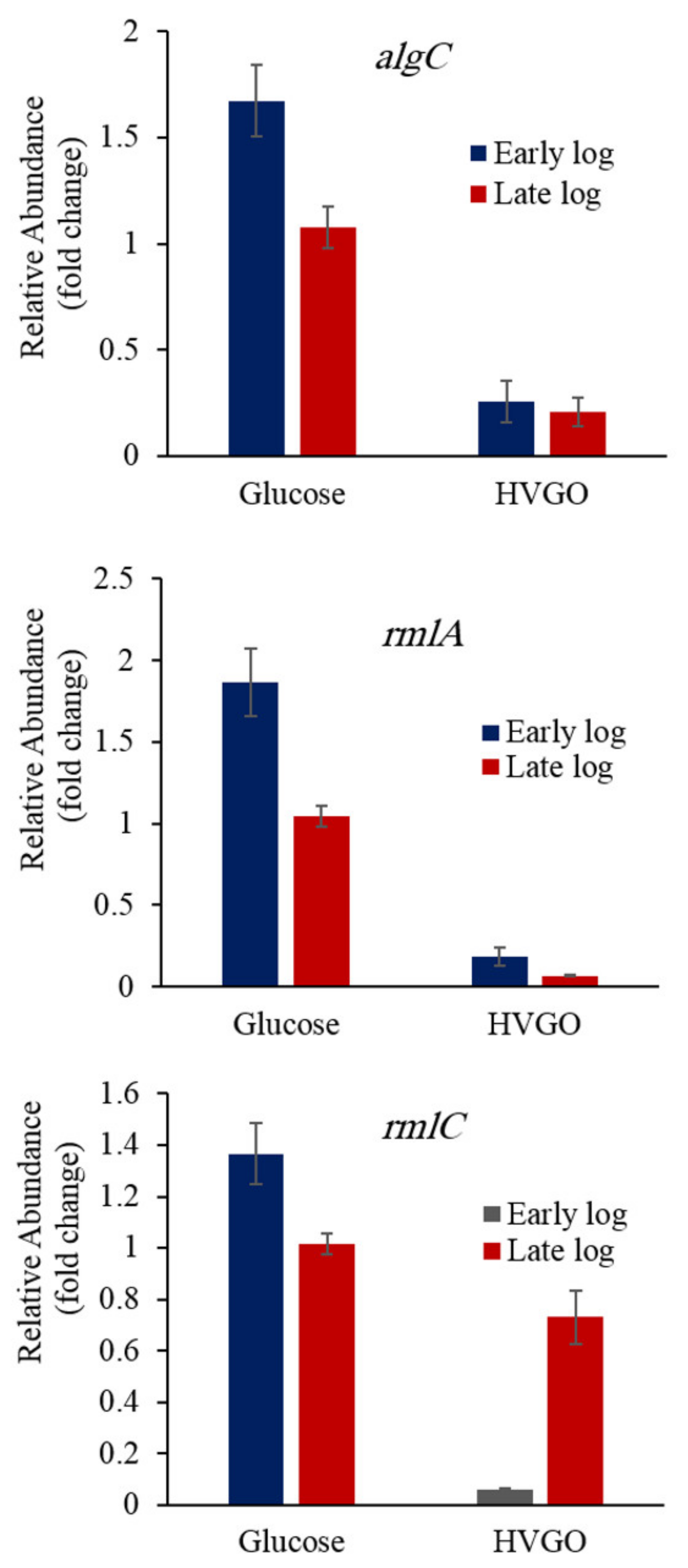
2.6. Downregulation of the Rhamnose Biosynthesis Genes in the HVGO Culture
2.7. HVGO Remodels the Expression of the Quorum Sensing Genes
3. Discussion
4. Materials and Methods
4.1. Bacteria
4.2. Culture Medium and Growth Conditions
4.3. Biosurfactants Production by the AK6U Strain
4.4. Surface Tension Measurement
4.5. Quantification of the Biosurfactants and Congener Analysis
4.6. Isolation of RNA from the AK6U Cultures
4.7. Synthesis of Complementary DNA
4.8. Quantification of Gene Expression by RT-qPCR
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Soberón-Chávez, G.; Maier, R.M. Biosurfactants: A general overview. In Biosurfactants; Soberón-Chávez, G., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1–11. [Google Scholar] [CrossRef]
- Sekhon, K.K.; Khanna, S.; Cameotra, S.S. Biosurfactant production and potential correlation with esterase activity. J. Pet. Environ. Biotechnol. 2012, 3, 133. [Google Scholar] [CrossRef] [Green Version]
- Perfumo, A.; Smyth, T.; Marchant, R.; Banat, I. Production and roles of biosurfactants and bioemulsifiers in accessing hydrophobic substrates. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1501–1512. [Google Scholar] [CrossRef]
- Akbari, S.; Abdurahman, N.H.; Yunus, R.M.; Fayaz, F.; Alara, O.R. Biosurfactants—A new frontier for social and environmental safety: A mini review. Biotechnol. Res. Innov. 2018, 2, 81–90. [Google Scholar] [CrossRef]
- Fenibo, E.O.; Ijoma, G.N.; Selvarajan, R.; Chikere, C.B. Microbial surfactants: The next generation multifunctional biomolecules for applications in the petroleum industry and its associated environmental remediation. Microorganisms 2019, 7, 581. [Google Scholar] [CrossRef] [Green Version]
- Płaza, G.; Achal, V. Biosurfactants: Eco-friendly and innovative biocides against biocorrosion. Int. J. Mol. Sci. 2020, 21, 2152. [Google Scholar] [CrossRef] [Green Version]
- Thakur, P.; Saini, N.K.; Thakur, V.K.; Gupta, V.K.; Saini, R.V.; Saini, A.K. Rhamnolipid the glycolipid biosurfactant: Emerging trends and promising strategies in the field of biotechnology and biomedicine. Microb. Cell. Fact. 2021, 20, 1. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Mawgoud, A.M.; Hausmann, R.; Lépine, F.; Müller, M.M.; Déziel, E. Rhamnolipids: Detection, Analysis, Biosynthesis, Genetic Regulation, and Bioengineering of Production. In Biosurfactants; Soberón-Chávez, G., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; Volume 20, pp. 14–55. [Google Scholar] [CrossRef]
- Banat, I.M.; Satpute, S.K.; Cameotra, S.S.; Patil, R.; Nyayanit, N.V. Cost effective technologies and renewable substrates for biosurfactants’ production. Front. Microbiol. 2014, 5, 697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Syldatk, C.; Lang, S.; Matulovic, U.; Wagner, F. Production of four interfacial active rhamnolipids from n-alkanes or glycerol by resting cells of Pseudomonas species DSM 2874. Z. Nat. C 1985, 40, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Déziel, E.; Lépine, F.; Dennie, D.; Boismenu, D.; Mamer, O.A.; Villemur, R. Liquid chromatography/mass spectrometry analysis of mixtures of rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochim. Biophys. Acta 1999, 1440, 244–252. [Google Scholar] [CrossRef]
- Reis, R.S.; Pereira, A.G.; Neves, B.C.; Freire, D.M. Gene regulation of rhamnolipid production in Pseudomonas aeruginosa—A review. Bioresour. Technol. 2011, 102, 6377–6384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Déziel, E.; Lépine, F.; Milot, S.; Villemur, R. Mass spectrometry monitoring of rhamnolipids from a growing culture of Pseudomonas aeruginosa strain 57RP. Biochim. Biophys. Acta 2000, 1485, 145–152. [Google Scholar] [CrossRef]
- Schmidt, V.K.D.O.; Carvalho, J.D.S.; De Oliveira, D.; De Andrade, C.J. Biosurfactant inducers for enhanced production of surfactin and rhamnolipids: An overview. World J. Microbiol. Biotechnol. 2021, 37, 21. [Google Scholar] [CrossRef]
- Makkar, R.S.; Cameotra, S.S.; Banat, I.M. Advances in utilization of renewable substrates for biosurfactant production. AMB Express 2011, 1, 5. [Google Scholar] [CrossRef] [Green Version]
- Gray, R.M. Heavy oil and residue properties and composition. In Upgrading Petroleum Residues and Heavy Oils; Marcel Dekker Inc.: New York, NY, USA, 1994; pp. 1–40. [Google Scholar]
- Thavasi, R.; Jayalakshmi, S.; Balasubramanian, T.; Banat, I.M. Biosurfactant production by Corynebacterium kutscheri from waste motor lubricant oil and peanut oil cake. Lett. Appl. Microbiol. 2007, 45, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, M.; Biswas, D.; Sana, S.; Datta, S. Utilization of waste engine oil by Ochrobactrum pseudintermedium strain C1 that secretes an exopolysaccharide as a bioemulsifier. Biocatal. Agric. Biotechnol. 2014, 3, 167–176. [Google Scholar] [CrossRef]
- Ismail, W.A.; Mohamed, M.E.; Awadh, M.N.; Obuekwe, C.; El Nayal, A.M. Simultaneous valorization and biocatalytic upgrading of heavy vacuum gas oil by the biosurfactant-producing Pseudomonas aeruginosa AK6U. Microb. Biotechnol. 2017, 10, 1628–1639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, O.P. Microbial biosurfactants and biodegradation. In Biosurfactants. Advances in Experimental Medicine and Biology; Sen, R., Ed.; Springer: New York, NY, USA, 2010; Volume 672, pp. 65–74. [Google Scholar] [CrossRef]
- Ismail, W.; El Nayal, A.M.; Ramadan, A.R.; Abotalib, N. Sulfur source-mediated transcriptional regulation of the rhlABC genes involved in biosurfactants production by Pseudomonas sp. strain AK6U. Front. Microbiol. 2014, 5, 423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ismail, W.; Shammary, S.A.; El-Sayed, W.S.; Obuekwe, C.; El Nayal, A.M.; Raheem, A.S.A.; Al-Humam, A. Stimulation of rhamnolipid biosurfactants production in Pseudomonas aeruginosa AK6U by organosulfur compounds provided as sulfur sources. Biotechnol. Rep. 2015, 7, 55–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mnif, S.; Chamkha, M.; Labat, M.; Sayadi, S. Simultaneous hydrocarbon biodegradation and biosurfactant production by oilfield-selected bacteria. J. Appl. Microbiol. 2011, 111, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Persson, A.; Österberg, E.; Dostalek, M. Biosurfactant production by Pseudomonas fluorescens 378: Growth and product characteristics. Appl. Microbiol. Biotechnol. 1988, 29, 1–4. [Google Scholar] [CrossRef]
- Khopade, A.; Biao, R.; Liu, X.; Mahadik, K.; Zhang, L.; Kokare, C. Production and stability studies of the biosurfactant isolated from marine Nocardiopsis sp. B4. Desalination 2012, 285, 198–204. [Google Scholar] [CrossRef]
- Guerra-Santos, L.H.; Käppeli, O.; Fiechter, A. Dependence of Pseudomonas aeruginosa continous culture biosurfactant production on nutritional and environmental factors. Appl. Microbiol. Biotechnol. 1986, 24, 443–448. [Google Scholar] [CrossRef]
- Santos, A.S.; Sampaio, A.P.W.; Vasquez, G.S.; Santa Anna, L.M.; Pereira, N., Jr.; Freire, D.M. Evaluation of different carbon and nitrogen sources in production of rhamnolipids by a strain of Pseudomonas aeruginosa. Appl. Biochem. Biotechnol. 2002, 98–100, 1025–1035. [Google Scholar] [CrossRef]
- Zhang, L.; Veres-Schalnat, T.A.; Somogyi, A.; Pemberton, J.E.; Maier, R.M. Fatty acid cosubstrates provide β-oxidation precursors for rhamnolipid biosynthesis in Pseudomonas aeruginosa, as evidenced by isotope tracing and gene expression assays. Appl. Environ. Microbiol. 2012, 78, 8611–8622. [Google Scholar] [CrossRef] [Green Version]
- Monteiro, S.A.; Sassaki, G.L.; de Souza, L.M.; Meira, J.A.; de Araújo, J.M.; Mitchell, D.A.; Ramos, L.P.; Krieger, N. Molecular and structural characterization of the biosurfactant produced by Pseudomonas aeruginosa DAUPE 614. Chem. Phys. Lipids 2007, 147, 1–13. [Google Scholar] [CrossRef]
- Zhang, L.; Pemberton, J.E.; Maier, R.M. Effect of fatty acid substrate chain length on Pseudomonas aeruginosa ATCC 9027 monorhamnolipid yield and congener distribution. Process Biochem. 2014, 49, 989–995. [Google Scholar] [CrossRef]
- Mata-Sandoval, J.C.; Karns, J.; Torrents, A. Effect of nutritional and environmental conditions on the production and composition of rhamnolipids by P. aeruginosa UG2. Microbiol. Res. 2001, 155, 249–256. [Google Scholar] [CrossRef]
- Schenk, T.; Schuphan, I.; Schmidt, B. High-performance liquid chromatographic determenation of the rhamnolipids produced by Pseudomonas aeruginosa. J. Chromatogr. A 1995, 693, 7–13. [Google Scholar] [CrossRef]
- Arino, S.; Marchal, R.; Vandecasteele, J.-P. Identification and production of a rhamnolipidic biosurfactant by a Pseudomonas species. Appl. Microbiol. Biotechnol. 1996, 45, 162–168. [Google Scholar] [CrossRef]
- Wittgens, A.; Kovacic, F.; Müller, M.M.; Gerlitzki, M.; Santiago-Schübel, B.; Hofmann, D.; Tiso, T.; Blank, L.M.; Henkel, M.; Hausmann, R.; et al. Novel insights into biosynthesis and uptake of rhamnolipids and their precursors. Appl. Microbiol. Biotechnol. 2017, 101, 2865–2878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguirre-Ramírez, M.; Medina, G.; González-Valdez, A.; Grosso-Becerra, V.; Soberón-Chávez, G. The Pseudomonas aeruginosa rmlBDAC operon, encoding dTDP-l-rhamnose biosynthetic enzymes, is regulated by the quorum-sensing transcriptional regulator RhlR and the alternative sigma factor σS. Microbiology 2012, 158, 908–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zdorovenko, E.L.; Shashkov, A.S.; Kadykova, A.A.; Kiseleva, E.P.; Savich, V.V.; Novik, G.I.; Knirel, Y.A. Structural analysis of the O-polysaccharide from the lipopolysaccharide of Pseudomonas putida BIM B-1100. Carbohydr. Res. 2018, 457, 8–13. [Google Scholar] [CrossRef]
- Woodcock, S.D.; Syson, K.; Little, R.H.; Ward, D.; Sifouna, D.; Brown, J.K.M.; Bornemann, S.; Malone, J.G. Trehalose and α-glucan mediate distinct abiotic stress responses in Pseudomonas aeruginosa. PLoS Genet. 2021, 17, e1009524. [Google Scholar] [CrossRef] [PubMed]
- Quilès, F.; Polyakov, P.; Humbert, F.; Francius, G. Production of extracellular glycogen by Pseudomonas fluorescens: Spectroscopic evidence and conformational analysis by biomolecular recognition. Biomacromolecules 2012, 13, 2118–2127. [Google Scholar] [CrossRef] [PubMed]
- Madigan, M.T.; Martinko, J.M.; Dunlap, P.V.; Clark, D.P. Brock Biology of Microorganisms, 12th ed.; Pearson: New York, NY, USA, 2009. [Google Scholar]
- Fuchs, G. Diversity of metabolic pathways. In Biology of the Prokaryotes; Lengeler, J.W., Drews, G., Schlegel, H.G., Eds.; Thieme Verlag: Stuttgart, Germany, 1999; pp. 202–2011. [Google Scholar]
- Müller, M.M.; Hausmann, R. Regulatory and metabolic network of rhamnolipid biosynthesis: Traditional and advanced engineering towards biotechnological production. Appl. Microbiol. Biotechnol. 2011, 91, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Kohlstedt, M.; Wittmann, C. GC-MS-based 13C metabolic flux analysis resolves the parallel and cyclic glucose metabolism of Pseudomonas putida KT2440 and Pseudomonas aeruginosa PAO1. Metab. Eng. 2019, 54, 35–53. [Google Scholar] [CrossRef]
- Dobler, L.; Vilela, L.F.; Almeida, R.V.; Neves, B.C. Rhamnolipids in perspective: Gene regulatory pathways, metabolic engineering, production and technological forecasting. N. Biotechnol. 2016, 33, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Bazire, A.; Diab, F.; Taupin, L.; Rodrigues, S.; Jebbar, M.; Dufour, A. Effects of osmotic stress on rhamnolipid synthesis and time-course production of cell-to-cell signal molecules by Pseudomonas aeruginosa. Open Microbiol. J. 2009, 3, 128–135. [Google Scholar] [CrossRef] [Green Version]
- Ochsner, U.A.; Reiser, J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1995, 92, 6424–6428. [Google Scholar] [CrossRef] [Green Version]
- Rahim, R.; Ochsner, U.A.; Olvera, C.; Graninger, M.; Messner, P.; Lam, J.S.; Soberón-Chávez, G. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol. Microbiol. 2001, 40, 708–718. [Google Scholar] [CrossRef]
- D’Argenio, D.A.; Wu, M.; Hoffman, L.R.; Kulasekara, H.D.; Déziel, E.; Smith, E.E.; Nguyen, H.; Ernst, R.K.; Freeman, T.J.L.; Spencer, D.H.; et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 2007, 64, 512–533. [Google Scholar] [CrossRef] [Green Version]
- Dekimpe, V.; Deziel, E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: The transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 2009, 155, 712–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina, G.; Juárez, K.; Díaz, R.; Soberón-Chávez, G. Transcriptional regulation of Pseudomonas aeruginosa rhlR, encoding a quorum-sensing regulatory protein. Microbiology 2003, 149, 3073–3081. [Google Scholar] [CrossRef] [Green Version]
- Bodour, A.A.; Drees, K.; Miller-Maier, R.M. Distribution of Biosurfactant-Producing Bacteria in Undisturbed and Contaminated Arid Southwestern Soils. Appl. Environ. Microbiol. 2003, 69, 3280–3287. [Google Scholar] [CrossRef] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| HAAs | Pseudo-Molecular Ion | Cultures # | |||
|---|---|---|---|---|---|
| Glucose(S) | HVGO(L) | HVGO(S) | |||
| 1 | C8–C8 | 301 | 76.01 | 42.98 | |
| 2 | C8–C10 | 329 | 14.11 | 8.11 | |
| 3 | C10–C8 | 329 | 9.55 | 6.06 | |
| 4 | C8–C12 | 357 | 1.57 | ||
| 5 | C12–C8 | 357 | 1.5 | ||
| 6 | C10–C10 | 357 | 0.79 | ||
| 7 | C10–C12 | 385 | 1.86 | ||
| 8 | C12–C10 | 385 | 1.73 | ||
| 9 | C12–C12 | 413 | 1.39 | ||
| 10 | C8–C12:1 | 355 | 9.71 | ||
| 11 | C12:1–C8 | 355 | 9.4 | ||
| 12 | C10–C12:1 | 383 | 7.45 | ||
| 13 | C12:1–C10 | 383 | 7.23 | ||
| 14 | C12–C12:1 | 411 | 0.35 | 0.29 | |
| 15 | C12:1–C12 | 411 | |||
| Total | 15 | 4 | 14 | ||
| Rhamnolipid Congener | Pseudo-Molecular Ion | Cultures # | |||
|---|---|---|---|---|---|
| Glucose(S) | HVGO(L) | HVGO(S) | |||
| 1 | Rha–C8 | 305 | 0.58 | 0.43 | |
| 2 | Rha–C10 | 333 | 0.08 | ||
| 3 | Rha–C12 | 361 | |||
| 4 | Rha–C8–C8 | 447 | 0.26 | ||
| 5 | Rha–Rha–C8 | 451 | 0.93 | 0.03 | |
| 6 | Rha–C8–C10 | 475 | 0.26 | 2.53 | 0.19 |
| 7 | Rha–C10–C8 | 475 | 2.42 | 0.17 | |
| 8 | Rha–Rha–C10 | 479 | 0.76 | 3.48 | |
| 9 | Rha–C10–C10 | 503 | 2.54 | 14.33 | 3.43 |
| 10 | Rha–Rha–C12:1 | 505 | 1.14 | 0.59 | |
| 11 | Rha–Rha–C12 | 507 | 1.18 | ||
| 12 | Rha–C10–C12:1 | 529 | 2.35 | 1.11 | 0.01 |
| 13 | Rha–C12:1–C10 | 529 | 0.74 | 1.01 | |
| 14 | Rha–C12–C10 | 531 | 0.97 | ||
| 15 | Rha–C10–C12 | 531 | 0.91 | ||
| 16 | Rha–Rha–C8–C8 | 593 | 86.44 | 1.36 | |
| 17 | Rha–Rha–C8–C10 | 621 | 2.42 | 2.5 | 1.91 |
| 18 | Rha–Rha–C10–C8 | 621 | 0.13 | 2.32 | 1.84 |
| 19 | Rha–Rha–C8–C12:1 | 647 | |||
| 20 | Rha–Rha–C12:1–C8 | 647 | |||
| 21 | Rha–Rha–C10–C10 | 649 | 0.58 | 50 | 69.29 |
| 22 | Rha–Rha–C10–C12:1 | 675 | 4.09 | 3.31 | |
| 23 | Rha–Rha–C12:1–C10 | 675 | 3.97 | 3.27 | |
| 24 | Rha–Rha–C12–C10 | 677 | 0.09 | 6.27 | 5.76 |
| 25 | Rha–Rha–C10–C12 | 677 | 6.21 | 5.59 | |
| 26 | Rha–Rha–C12:1–C12 | 703 | 0.44 | 0.01 | |
| 27 | Rha–Rha–C10–C14:1 | 703 | |||
| 28 | Rha–Rha–C12–C12 | 705 | |||
| Total | 28 | 15 | 17 | 16 | |
| Gene | Locus | Primers | Sequence (5′-3′) | Amplicon Size (bp) | Source |
|---|---|---|---|---|---|
| rhlI | PA3476 | RhlI-F RhlI-R | CTCTCTGAATCGCTGGAAGG GATGGTCGAACTGGTCGAAT | 145 | This study |
| pqsE | PA1000 | PqsE-F PqsE-R | GACATGGAGGCTTACCTGGA CTCAGTTCGTCGAGGGATTC | 197 | This study |
| lasI | PA1432 | LasI-F LasI-R | GGCTGGGACGTTAGTGTCAT GGCACGGATCATCATCTTCT | 372 | This study |
| rmlA | PA5163 | RmlA-F RmlA-R | CCATCAGCCTGGAAGAGAAG GCTTGAGGTATTGGCCGTAG | 390 | This study |
| rmlC | PA5164 | RmlC-F RmlC-R | CTTCGTCCAGGACAACCATT CACATCTGGCGCTTGTTCT | 201 | This study |
| algC | PA5322 | AlgC-F AlgC-R | CCTACCCCGGTGCTGTACTA AGTCGACCACCACCTTCATC | 280 | This study |
| rhlA | PA3479 | RhlA-F RhlA-R | TGGACTCCAGGTCGAGGAAA GAAAGCCAGCAACCATCAGC | 263 | [21] |
| rhlB | Kpd1 Kpd2 | GCCCACGACCAGTTCGAC CATCCCCCTCCCTATGAC | 226 | [50] | |
| rhlC | PA1130 | RhlC-F2 RhlC-R2 | GTCGAGTCCCTGGTTGAAGG CGTGCTGGTGGTACTGTTCA | 211 | [21] |
| 16S rRNA | 16S-F 16S-R | CACCGGCAGTCTCCTTAGAG AAGCAACGCGAAGAACCTTA | 203 | [21] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alkhalaf, S.A.; Ramadan, A.R.; Obuekwe, C.; El Nayal, A.M.; Abotalib, N.; Ismail, W. Heavy Vacuum Gas Oil Upregulates the Rhamnosyltransferases and Quorum Sensing Cascades of Rhamnolipids Biosynthesis in Pseudomonas sp. AK6U. Molecules 2021, 26, 4122. https://doi.org/10.3390/molecules26144122
Alkhalaf SA, Ramadan AR, Obuekwe C, El Nayal AM, Abotalib N, Ismail W. Heavy Vacuum Gas Oil Upregulates the Rhamnosyltransferases and Quorum Sensing Cascades of Rhamnolipids Biosynthesis in Pseudomonas sp. AK6U. Molecules. 2021; 26(14):4122. https://doi.org/10.3390/molecules26144122
Chicago/Turabian StyleAlkhalaf, Sarah A., Ahmed R. Ramadan, Christian Obuekwe, Ashraf M. El Nayal, Nasser Abotalib, and Wael Ismail. 2021. "Heavy Vacuum Gas Oil Upregulates the Rhamnosyltransferases and Quorum Sensing Cascades of Rhamnolipids Biosynthesis in Pseudomonas sp. AK6U" Molecules 26, no. 14: 4122. https://doi.org/10.3390/molecules26144122






