In Silico Prediction, Molecular Docking and Dynamics Studies of Steroidal Alkaloids of Holarrhena pubescens Wall. ex G. Don to Guanylyl Cyclase C: Implications in Designing of Novel Antidiarrheal Therapeutic Strategies
Abstract
1. Introduction
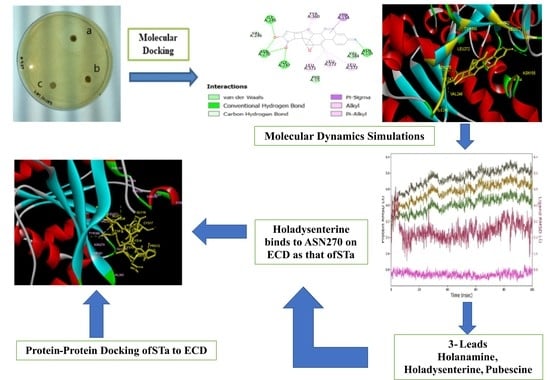
2. Results and Discussion
2.1. Antimicrobial Activity
2.2. Sequence Analysis and Model Generation
2.3. Validation of Homology Model
2.4. Active Site Prediction
2.5. Docking Study
2.6. Drug-Likeness Prediction
2.7. ADMET Prediction
2.8. Molecular Dynamics Analysis
2.9. Molecular Interaction of Ligands with Amino Acids of the Target Protein
3. Materials and Methods
3.1. Plant Material
3.2. Test Strain
3.3. Extraction and Preparation of Alkaloid Rich Fraction
3.4. Antimicrobial Activity
3.5. Protein Model Generation
3.6. Docking Studies
3.7. Ligand and Protein Preparation
3.8. Drug-Likeness Prediction
3.9. ADMET Screening
3.10. Molecular Dynamics Simulation
3.11. Protein–Protein Docking
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADMET | Absorption: distribution, metabolism, excretion and toxicity |
| BBB | Blood brain barrier |
| CFTR | Cystic fibrosis transmembrane conductance regulator |
| ECD | Extracellular domain |
| ECDGC-C | Extracellular domain of Guanylyl cyclase c |
| ETEC | Enterotoxigenic Escherichia coli |
| GC-C | Guanylyl cyclase c |
| GCs | Guanylyl cyclases |
| HBA | Hydrogen bond acceptor |
| HBD | Hydrogen bond donor |
| HIA | Human Intestinal Absorption |
| IBD | Inflammatory bowel disease |
| IBS | Irritable bowel syndrome |
| MLCK | Myosin light chain kinase |
| NHE3 | Na+/H+ exchanger isotype 3 |
| NPR-A | atrial brain natriuretic peptide |
| NPR-B | brain natriuretic peptide |
| NPR-C | Natriuretic Peptide Receptor-C |
| PDB | Protein Data Bank |
| P-gp | P- glycoprotein |
| PSA | Polar surface area |
| RCSB | Research Collaboratory for Structural Bioinformatics |
| RMSD | Root mean square deviation |
| RMSF | Root mean square fluctuation |
| STa | Heats stable enterotoxin |
| TJ | Tight junction |
Sample Availability
References
- Qadri, F.; Das, S.K.; Faruque, A.S.; Fuchs, G.J.; Albert, M.J.; Sack, R.B. Prevalence of toxin types and colonization factors in enterotoxigenic Escherichia coli isolated during a 2-year period from diarrheal patients in Bangladesh. J. Clin. Microbiol. 2000, 38, 27–31. [Google Scholar] [CrossRef]
- Harris, A.M.; Chowdhury, F.; Ara Begum, Y.; Khan, A.I.; Harris, J.B. Shifting prevalence of major diarrheal pathogens in patients seeking hospital care during floods in 1998, 2004 and 2007 in Dhaka, Bangladesh. Am. J. Trop. Med. Hyg. 2008, 79, 708–714. [Google Scholar] [CrossRef]
- Estrada-Garcia, T.; Lopez-Saucedo, C.; Thompson-Bonilla, R.; Abonce, M.; Lopez-Hernandez, D. Association of Diarrheagenic Escherichia coli Pathotypes with Infection and Diarrhea among Mexican Children and Association of Atypical Enteropathogenic E. coli with Acute Diarrhea. J. Clin. Microbiol. 2009, 47, 93–98. [Google Scholar] [CrossRef]
- Rao, C.D.; Maiya, P.P.; Babu, M.A. Non-diarrhoeal increased frequency of bowel movements (IFoBM-ND): Enterovirus association with the symptoms in children. BMJ Open Gastroenterol. 2014, 1, e000011. [Google Scholar] [CrossRef]
- Sack, R.B.; Gorbach, S.L.; Banwell, J.B.; Jacob, B.; Chatterjee, B.D. Enterotoxigenic Escherichia coli isolated from patients with severe cholera-like disease. J. Infec. Dis. 1971, 123, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhong, Z.; Luo, Y.; Cox, E.; Devrident, B. Heat-Stable Enterotoxins of Enterotoxi-genic Escherichia coli and Their Impact on Host Immunity. Toxins 2019, 11, 24. [Google Scholar] [CrossRef]
- Wenneras, C.; Erling, V. Prevalence of enterotoxigenic Escherichia coli-associated diarrhoea and carrier state in the developing world. J. Health Popul. Nutr. 2004, 22, 370–382. [Google Scholar] [PubMed]
- Potter, L.R. Guanylyl cyclase structure, function and regulation. Cell Signal. 2011, 23, 1921–1926. [Google Scholar] [CrossRef]
- De Sauvage, F.J.; Camerato, T.R.; Goeddel, D.V. Primary Structure and Functional Expression of the Human Receptor for Escherichia coli Heat-Stable Enterotoxin. J. Biol. Chem. 1991, 266, 17912–17918. [Google Scholar] [CrossRef]
- Weiglmeier, P.R.; Rösch, P.; Berkner, H. Cure and Curse: E. coli Heat-Stable Enterotoxin and Its Receptor Guanylyl Cyclase C. Toxins 2010, 2, 2213–2229. [Google Scholar] [CrossRef]
- Saha, S.; Biswas, K.H.; Kondapalli, C.; Isloor, N.; Visweswariah, S.S. The linker region in receptor guanylyl cyclases is a key regulatory module. Mutational analysis of guanylyl cyclase C. J. Biol. Chem. 2009, 284, 27135–27145. [Google Scholar] [CrossRef] [PubMed]
- Garbers, D.L. Guanylyl cyclase receptors and their endocrine, paracrine, and autocrine ligands. Cell 1992, 71, 1–4. [Google Scholar] [CrossRef]
- Pitari, G.M. Pharmacology and clinical potential of guanylyl cyclase C agonists in the treatment of ulcerative colitis. Drug Des. Devel. Ther. 2013, 7, 351–360. [Google Scholar] [CrossRef]
- Lima, A.A.M.; Fonteles, M.C. From Escherichia coli heat-stable enterotoxin to mammalian endogenous guanylin hormones. Braz. J. Med. Biol. Res. 2014, 47, 179–191. [Google Scholar] [CrossRef]
- Wolfe, H.R.; Waldman, S.A. A comparative molecular field analysis (COMFA) of the structural determinants of heat-stable enterotoxins mediating activation of guanylyl cyclase C. J. Med. Chem. 2002, 45, 1731–1734. [Google Scholar] [CrossRef]
- Vaandrager, A.B. Structure and function of the heat-stable enterotoxin receptor/guanylyl cyclase C. Mol. Cell. Biochem. 2002, 230, 73–83. [Google Scholar] [CrossRef]
- Field, M. Intestinal ion transport and the pathophysiology of diarrhea. J. Clin. Investig. 2003, 111, 931–943. [Google Scholar] [CrossRef]
- Han, X.; Mann, E.; Gilbert, S.; Guan, Y.; Steinbrecher, K.A.; Montrose, M.A.; Cohen, M.B. Loss of Guanylyl Cyclase C (GCC) Signaling Leads to Dysfunctional Intestinal Barrier. PLoS ONE 2011, 6, e16139. [Google Scholar] [CrossRef]
- Bardhan, P.K. Improving the ORS: Does glutamine have a role? J. Health Popul. Nutr. 2007, 25, 263–266. [Google Scholar]
- Bhutta, T.I.; Tahir, K.I. Loperamide poisoning in children. Lancet 1990, 335, 363. [Google Scholar] [CrossRef]
- Schwartz, R.H.; Rodriguez, W.J. Toxic delirium possibly caused by loperamide. J. Pediatr. 1991, 118, 656–657. [Google Scholar] [CrossRef]
- Aboubaker, S. The Integrated Global Action Plan for the Prevention and Control of Pneumonia and Diarrhoea (GAPPD) in Ending Preventable Child Deaths from Pneumonia and Diarrhoea by 2025, World Health Organization/The United Nations Children’s Fund (UNICEF). 2013. Available online: www.who.int/maternal_child_adolescent/documents/global...plan...diarrhoea/en/ (accessed on February 2018).
- Patwardhan, B.; Vaidya, A.D.B.; Chorghade, M. Ayurveda and natural products drug discovery. Curr. Sci. 2004, 86, 789–799. [Google Scholar]
- Ballal, M.; Srujan, D.; Bhat, K.K.; Shirwaikar, A.; Shivananda, P.G. Antibacterial activity of Holarrhena antidysenterica (Kurchi) against the enteric pathogens. Ind. J. Pharmacol. 2000, 32, 392–393. [Google Scholar]
- Sharma, D.K.; Gupta, V.K.; Kumar, S.; Joshi, V.; Singh, M. Evaluation of antidiarrheal activity of ethanolic extract of Holarrhena antidysenterica seeds in rats. Vet. World 2015, 8, 1392–1395. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, D.; Shilpa, P.N.; Devraj, S.N. Antibacterial and antidiarrheal effects of alkaloids Holarrhena antidysenetrica WALL. Ind. J. Exp. Biol. 2004, 42, 589–594. [Google Scholar]
- Steinbrecher, K.A.; Hermal-Laws, E.; Garin-Laflam, M.P.; Mann, E.A.; Bezerra, L.D.; Hogan, S.P. Murine Guanylate Cyclase C regulates colonic injury and inflammation. J. Immunol. 2011, 186, 7205–7214. [Google Scholar] [CrossRef] [PubMed]
- Camilleri, M. Guanylate cyclase C agonists: Emerging gastrointestinal therapies and actions. Gastroenterology 2015, 148, 483–487. [Google Scholar] [CrossRef] [PubMed]
- Waldman, S.A.; Camilleri, M. Guanylate cyclase-C as a therapeutic target in gastrointestinal disorders. Gut 2018, 67, 1543–1552. [Google Scholar] [CrossRef] [PubMed]
- Voravuthikunchai, S.; Lortheeranuwat, A.; Jeeju, W.; Sririak, T.; Pongpaichit, S.; Supawita, T. Effective medicinal plants against enterohaemorrhagic Escherichia coli O157:H7. J. Ethnopharmacol. 2004, 94, 49–54. [Google Scholar] [CrossRef]
- Bijvelds, M.J.C.; Loos, M.; Hellemans, A.; Bongartz, J.P.; Donck, L.V.; Cos, E.; de Jonge, H. Inhibition of Heat-Stable Toxin–Induced Intestinal Salt and Water Secretion by a Novel Class of Guanylyl Cyclase C Inhibitors. J. Infect. Dis. 2015, 212, 1806–1815. [Google Scholar] [CrossRef]
- Kumar, N.; Singh, B.; Bhandari, P.; Gupta, A.P.; Kaul, V.K. Steroidal Alkaloids from Holarrhena antidysenterica (L.) WALL. Chem. Pharm. Bull. 2007, 55, 912–914. [Google Scholar] [CrossRef]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef]
- Singh, S.; Singh, G.; Heim, J.M.; Gerzer, R. Isolation and expression of a guanylate cyclase-coupled heat stable enterotoxin receptor cDNA from a human colonic cell line. Biochem. Biophys. Res. Commun. 1991, 179, 1455–1463. [Google Scholar] [CrossRef]
- Wada, A.; Hirayama, T.; Kitaura, H.; Fujisawa, J.; Hasegawa, M.; Hidaka, Y.; Shimonishi, Y. Identification of ligand recognition sites in heat-stable enterotoxin receptor, membrane-associated guanylyl cyclase C by site-directed mutational analysis. Infec. Immun. 1996, 64, 5144–5150. [Google Scholar] [CrossRef]
- Potter, L.; Hunter, T. Guanylyl Cyclase-linked Natriuretic Peptide Receptors: Structure and Regulation. J. Biol. Chem. 2001, 276, 6057–6060. [Google Scholar] [CrossRef]
- He, X.L.; Chow, D.C.; Martick, M.M.; Garcia, K.C. Allosteric activation of a spring-loaded natriuretic peptide receptor dimer by hormone. Science 2001, 293, 1657–1662. [Google Scholar] [CrossRef] [PubMed]
- He, X.L.; Dukkipati, A.; Garcia, K.C. Structural determinants of natriuretic peptide receptor specificity and degeneracy. J. Mol. Biol. 2006, 361, 698–714. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, H.; Qiu, Y.; Ogata, C.M.; Misono, K.S. Crystal structure of hormone bound atrial natriuretic peptide receptor extracellular domain: Rotation mechanism for transmembrane signal transduction. J. Biol. Chem. 2004, 279, 28625–28631. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogra. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of non-bonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, D.; Luthy, R.; Bowie, J.U. VERIFY 3D: Assessment of protein models with three-dimensional profile. Methods Enzymol. 1991, 277, 396. [Google Scholar]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed Atlas of Surface Topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; William, E.H.; Belew, R.K.; Olson, A.J. Automated Docking Using a Lamarckian Genetic Algorithm and an Empirical Binding Free Energy Function. J. Comput. Chem. 1992, 19, 1639–1662. [Google Scholar] [CrossRef]
- Biovia, D.S. Discovery Studio Visualizer, v17.2.0.16349; Dassault Systèmes: San Diego, CA, USA, 2016. [Google Scholar]
- Patil, R.; Das, S.; Stanley, A.; Yadav, L.; Sudhakar, A.; Verma, A.K. Optimized Hydrophobic Interactions and Hydrogen Bonding at the Target-Ligand Interface Leads the Pathways of Drug-Designing. PLoS ONE 2010, 5, e12029. [Google Scholar] [CrossRef] [PubMed]
- Berg, L. Exploring Non-Covalent Interactions between Drug-Like Molecules and the Protein Acetylcholinesterase. Ph.D. Thesis, Department of Chemistry Umeå University, Umea, Sweden, 2017; pp. 1–76. [Google Scholar]
- Di Bosco, A.M.; Grieco, P.; Diurno, M.V.; Campiglia, P.; Novellino, E.; Mazzoni, O. Binding site of loperamide: Automated docking of loperamide in human mu- and delta-opioid receptors. Chem. Biol. Drug. Des. 2008, 71, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliver. Rev. 1997, 23, 4–25. [Google Scholar] [CrossRef]
- Rappaport, J.A.; Waldman, S.A. The Guanylate Cyclase C—cGMP Signaling Axis opposes Intestinal Epithelial Injury and Neoplasia. Front. Oncol. 2018, 8, 299. [Google Scholar] [CrossRef] [PubMed]
- Charmot, D. Non-systemic drugs: A critical review. Curr. Pharm. Des. 2012, 18, 1434–1445. [Google Scholar] [CrossRef] [PubMed]
- Lääveri, T.; Sterne, J.; Rombo, L.; Kantele, A. Systematic review of loperamide: No proof of antibiotics being superior to loperamide in treatment of mild/moderate travellers’ diarrhea. Travel Med. Infect. Dis. 2016, 14, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Waring, M.J.; Arrowsmith, J.; Leach, A.R.; Leeson, P.D.; Mandrell, S.; Owen, R.M.; Pairaudeau, G. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat. Rev. Drug Discov. 2015, 14, 475–486. [Google Scholar] [CrossRef]
- Artursson, P.; Karlsson, J. Correlation between oral drug absorption in humans and apparent drug permeability coefficients in human intestinal epithelial (Caco-2) cells. Biochem. Biophys. Res. Commun. 1991, 175, 880–885. [Google Scholar] [CrossRef]
- Lazerwith, S.E. Optimization of pharmacokinetics through manipulation of physicochemical properties in a series of HCV inhibitors. ACS Med. Chem. Lett. 2011, 2, 715–719. [Google Scholar] [CrossRef][Green Version]
- Di, L.; Kern, E.H. Effects of Properties on Biological Assays, In Drug Like Properties: Concepts, Structure Design and Methods from ADME to Toxicity Optimization, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 487–496. [Google Scholar]
- Hughes, J.D.; Blagg, J.; Price, D.A.; Bailey, S.; Devraj, R.V.; Ellsworth, E.; Gibbs, M.E. Physiochemical drug properties associated with in vivo toxicological outcomes. Bioorg. Med. Chem. Lett. 2008, 18, 4872–4875. [Google Scholar] [CrossRef] [PubMed]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Feng, R.M. Assessment of blood brain barrier penetration: In silico, in vitro and in vivo. Curr. Drug Metab. 2002, 3, 647–657. [Google Scholar] [CrossRef]
- Dean, M.; Hamon, Y.; Chimini, G. The human ATP-binding cassette (ABC) transporter superfamily. J. Lipid Res. 2001, 42, 1007–1017. [Google Scholar] [CrossRef]
- Anwar-Mohamed, A.; El-Kadi, A. P-glycoprotein effects on drugs pharmacokinetics and drug-drug-interactions and their clinical implications. Libyan J. Pharm. Clin. Pharmacol. 2012, 1, 48154. [Google Scholar] [CrossRef]
- Sharom, F.J. The P-glycoprotein multidrug transport. Essays Biochem. 2011, 50, 161–178. [Google Scholar] [CrossRef]
- Nebert, D.W.; Russell, D.W. Clinical importance of the Cytochrome P450. Lancet 2002, 360, 1155–1162. [Google Scholar] [CrossRef]
- Danisov, I.G.; Makris, T.M.; Sligar, S.G.; Schlichting, I. Structure and chemistry of Cytochrome P450. Chem. Rev. 2005, 105, 2253–2277. [Google Scholar] [CrossRef]
- Ogu, C.C.; Maxa, J.L. Drug interactions due to cytochrome P450. BUMC Proc. 2000, 13, 421–423. [Google Scholar] [CrossRef]
- Beijnen, J.H.; Schellens, J.H. Drug interactions in oncology. Lancet Oncol. 2004, 5, 489–496. [Google Scholar] [CrossRef]
- Scripture, C.D.; Sparreboom, A.; Figg, W.D. Modulation of cytochrome P450 activity: Implications for cancer therapy. Lancet Oncol. 2005, 6, 780–789. [Google Scholar] [CrossRef]
- Wang, Y.; Xing, J.; Xu, Y.; Zhou, N.; Peng, J.; Xiong, Z.; Liu, X.; Luo, X.; Luo, C. In silico ADME/T modelling for rational drug design. Q. Rev. Biophys. 2015, 48, 488–515. [Google Scholar] [CrossRef]
- Cheng, F.; Yu, Y.; Shen, J.; Yan, L.; Li, W.; Liu, G.; Lee, P.W.; Tang, Y. Classification of cytochrome P450 inhibitors and noninhibitors using combined classifiers. J. Chem. Inf. Model. 2011, 51, 996–1011. [Google Scholar] [CrossRef]
- Jayaraj, J.M.; Krishnasamy, G.; Lee, J.K.; Muthusamy, K. In silico identification and screening of CYP24A1 inhibitors: 3D QSR pharmacophore mapping and moecular dynamics analysis. J. Biomol. Struct. Dyn. 2019, 37, 1700–1714. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Hidaka, Y.; Matsumoto, Y.; Sanni, T.; Shimonshi, Y. Determination of the Binding Site on the Extracellular Domain of Guanylyl Cyclase C to Heat-stable Enterotoxin. J. Biol. Chem. 1999, 274, 31713–31718. [Google Scholar] [CrossRef]
- Kozakov, D.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Vajda, S. How good is automated protein docking? Proteins Struct. Funct. Bioinform. 2013, 81, 2159–2166. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Nagarjuna, D.; Mittal, G.; Dhanda, R.S.; Rajni, G.; Manisha, Y. Alarming levels of antimicrobial resistance among sepsis patients admitted to ICU in a tertiary care hospital in India—A case control retrospective study. Antimicrob. Resist. Infect. Control 2018, 7, 150. [Google Scholar] [CrossRef]
- Nnadi, C.O.; Nwodo, N.J.; Kaiser, M.; Brun, R.; Schmidt, T.J. Steroid Alkaloids from Holarrhena africana with Strong Activity against Trypanosoma brucei rhodesiense. Molecules 2017, 22, 1129. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Technical Bulletin of the Registry of Medical Technologists. Am. J. Clin. Pathol. 1966, 36, 49–52. [Google Scholar]
- Singh, K.D.; Muthusamy, K. Molecular modeling, quantum polarized ligand docking and structure-based 3D-QSAR analysis of the imidazole series as dual AT(1) and ET(A) receptor antagonists. Acta Pharmacol. Sin. 2013, 34, 1592–1606. [Google Scholar] [CrossRef]
- Loganathan, L.; Muthusamy, K.; Jayaraj, J.M.; Kajamaideen, A.; Balthasar, J.J. In silico insights on tankyrase protein: A potential target for colorectal cancer. J. Biomol. Struct. Dyn. 2019, 37, 3637–3648. [Google Scholar] [CrossRef] [PubMed]









| Enterotoxigenic E. coli (ETEC) | ||||
|---|---|---|---|---|
| A | B | |||
| Treatment | Concentration | Dose/Disc | Zone of Inhibition |  |
| Alkaloid Rich Fraction (mg/mL) | 100 mg/mL | 1 mg | 16 ± 0.38 mm | |
| 50 mg/mL | 0.5 mg | 14 ± 0.53 mm | ||
| 25 mg/mL | 0.25 mg | 0.0 ± 0.0 | ||
| 12.5 mg/mL | 0.125 mg | 0.00 ± 0.0 | ||
| 6.25 mg/mL | 0.625 mg | 0.00 ± 0.0 | ||
| Positive control (Gentamycin) | 10 µg | 35 ± 0.707 mm | ||
| Negative Control | - | Nil | ||
| Solvent Control | - | Nil | ||
| Ligand | PubChem ID | Binding Energy a kcal/mol | H Bond b | Structure |
|---|---|---|---|---|
| Pubescine | 72313 | −8.05 | 2 |  |
| Kurchessine | 442979 | −8.52 | NA |  |
| Holadienine | 12310532 | −8.54 | NA |  |
| Conessimine | 12303831 | −8.59 | NA |  |
| Conessine | 441082 | −9.00 | NA |  |
| Holadysenterine | 16742955 | −8.06 | 5 |  |
| Isoconessimine | 11772257 | −9.05 | NA |  |
| Kurchine | 551434 | −9.05 | NA |  |
| Holanamine | 6869-29-0 | −8.44 | 1 |  |
| Loperamide | 3955 | −8.05 | NA |  |
| Ligand | Mol wt. | No. of HBA | No. of HBD | MolLogP | MolPSA |
|---|---|---|---|---|---|
| Kurchessine | 372.35 | 2 | 0 | 6.10 | 5.76 Å2 |
| Conessine | 356.32 | 2 | 0 | 5.16 | 6.03 Å2 |
| Isoconessimine | 342.30 | 2 | 1 | 4.61 | 15.09 Å2 |
| Pubescine | 382.19 | 5 | 1 | 2.28 | 50.49 Å2 |
| Holadienine | 325.24 | 2 | 0 | 4.25 | 16.93 Å2 |
| Holanamine | 325.20 | 3 | 1 | 3.37 | 39.05 Å2 |
| Conessimine | 342.30 | 2 | 1 | 4.67 | 14.99 Å2 |
| Holadysenterine | 390.29 | 4 | 4 | 2.59 | 74.72 Å2 |
| Kurchine | 342.30 | 2 | 1 | 4.61 | 15.09 Å2 |
| Loperamide | 476.22 | 3 | 1 | 5.39 | 34.51 Å2 |
| Ligand | Blood Brain Barrier | Caco-2 Permeability | Human Intestinal Absorption | P-Glycoprotein Substrate |
|---|---|---|---|---|
| Kurchessine | BBB+ | Caco2+ | HIA+ | Substrate, Inhibitor |
| Conessine | BBB+ | Caco2+ | HIA+ | Substrate, Inhibitor |
| Isoconessimine | BBB+ | Caco2+ | HIA+ | Substrate, Inhibitor |
| Pubescine | BBB+ | Caco2+ | HIA+ | Substrate, Inhibitor |
| Holadienine | BBB+ | Caco2+ | HIA+ | Substrate, Inhibitor |
| Holanamine | BBB+ | Caco2+ | HIA+ | Substrate, Non-inhibitor |
| Conessimine | BBB+ | Caco2+ | HIA+ | Substrate, Inhibitor |
| Holadysenterine | BBB+ | Caco2- | HIA+ | Substrate, Non-inhibitor |
| Kurchine | BBB+ | Caco2+ | HIA+ | Substrate, Inhibitor |
| Loperamide | BBB+ | Caco2+ | HIA+ | Substrate, Inhibitor |
| Ligand. | CYP2C9 Substrate | CYP2D6 Substrate | CYP4503 A4 Substrate | CYP450 1A2 Inhibitor | CYP4502C9 Inhibitor | CYP4502D6 Inhibitor | CYP450 3A4 Inhibitor |
|---|---|---|---|---|---|---|---|
| Kurchessine | Non substrate | Non-Substrate | Substrate | Non-inhibitor | Non-inhibitor | Non inhibitor | Non inhibitor |
| Conessine | Non substrate | Non Substrate | Substrate | Non inhibitor | Non inhibitor | Non inhibitor | Non inhibitor |
| Isoconessimine | Non substrate | Non substrate | Substrate | Non-inhibitor | Non-inhibitor | Non inhibitor | Non inhibitor |
| Pubescine | Non substrate | Non substrate | Substrate | Inhibitor | Non inhibitor | Inhibitor | Non inhibitor |
| Holadienine | Non substrate | Non substrate | Substrate | Non inhibitor | Non inhibitor | Non inhibitor | Non inhibitor |
| Holanamine | Non substrate | Non substrate | Substrate | Non inhibitor | Non inhibitor | Non inhibitor | Non inhibitor |
| Conessimine | Non substrate | Non Substrate | Substrate | Non inhibitor | Non inhibitor | Non inhibitor | Non inhibitor |
| Holadysenterine | Non substrate | Non substrate | Substrate | Non inhibitor | Non inhibitor | Non inhibitor | Inhibitor |
| Kurchine | Non substrate | Non Substrate | Substrate | Non inhibitor | Non inhibitor | Non inhibitor | Non inhibitor |
| Loperamide | Non substrate | Non substrate | Substrate | Non inhibitor | Non inhibitor | Inhibitor | Non inhibitor |
| Ligand | AMES toxicity | Carcinogenicity | Rat Acute Toxicity (mol/kg) |
|---|---|---|---|
| Kurchessine | Non AMES toxic | Non-carcinogens | 2.5575 |
| Conessine | Non AMES toxic | Non-carcinogens | 2.6198 |
| Isoconessimine | Non AMES toxic | Non-carcinogens | 2.6781 |
| Pubescine | Non AMES toxic | Non-carcinogens | 2.9255 |
| Holadienine | Non AMES toxic | Non-carcinogens | 2.5022 |
| Holanamine | Non AMES toxic | Non-carcinogens | 2.1900 |
| Conessimine | Non AMES toxic | Non-carcinogens | 2.6437 |
| Holadysenterine | Non AMES toxic | Non carcinogens | 2.4973 |
| Kurchine | Non AMES toxic | Non carcinogens | 2.6781 |
| Loperamide | Non AMES toxic | Non carcinogens | 3.6560 |
| S.No | Heatstable Enterotoxin STa | Extacellular Domain ECD | Bond Length (Å) |
|---|---|---|---|
| 1- | ALA15 | GLU243 | 2.09 |
| 2- | CYS6 | ASN270 | 2.87 |
| 3- | CYS6 | TYR360 | 2.01 |
| 4- | GLU7 | THR154 | 2.51 |
| 5- | CYS14 | GLU243 | 2.22 |
| 6- | CYS17 | LYS160 | 1.68 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, N.; Choudhary, S.K.; Bhagat, N.; Karthikeyan, M.; Chaturvedi, A. In Silico Prediction, Molecular Docking and Dynamics Studies of Steroidal Alkaloids of Holarrhena pubescens Wall. ex G. Don to Guanylyl Cyclase C: Implications in Designing of Novel Antidiarrheal Therapeutic Strategies. Molecules 2021, 26, 4147. https://doi.org/10.3390/molecules26144147
Gupta N, Choudhary SK, Bhagat N, Karthikeyan M, Chaturvedi A. In Silico Prediction, Molecular Docking and Dynamics Studies of Steroidal Alkaloids of Holarrhena pubescens Wall. ex G. Don to Guanylyl Cyclase C: Implications in Designing of Novel Antidiarrheal Therapeutic Strategies. Molecules. 2021; 26(14):4147. https://doi.org/10.3390/molecules26144147
Chicago/Turabian StyleGupta, Neha, Saurav Kumar Choudhary, Neeta Bhagat, Muthusamy Karthikeyan, and Archana Chaturvedi. 2021. "In Silico Prediction, Molecular Docking and Dynamics Studies of Steroidal Alkaloids of Holarrhena pubescens Wall. ex G. Don to Guanylyl Cyclase C: Implications in Designing of Novel Antidiarrheal Therapeutic Strategies" Molecules 26, no. 14: 4147. https://doi.org/10.3390/molecules26144147
APA StyleGupta, N., Choudhary, S. K., Bhagat, N., Karthikeyan, M., & Chaturvedi, A. (2021). In Silico Prediction, Molecular Docking and Dynamics Studies of Steroidal Alkaloids of Holarrhena pubescens Wall. ex G. Don to Guanylyl Cyclase C: Implications in Designing of Novel Antidiarrheal Therapeutic Strategies. Molecules, 26(14), 4147. https://doi.org/10.3390/molecules26144147