NMR Spectroscopy for Protein Higher Order Structure Similarity Assessment in Formulated Drug Products
Abstract
:1. Introduction
2. Results
2.1. 1D 1H NMR Spectroscopy
2.1.1. Excipients
2.1.2. Process-Related Impurities
2.1.3. Protein HOS
2.1.4. Similarity Metrics of DM
2.2. 2D 1H-13C Spectroscopy
2.2.1. Protein Specificity
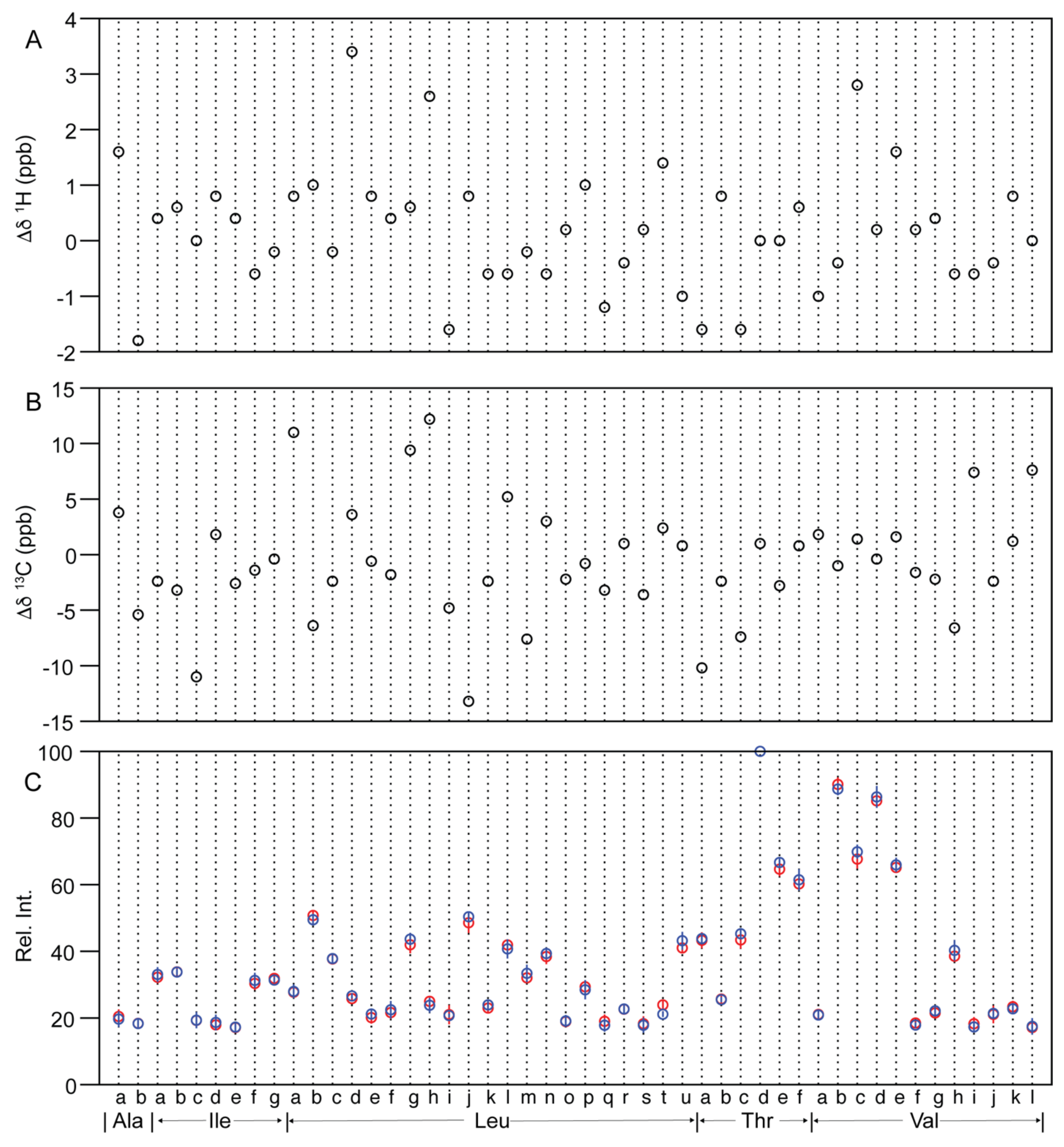
2.2.2. Similarity Metrics of Δδ
2.2.3. Methyl Peak Profile
2.3. 2D 1H-15N Spectroscopy
3. Materials and Methods
3.1. Drug Product NMR Samples
3.2. NMR Spectrsocopy
3.2.1. 1D 1H NMR Spectra Collection and Processing
3.2.2. 2D 1H-13C NMR Spectra Collection and Processing
3.2.3. 2D 1H-15N NMR Spectra Collection and Processing
3.3. Calculation of Similarity Metrics
3.3.1. Mahalanobis Distance (DM) between 1D Spectra
3.3.2. Chemical Shift Difference (Δδ) between 2D Spectra
3.3.3. Methyl Peak Profile between 2D Spectra
4. Discussion and Conclusions
4.1. HOS Inferrred from 1D and 2D Spectra
4.2. HOS Similarity Metrics Calculated from 1D and 2D Spectra
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
Sample Availability
References
- Zhang, L.; Lionberger, R.A. Generics 2030: Where Are We Heading in 2030 for Generic Drug Science, Research, and Regulation? Clin. Pharmacol. Ther. 2020, 107, 1293–1295. [Google Scholar] [CrossRef] [PubMed]
- Christl, L.A.; Woodcock, J.; Kozlowski, S. Biosimilars: The US Regulatory Framework. Annu. Rev. Med. 2017, 68, 243–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, A.C.; Lee, S.L.; Harris, D.P.; Buhse, L.; Kozlowski, S.; Yu, L.; Kopcha, M.; Woodcock, J. Advancing pharmaceutical quality: An overview of science and research in the US FDA’s Office of Pharmaceutical Quality. Int. J. Pharm. 2016, 515, 390–402. [Google Scholar] [CrossRef] [PubMed]
- Kozlowski, S.; Woodcock, J.; Midthun, K.; Behrman Sherman, R. Developing the Nation’s Biosimilars Program. N. Engl. J. Med. 2011, 365, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Moussa, E.M.; Panchal, J.P.; Moorthy, B.S.; Blum, J.S.; Joubert, M.K.; Narhi, L.O.; Topp, E.M. Immunogenicity of Therapeutic Protein Aggregates. J. Pharm. Sci. 2016, 105, 417–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruia, F.; Du, J.; Santacroce, P.V.; Remmele, R.L.; Bee, J.S. Technical Decision Making with Higher Order Structure Data: Impact of a Formulation Change on the Higher Order Structure and Stability of a mAb. J. Pharm. Sci. 2015, 104, 1539–1542. [Google Scholar] [CrossRef] [PubMed]
- Aubin, Y.; Hodgson, D.J.; Thach, W.B.; Gingras, G.; Sauvé, S. Monitoring Effects of Excipients, Formulation Parameters and Mutations on the High Order Structure of Filgrastim by NMR. Pharm. Res. 2015, 32, 3365–3375. [Google Scholar] [CrossRef] [PubMed]
- Bramham, J.E.; Podmore, A.; Davies, S.A.; Golovanov, A.P. Comprehensive Assessment of Protein and Excipient Stability in Biopharmaceutical Formulations Using 1H NMR Spectroscopy. Acs Pharmacol. Transl. Sci. 2020, 4, 288–295. [Google Scholar]
- Panjwani, N.; Hodgson, D.J.; Sauvé, S.; Aubin, Y. Assessment of the Effects of pH, Formulation and Deformulation on the Conformation of Interferon Alpha-2 by NMR. J. Pharm. Sci. 2010, 99, 3334–3342. [Google Scholar] [CrossRef]
- Shah, D.D.; Singh, S.M.; Mallela, K.M.G. Effect of Chemical Oxidation on the Higher Order Structure, Stability, Aggregation, and Biological Function of Interferon Alpha-2a: Role of Local Structural Changes Detected by 2D NMR. Pharm. Res. 2018, 35, 17. [Google Scholar] [CrossRef]
- Hodgson, D.J.; Aubin, Y. Assessment of the structure of pegylated-recombinant protein therapeutics by the NMR fingerprint assay. J. Pharm. Biomed. Anal. 2017, 138, 351–356. [Google Scholar] [CrossRef]
- Casagrande, F.; Dégardin, K.; Ross, A. Protein NMR of biologicals: Analytical support for development and marketed products. J. Biomol. NMR 2020, 74, 657–671. [Google Scholar] [CrossRef]
- Weiss, W.F.; Gabrielson, J.P.; Al-Azzam, W.; Chen, G.; Davis, D.L.; Das, T.K.; Hayes, D.B.; Houde, D.; Singh, S.K. Technical Decision Making with Higher Order Structure Data: Perspectives on Higher Order Structure Characterization from the Biopharmaceutical Industry. J. Pharm. Sci. 2016, 105, 3465–3470. [Google Scholar] [CrossRef]
- Keire, D.A. Analytical Tools for Physicochemical Characterization and Fingerprinting. In Science and Regulations of Naturally Derived Complex Drugs; Sasisekharan, R., Lee, S.L., Rosenberg, A., Walker, L.A., Eds.; Springer International Publishing Ag: Cham, Switzerland, 2019; Volume 32, pp. 91–113. [Google Scholar]
- Guerrini, M.; Rudd, T.R.; Yates, E.A. NMR in the Characterization of Complex Mixture Drugs. In Science and Regulations of Naturally Derived Complex Drugs; Sasisekharan, R., Lee, S.L., Rosenberg, A., Walker, L.A., Eds.; Springer International Publishing Ag: Cham, Switzerland, 2019; Volume 32, pp. 115–137. [Google Scholar]
- Amezcua, C.A.; Szabo, C.M. Assessment of Higher Order Structure Comparability in Therapeutic Proteins Using Nuclear Magnetic Resonance Spectroscopy. J. Pharm. Sci. 2013, 102, 1724–1733. [Google Scholar] [CrossRef]
- Arbogast, L.W.; Brinson, R.G.; Formolo, T.; Hoopes, J.T.; Marino, J.P. 2D 1HN, 15N Correlated NMR Methods at Natural Abundance for Obtaining Structural Maps and Statistical Comparability of Monoclonal Antibodies. Pharm. Res. 2016, 33, 462–475. [Google Scholar] [CrossRef]
- Arbogast, L.W.; Brinson, R.G.; Marino, J.P. Mapping Monoclonal Antibody Structure by 2D 13C NMR at Natural Abundance. Anal. Chem. 2015, 87, 3556–3561. [Google Scholar] [CrossRef]
- Kiss, R.; Fizil, Á.; Szántay, C. What NMR can do in the biopharmaceutical industry. J. Pharm. Biomed. Anal. 2018, 147, 367–377. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Long, D.S.; Lute, S.C.; Levy, M.J.; Brorson, K.A.; Keire, D.A. Simple NMR methods for evaluating higher order structures of monoclonal antibody therapeutics with quinary structure. J. Pharm. Biomed. Anal. 2016, 128, 398–407. [Google Scholar] [CrossRef]
- Hodgson, D.J.; Ghasriani, H.; Aubin, Y. Assessment of the higher order structure of Humira®, Remicade®, Avastin®, Rituxan®, Herceptin®, and Enbrel® by 2D-NMR fingerprinting. J. Pharm. Biomed. Anal. 2019, 163, 144–152. [Google Scholar] [CrossRef]
- Japelj, B.; Ilc, G.; Marušič, J.; Senčar, J.; Kuzman, D.; Plavec, J. Biosimilar structural comparability assessment by NMR: From small proteins to monoclonal antibodies. Sci. Rep. 2016, 6, 32201. [Google Scholar] [CrossRef]
- Elliott, K.W.; Ghasriani, H.; Wikström, M.; Giddens, J.P.; Aubin, Y.; Delaglio, F.; Marino, J.P.; Arbogast, L.W. Comparative Analysis of One-Dimensional Protein Fingerprint by Line Shape Enhancement and Two-Dimensional 1H,13C Methyl NMR Methods for Characterization of the Higher Order Structure of IgG1 Monoclonal Antibodies. Anal. Chem. 2020, 92, 6366–6373. [Google Scholar] [CrossRef] [PubMed]
- Franks, J.; Glushka, J.N.; Jones, M.T.; Live, D.H.; Zou, Q.; Prestegard, J.H. Spin Diffusion Editing for Structural Fingerprints of Therapeutic Antibodies. Anal. Chem. 2016, 88, 1320–1327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poppe, L.; Jordan, J.B.; Lawson, K.; Jerums, M.; Apostol, I.; Schnier, P.D. Profiling Formulated Monoclonal Antibodies by 1H NMR Spectroscopy. Anal. Chem. 2013, 85, 9623–9629. [Google Scholar] [CrossRef] [PubMed]
- Ghasriani, H.; Hodgson, D.J.; Brinson, R.G.; McEwen, I.; Buhse, L.F.; Kozlowski, S.; Marino, J.P.; Aubin, Y.; Keire, D.A. Precision and robustness of 2D-NMR for structure assessment of filgrastim biosimilars. Nat. Biotechnol. 2016, 34, 139. [Google Scholar] [CrossRef]
- Wang, D.; Park, J.; Patil, S.M.; Smith, C.J.; Leazer, J.L., Jr.; Keire, D.A.; Chen, K. An NMR-Based Similarity Metric for Higher Order Structure Quality Assessment Among, U.S. Marketed Insulin Therapeutics. J. Pharm. Sci. 2020, 109, 1519–1528. [Google Scholar] [CrossRef]
- Ionova, Y.; Wilson, L. Biologic excipients: Importance of clinical awareness of inactive ingredients. PLoS ONE 2020, 15, e0235076. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 2009, 37, 603–610. [Google Scholar] [CrossRef]
- Ulrich, E.L.; Akutsu, H.; Doreleijers, J.F.; Harano, Y.; Ioannidis, Y.E.; Lin, J.; Livny, M.; Mading, S.; Maziuk, D.; Miller, Z.; et al. BioMagResBank. Nucleic Acids Res. 2008, 36, 402–408. [Google Scholar] [CrossRef] [Green Version]
- Patil, S.M.; Li, V.; Peng, J.; Kozak, D.; Xu, J.; Cai, B.; Keire, D.A.; Chen, K. A Simple and Noninvasive DOSY NMR Method for Droplet Size Measurement of Intact Oil-In-Water Emulsion Drug Products. J. Pharm. Sci. 2019, 108, 815–820. [Google Scholar] [CrossRef]
- Wang, W.; Ignatius, A.A.; Thakkar, S.V. Impact of Residual Impurities and Contaminants on Protein Stability. J. Pharm. Sci. 2014, 103, 1315–1330. [Google Scholar] [CrossRef]
- Gottlieb, H.E.; Kotlyar, V.; Nudelman, A. NMR Chemical Shifts of Common Laboratory Solvents as Trace Impurities. J. Org. Chem. 1997, 62, 7512–7515. [Google Scholar] [CrossRef]
- Malmstrøm, J. Quantification of Silicone Oil and Its Degradation Products in Aqueous Pharmaceutical Formulations by 1H-NMR Spectroscopy. J. Pharm. Sci. 2019, 108, 1512–1520. [Google Scholar] [CrossRef]
- Suh, M.S.; Patil, S.M.; Kozak, D.; Pang, E.; Choi, S.; Jiang, X.; Rodriguez, J.D.; Keire, D.A.; Chen, K. An NMR Protocol for In Vitro Paclitaxel Release from an Albumin-Bound Nanoparticle Formulation. AAPS Pharmscitech 2020, 21, 136. [Google Scholar] [CrossRef]
- Skidmore, K.; Hewitt, D.; Kao, Y.-H. Quantitation and characterization of process impurities and extractables in protein-containing solutions using proton NMR as a general tool. Biotechnol. Prog. 2012, 28, 1526–1533. [Google Scholar] [CrossRef]
- Palmer, A.G.; Kroenke, C.D.; Patrick Loria, J. Nuclear Magnetic Resonance Methods for Quantifying Microsecond-to-Millisecond Motions in Biological Macromolecules. In Methods in Enzymology; James, T.L., Dötsch, V., Schmitz, U., Eds.; Academic Press: Cambridge, MA, USA, 2001; Volume 339, pp. 204–238. [Google Scholar]
- Chen, K.; Park, J.; Li, F.; Patil, S.M.; Keire, D.A. Chemometric Methods to Quantify 1D and 2D NMR Spectral Differences among Similar Protein Therapeutics. AAPS Pharmscitech 2018, 19, 1011–1019. [Google Scholar] [CrossRef]
- Chang, X.; Jørgensen, A.M.M.; Bardrum, P.; Led, J.J. Solution Structures of the R6 Human Insulin Hexamer. Biochemistry 1997, 36, 9409–9422. [Google Scholar] [CrossRef]
- Quinternet, M.; Starck, J.P.; Delsuc, M.A.; Kieffer, B. Heteronuclear NMR provides an accurate assessment of therapeutic insulin’s quality. J. Pharm. Biomed. Anal. 2013, 78, 252–254. [Google Scholar] [CrossRef]
- Brinson, R.G.; Marino, J.P.; Delaglio, F.; Arbogast, L.W.; Evans, R.M.; Kearsley, A.; Gingras, G.; Ghasriani, H.; Aubin, Y.; Pierens, G.K.; et al. Enabling adoption of 2D-NMR for the higher order structure assessment of monoclonal antibody therapeutics. mAbs 2019, 11, 94–105. [Google Scholar] [CrossRef] [Green Version]
- Nishizaki, Y.; Lankin, D.C.; Chen, S.-N.; Pauli, G.F. Accurate and Precise External Calibration Enhances the Versatility of Quantitative NMR (qNMR). Anal. Chem. 2021, 93, 2733–2741. [Google Scholar] [CrossRef]
- Schanda, P.; Brutscher, B. Very Fast Two-Dimensional NMR Spectroscopy for Real-Time Investigation of Dynamic Events in Proteins on the Time Scale of Seconds. J. Am. Chem. Soc. 2005, 127, 8014–8015. [Google Scholar] [CrossRef]
- Zhuo, Y.; Keire, D.A.; Chen, K. Minor N-Glycan Mapping of Monoclonal Antibody Therapeutics Using Middle-Down NMR Spectroscopy. Mol. Pharm. 2021, 18, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. Nmrpipe-a Multidimensional Spectral Processing System Based on Unix Pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Bigam, C.G.; Yao, J.; Abildgaard, F.; Dyson, H.J.; Oldfield, E.; Markley, J.L.; Sykes, B.D. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 1995, 6, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Brereton, R.G. The Mahalanobis distance and its relationship to principal component scores. J. Chemom. 2015, 29, 143–145. [Google Scholar] [CrossRef]
- Aubin, Y.; Gingras, G.; Sauvé, S. Assessment of the Three-Dimensional Structure of Recombinant Protein Therapeutics by NMR Fingerprinting: Demonstration on Recombinant Human Granulocyte Macrophage-Colony Stimulation Factor. Anal. Chem. 2008, 80, 2623–2627. [Google Scholar] [CrossRef]
- Chen, K.; Freedberg, D.I.; Keire, D.A. NMR profiling of biomolecules at natural abundance using 2D 1H–15N and 1H–13C multiplicity-separated (MS) HSQC spectra. J. Magn. Reson. 2015, 251, 65–70. [Google Scholar] [CrossRef]
- Arbogast, L.W.; Delaglio, F.; Schiel, J.E.; Marino, J.P. Multivariate Analysis of Two-Dimensional 1H, 13C Methyl NMR Spectra of Monoclonal Antibody Therapeutics To Facilitate Assessment of Higher Order Structure. Anal. Chem. 2017, 89, 11839–11845. [Google Scholar] [CrossRef] [Green Version]
- Haxholm, G.W.; Petersen, B.O.; Malmstrøm, J. Higher-Order Structure Characterization of Pharmaceutical Proteins by 2D Nuclear Magnetic Resonance Methyl Fingerprinting. J. Pharm. Sci. 2019, 108, 3029–3035. [Google Scholar] [CrossRef]
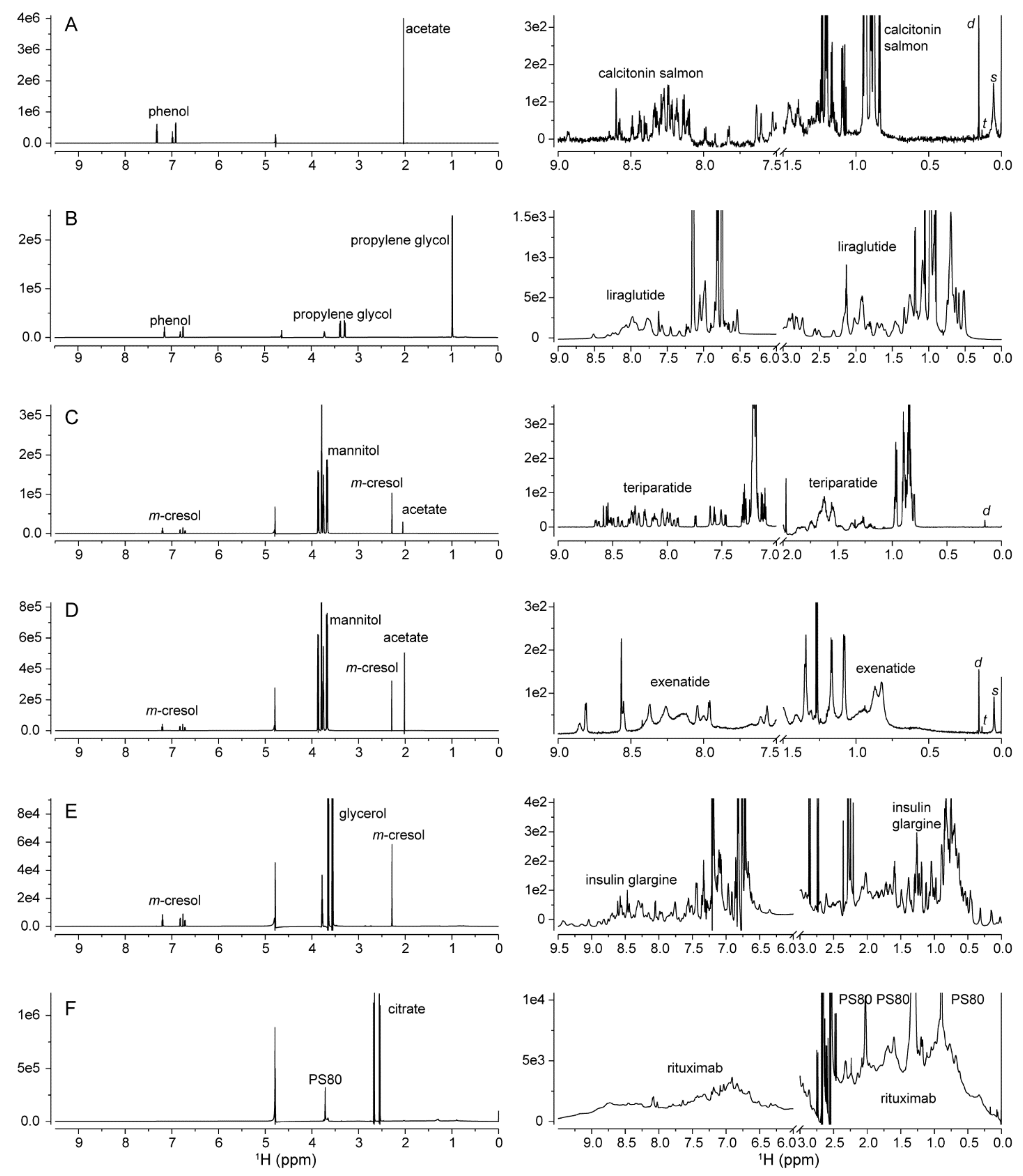
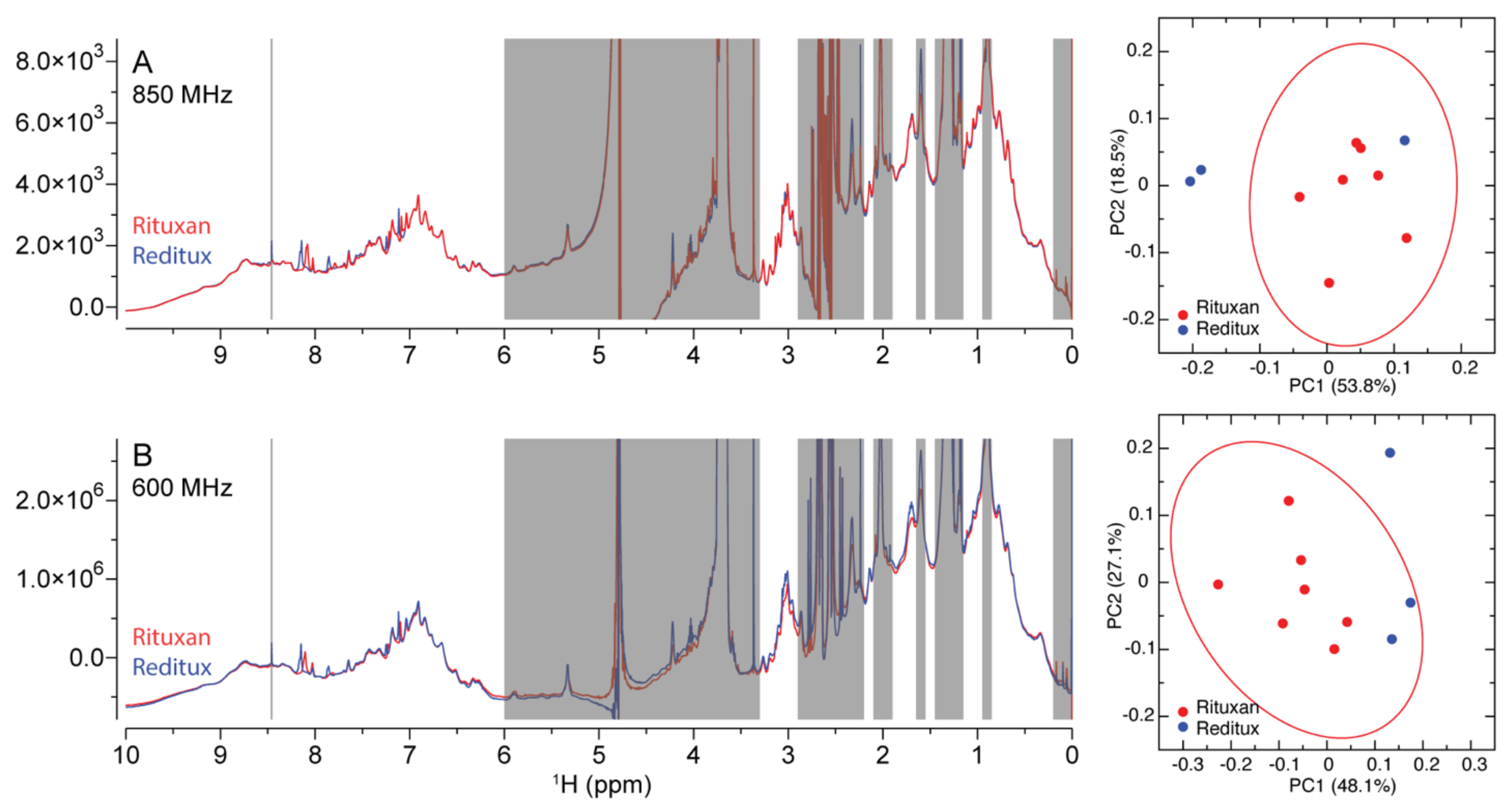
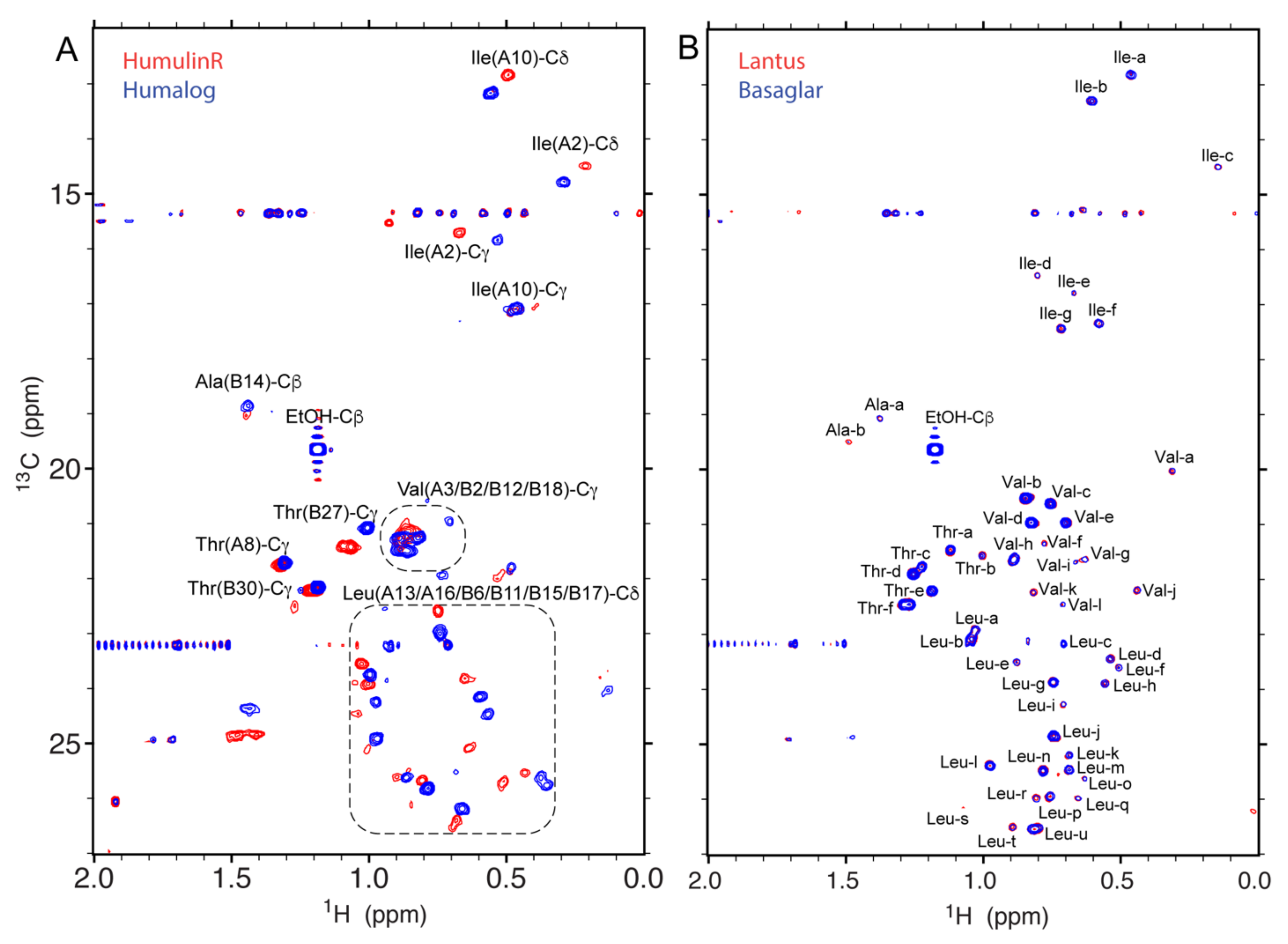

| Drug Product | Drug Substance | Number of Amino Acids | M.W. (kDa) | Concentration (mM) | pH |
|---|---|---|---|---|---|
| Miacalcin® | Calcitonin-Salmon | 32 | 3.43 | 0.0097 1 | n/a |
| Saxenda® | Liraglutide | 38 | 3.75 | 1.6 | 8.15 |
| Forteo® | Teriparatide | 34 | 4.12 | 0.061 | 4 |
| Byetta® | Exenatide | 39 | 4.19 | 0.060 | 4.5 |
| HumulinR® | Insulin Human | 51 | 5.81 | 0.60 2 | n/a |
| Humalog® | Insulin Lispro | 51 | 5.81 | 0.60 2 | 7.0–7.8 |
| Lantus®Basaglar® | Insulin Glargine | 53 | 6.06 | 0.60 | 4 |
| Rituxan®Reditux® | Rituximab | 1328 | 145 | 0.069 | 6.5 |
| Peak | Antus® | Basaglar® | p Value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lot 1 | Lot 2 | Lot 3 | Lot 4 | Lot 5 | Lot 1 | Lot 2 | Lot 3 | Lot 4 | Lot 5 | ||
| Ala-a | 17.5 | 20.7 | 19.8 | 20.8 | 23.4 | 20.2 | 17.0 | 22.7 | 20.8 | 17.6 | 0.61 |
| Ala-b | 18.0 | 19.0 | 17.4 | 18.1 | 19.5 | 16.3 | 18.8 | 17.3 | 18.7 | 20.7 | 0.96 |
| Ile-a | 30.2 | 34.7 | 32.7 | 30.6 | 32.8 | 34.5 | 33.2 | 29.6 | 35.5 | 32.4 | 0.54 |
| Ile-b | 35.3 | 34.5 | 33.6 | 32.5 | 33.3 | 34.9 | 34.3 | 32.2 | 34.1 | 33.3 | 0.92 |
| Ile-c | 20.1 | 21.9 | 19.2 | 17.8 | 17.4 | 20.9 | 21.1 | 15.5 | 18.3 | 21.0 | 0.96 |
| Ile-d | 20.0 | 16.8 | 17.8 | 16.9 | 18.0 | 19.8 | 20.8 | 17.2 | 15.2 | 20.2 | 0.57 |
| Ile-e | 16.9 | 15.9 | 16.4 | 17.3 | 19.2 | 19.3 | 15.7 | 16.5 | 16.7 | 17.9 | 0.93 |
| Ile-f | 29.8 | 32.3 | 27.4 | 30.4 | 31.9 | 30.6 | 31.4 | 35.0 | 28.7 | 30.7 | 0.52 |
| Ile-g | 32.0 | 31.4 | 30.6 | 30.9 | 34.6 | 31.0 | 32.6 | 31.8 | 31.0 | 30.4 | 0.54 |
| Leu-a | 27.6 | 26.4 | 26.7 | 27.2 | 30.7 | 29.1 | 27.9 | 25.4 | 26.6 | 30.8 | 0.84 |
| Leu-b | 52.8 | 52.0 | 48.8 | 49.4 | 50.7 | 50.9 | 49.3 | 50.3 | 46.5 | 50.3 | 0.28 |
| Leu-c | 36.9 | 37.6 | 36.3 | 39.3 | 38.4 | 39.9 | 35.9 | 38.0 | 38.3 | 37.2 | 0.88 |
| Leu-d | 25.7 | 25.9 | 26.7 | 24.8 | 25.8 | 26.6 | 25.3 | 27.0 | 27.0 | 27.2 | 0.11 |
| Leu-e | 22.0 | 17.7 | 18.8 | 20.9 | 21.1 | 23.4 | 20.9 | 22.3 | 19.4 | 20.0 | 0.35 |
| Leu-f | 22.5 | 18.8 | 21.6 | 21.2 | 24.0 | 23.6 | 25.7 | 22.1 | 18.5 | 22.3 | 0.58 |
| Leu-g | 41.1 | 41.1 | 39.6 | 41.7 | 46.2 | 44.7 | 43.6 | 43.5 | 40.8 | 45.7 | 0.26 |
| Leu-h | 23.3 | 26.6 | 23.6 | 25.6 | 25.8 | 25.9 | 24.6 | 21.2 | 21.6 | 25.7 | 0.35 |
| Leu-i | 20.1 | 22.2 | 16.9 | 21.1 | 24.8 | 21.3 | 21.0 | 21.1 | 20.2 | 19.9 | 0.83 |
| Leu-j | 46.4 | 50.1 | 44.5 | 49.9 | 52.0 | 51.8 | 50.8 | 48.3 | 49.6 | 51.1 | 0.28 |
| Leu-k | 21.7 | 25.4 | 21.7 | 23.1 | 23.0 | 26.2 | 22.1 | 23.7 | 24.9 | 22.6 | 0.38 |
| Leu-l | 42.4 | 41.8 | 41.6 | 41.3 | 42.3 | 43.9 | 41.4 | 37.5 | 38.0 | 42.9 | 0.43 |
| Leu-m | 31.2 | 31.0 | 32.7 | 31.2 | 33.9 | 34.2 | 34.6 | 29.1 | 32.9 | 36.0 | 0.34 |
| Leu-n | 36.2 | 38.2 | 37.2 | 38.4 | 42.3 | 40.8 | 37.6 | 37.3 | 40.5 | 40.5 | 0.52 |
| Leu-o | 18.1 | 17.6 | 18.8 | 21.4 | 18.5 | 18.8 | 17.3 | 20.2 | 21.6 | 17.7 | 0.85 |
| Leu-p | 30.6 | 30.6 | 29.4 | 28.0 | 28.1 | 31.7 | 28.7 | 24.7 | 26.4 | 30.8 | 0.57 |
| Leu-q | 19.1 | 21.8 | 19.7 | 16.1 | 18.3 | 18.6 | 19.9 | 15.1 | 16.7 | 18.7 | 0.38 |
| Leu-r | 24.2 | 22.7 | 23.0 | 21.9 | 21.5 | 22.6 | 24.7 | 23.5 | 20.5 | 22.2 | 0.96 |
| Leu-s | 15.3 | 19.1 | 18.4 | 17.3 | 21.4 | 15.7 | 17.9 | 15.6 | 18.9 | 21.0 | 0.75 |
| Leu-t | 23.0 | 25.3 | 23.7 | 25.3 | 22.4 | 21.7 | 21.5 | 21.6 | 19.6 | 21.3 | 0.0055 |
| Leu-u | 42.5 | 38.8 | 40.1 | 41.6 | 42.2 | 43.2 | 40.2 | 42.2 | 42.8 | 47.5 | 0.17 |
| Thr-a | 45.8 | 45.8 | 41.2 | 40.9 | 42.8 | 43.8 | 41.9 | 44.8 | 42.4 | 46.0 | 0.73 |
| Thr-b | 27.2 | 27.0 | 24.0 | 25.0 | 25.0 | 24.1 | 25.8 | 26.1 | 25.9 | 25.5 | 0.83 |
| Thr-c | 39.4 | 45.2 | 42.5 | 43.8 | 46.1 | 45.9 | 44.2 | 47.3 | 44.7 | 44.3 | 0.21 |
| Thr-d | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | n/a |
| Thr-e | 64.0 | 65.5 | 61.5 | 64.8 | 67.4 | 68.5 | 68.0 | 64.5 | 65.5 | 66.9 | 0.13 |
| Thr-f | 60.2 | 61.1 | 59.0 | 58.8 | 61.9 | 65.1 | 64.5 | 58.8 | 58.1 | 60.7 | 0.46 |
| Val-a | 22.5 | 21.6 | 22.1 | 19.1 | 20.0 | 22.3 | 21.7 | 18.7 | 20.6 | 21.3 | 0.85 |
| Val-b | 92.1 | 91.1 | 87.1 | 88.6 | 91.4 | 91.2 | 86.6 | 87.4 | 88.4 | 89.6 | 0.29 |
| Val-c | 67.2 | 65.7 | 64.6 | 68.9 | 71.8 | 73.0 | 70.7 | 67.1 | 69.2 | 69.6 | 0.2 |
| Val-d | 84.8 | 84.8 | 82.9 | 87.1 | 86.3 | 89.3 | 90.4 | 83.1 | 85.1 | 83.9 | 0.5 |
| Val-e | 64.2 | 65.2 | 63.6 | 67.1 | 65.6 | 68.1 | 67.2 | 62.5 | 66.4 | 65.3 | 0.52 |
| Val-f | 20.0 | 17.8 | 19.3 | 18.0 | 17.4 | 18.4 | 17.2 | 17.3 | 19.2 | 17.2 | 0.34 |
| Val-g | 20.9 | 20.2 | 21.8 | 21.3 | 23.5 | 20.9 | 21.6 | 21.1 | 22.1 | 25.1 | 0.52 |
| Val-h | 40.0 | 37.7 | 37.8 | 37.6 | 39.6 | 43.3 | 36.4 | 40.0 | 38.3 | 43.3 | 0.29 |
| Val-i | 15.9 | 20.7 | 17.5 | 17.3 | 20.0 | 18.1 | 19.3 | 16.4 | 15.4 | 17.4 | 0.42 |
| Val-j | 24.0 | 20.8 | 21.9 | 16.8 | 21.7 | 22.9 | 23.2 | 20.9 | 20.3 | 19.4 | 0.83 |
| Val-k | 24.2 | 21.1 | 25.7 | 22.8 | 23.0 | 21.5 | 21.9 | 24.0 | 24.3 | 22.0 | 0.55 |
| Val-l | 15.8 | 19.9 | 14.5 | 17.9 | 17.7 | 20.2 | 13.5 | 17.1 | 18.6 | 18.0 | 0.84 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Zhuo, Y.; Karfunkle, M.; Patil, S.M.; Smith, C.J.; Keire, D.A.; Chen, K. NMR Spectroscopy for Protein Higher Order Structure Similarity Assessment in Formulated Drug Products. Molecules 2021, 26, 4251. https://doi.org/10.3390/molecules26144251
Wang D, Zhuo Y, Karfunkle M, Patil SM, Smith CJ, Keire DA, Chen K. NMR Spectroscopy for Protein Higher Order Structure Similarity Assessment in Formulated Drug Products. Molecules. 2021; 26(14):4251. https://doi.org/10.3390/molecules26144251
Chicago/Turabian StyleWang, Deyun, You Zhuo, Mike Karfunkle, Sharadrao M. Patil, Cameron J. Smith, David A. Keire, and Kang Chen. 2021. "NMR Spectroscopy for Protein Higher Order Structure Similarity Assessment in Formulated Drug Products" Molecules 26, no. 14: 4251. https://doi.org/10.3390/molecules26144251
APA StyleWang, D., Zhuo, Y., Karfunkle, M., Patil, S. M., Smith, C. J., Keire, D. A., & Chen, K. (2021). NMR Spectroscopy for Protein Higher Order Structure Similarity Assessment in Formulated Drug Products. Molecules, 26(14), 4251. https://doi.org/10.3390/molecules26144251






