Oxazolidinone Antibiotics: Chemical, Biological and Analytical Aspects
Abstract
:1. Introduction
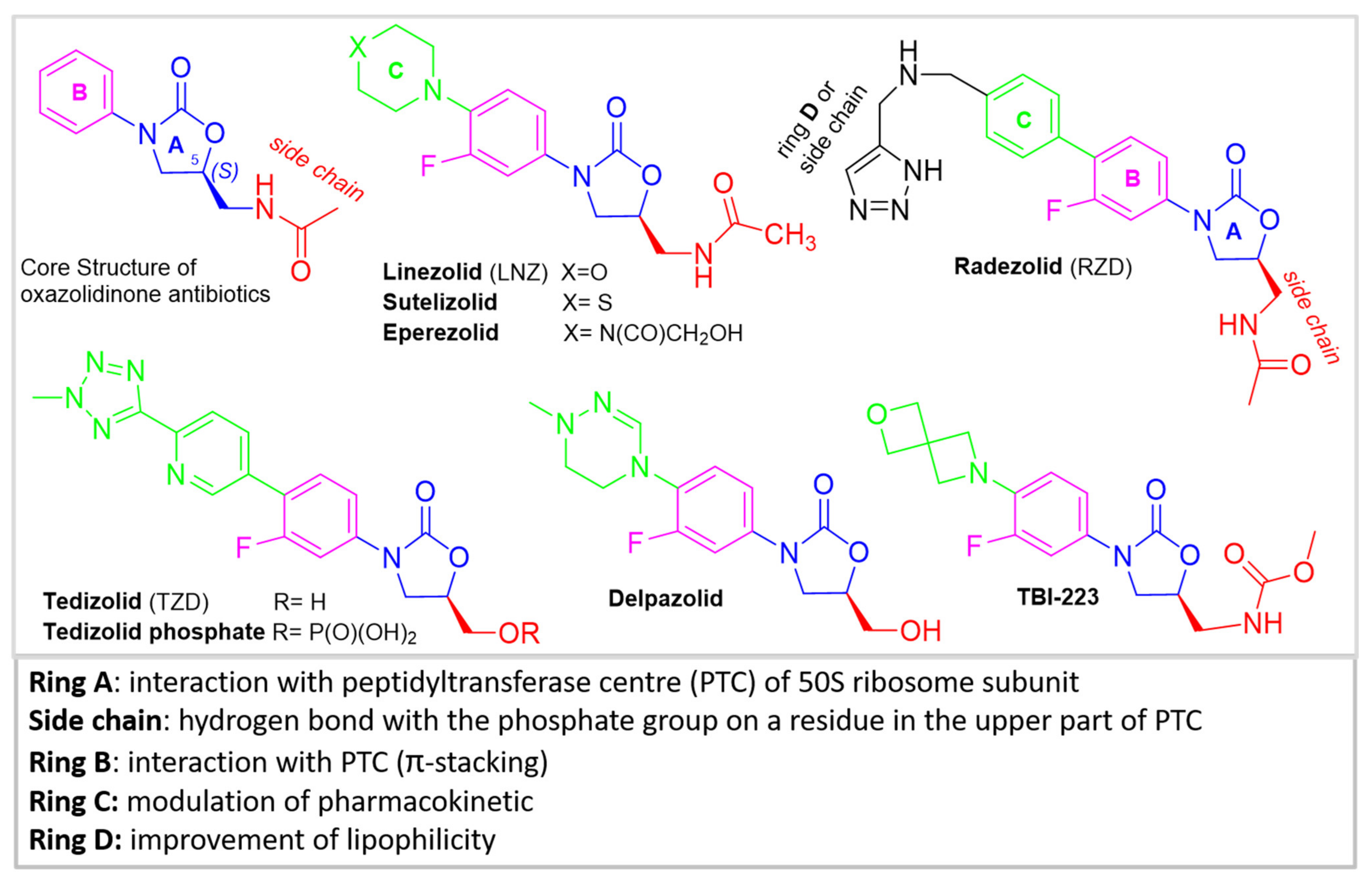
2. Chemistry of Oxazolidinone Antibiotics
3. Analytical Determination
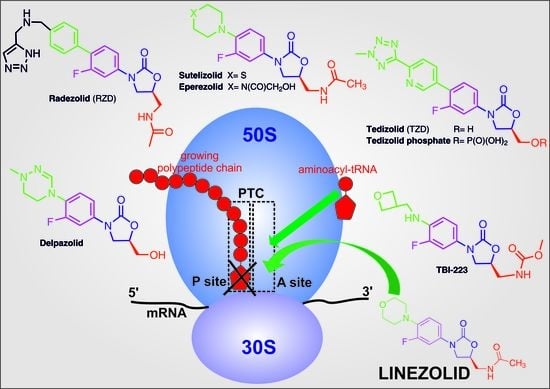
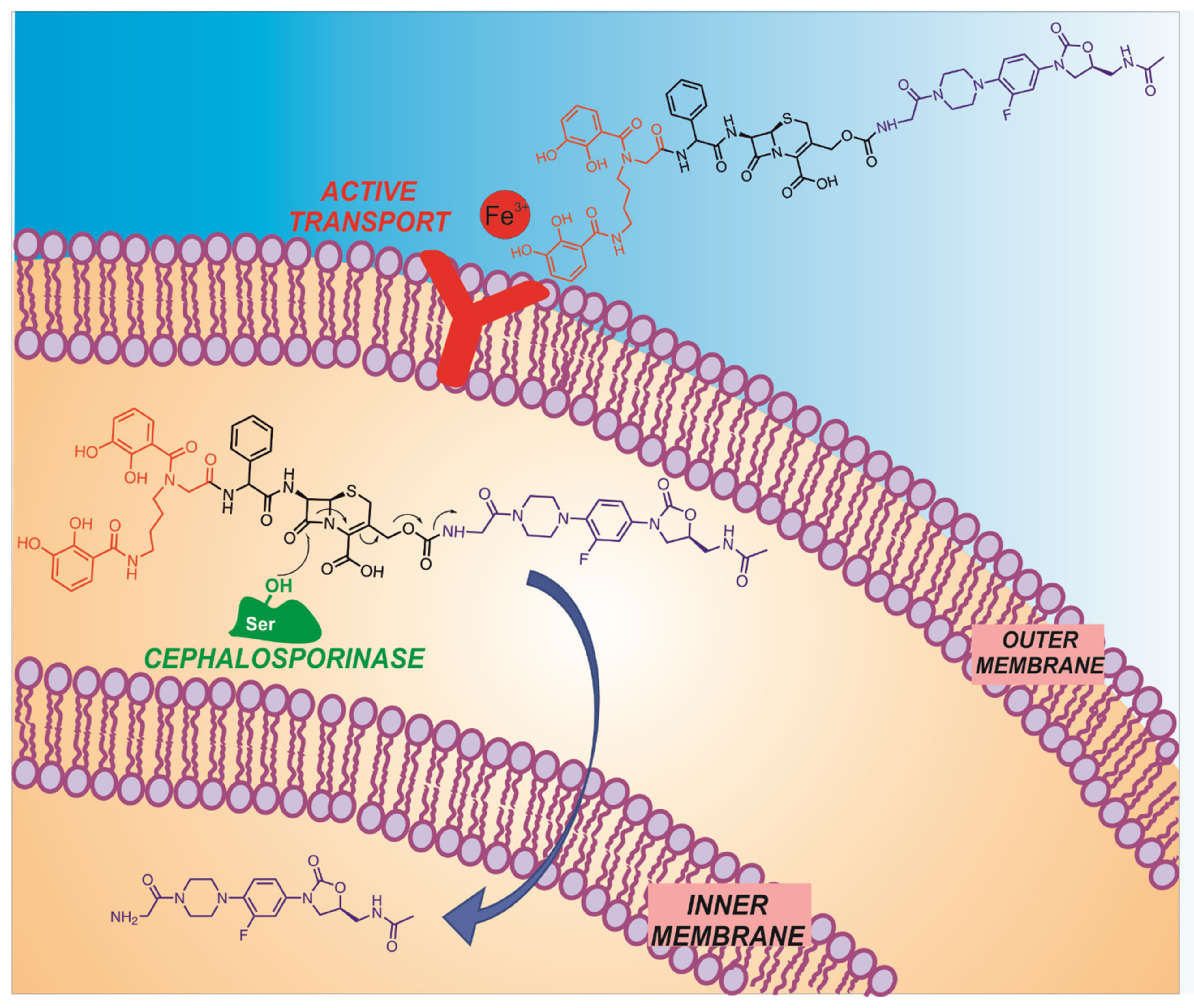
4. Biological Activity of Oxazolidinones and Delivery Systems
5. Perspectives, Challenges and Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Clardy, J.; Fischbach, M.; Currie, C.R. The natural history of antibiotics. Curr. Biol. 2009, 19, R437–R441. [Google Scholar] [CrossRef] [Green Version]
- Da Cunha, B.R.; Fonseca, L.P.; Calado, C.R.C. Antibiotic Discovery: Where Have We Come from, Where Do We Go? Antibiotics 2019, 8, 45. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Ayukekbong, J.A.; Ntemgwa, M.; Atabe, A.N. The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob. Resist. Infect. Control 2017, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jampilek, J. Design and Discovery of New Antibacterial Agents: Advances, Perspectives, Challenges. Curr. Med. Chem. 2019, 25, 4972–5006. [Google Scholar] [CrossRef]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Zignol, M.; Dean, A.S.; Falzon, D.; Van Gemert, W.; Wright, A.; Van Deun, A.; Portaels, F.; Laszlo, A.; Espinal, M.A.; Pablos-Méndez, A.; et al. Twenty Years of Global Surveillance of Antituberculosis-Drug Resistance. N. Engl. J. Med. 2016, 375, 1081–1089. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Tuberculosis Report 2015: WHO/HTM/TB/2015.22; World Health Organization (WHO): Geneva, Switzerland, 2015. [Google Scholar]
- Ministero della Salute. Piano Nazionale di Contrasto dell’Antimicrobico-Resistenza (PNCAR) 2017–2020; Ministero della Salute: Rome, Italy, 2017.
- Jiang, J.; Hou, Y.; Duan, M.; Wang, B.; Wu, Y.; Ding, X.; ZhaoJiang, Y. Design, synthesis and antibacterial evaluation of novel oxazolidinone derivatives nitrogen-containing fused heterocyclic moiety. Bioorg. Med. Chem. Lett. 2021, 32, 127660. [Google Scholar] [CrossRef] [PubMed]
- Bozdogan, B.; Appelbaum, P.C. Oxazolidinones: Activity, mode of action, and mechanism of resistance. Int. J. Antimicrob. Agents 2004, 23, 113–119. [Google Scholar] [CrossRef]
- Kokilambigai, K.S.; Lakshmi, K.S.; Sai Susmitha, A.; Seetharaman, R.; Kavitha, J. Linezolid: A Review of Analytical Methods in Pharmaceuticals and Biological Matrices. Crit. Rev. Anal. Chem. 2020, 50, 179–188. [Google Scholar] [CrossRef]
- Dorn, C.; Schießer, S.; Wulkersdorfer, B.; Hitzenbichler, F.; Kees, M.G.; Zeitlinger, M. Determination of free clindamycin, flucloxacillin or tedizolid in plasma: Pay attention to physiological conditions when using ultrafiltration. Biomed. Chromatogr. 2020, 34, e4820. [Google Scholar] [CrossRef] [Green Version]
- Makarov, G.; Reshetnikova, R. Investigation of radezolid interaction with non-canonical chloramphenicol binding site by molecular dynamics simulations. J. Mol. Graph. Model. 2021, 105, 107902. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, D.T.; Liu, J.; Lee, R.B.; Lee, R.E. New agents for the treatment of drug-resistant Mycobacterium tuberculosis. Adv. Drug Deliv. Rev. 2016, 102, 55–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michalska, K.; Lewandowska, K.; Mizera, M.; Bocian, W.; Pałys, B.; Cielecka-Piontek, J. Spectroscopic identification of intermediates and final products of the chiral pool synthesis of sutezolid. J. Mol. Struct. 2020, 1217, 128396. [Google Scholar] [CrossRef]
- Takrouri, K.; Cooper, H.D.; Spaulding, A.; Zucchi, P.; Koleva, B.; Cleary, D.C.; Tear, W.; Beuning, P.J.; Hirsch, E.B.; Aggen, J.B. Progress against Escherichia coli with the Oxazolidinone Class of Antibacterials: Test Case for a General Approach To Improving Whole-Cell Gram-Negative Activity. ACS Infect. Dis. 2016, 2, 405–426. [Google Scholar] [CrossRef] [PubMed]
- Hampton, T. Report Reveals Scope of US Antibiotic Resistance Threat. JAMA 2013, 310, 1661–1663. [Google Scholar] [CrossRef]
- Nikaido, H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ang, W.; Ye, W.; Sang, Z.; Liu, Y.; Yang, T.; Deng, Y.; Luo, Y.; Wei, Y. Discovery of novel bis-oxazolidinone compounds as potential potent and selective antitubercular agents. Bioorg. Med. Chem. Lett. 2014, 24, 1496–1501. [Google Scholar] [CrossRef] [PubMed]
- Pınar, P.T.; Şentürk, Z. Electrochemical and analytical performance of cathodically pretreated boron-doped diamond electrode for the determination of oxazolidinone antibiotic linezolid in cationic surfactant media. J. Electroanal. Chem. 2020, 878, 114681. [Google Scholar] [CrossRef]
- Barbachyn, M.R.; Ford, C.W. Oxazolidinone Structure-Activity Relationships Leading to Linezolid. Angew. Chem. Int. Ed. 2003, 42, 2010–2023. [Google Scholar] [CrossRef]
- Mukhtar, T.A.; Wright, G.D. Streptogramins, Oxazolidinones, and Other Inhibitors of Bacterial Protein Synthesis. Chem. Rev. 2005, 105, 529–542. [Google Scholar] [CrossRef]
- Chellat, M.F.; Raguž, L.; Riedl, R. Targeting Antibiotic Resistance. Angew. Chem. Int. Ed. 2016, 55, 6600–6626. [Google Scholar] [CrossRef] [PubMed]
- Ippolito, J.A.; Kanyo, Z.F.; Wang, D.; Franceschi, F.J.; Moore, P.B.; Steitz, T.A.; Duffy, E.M. Crystal Structure of the Oxazolidinone Antibiotic Linezolid Bound to the 50S Ribosomal Subunit. J. Med. Chem. 2008, 51, 3353–3356. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.; Schluenzen, F.; Harms, J.M.; Starosta, A.; Connell, S.; Fucini, P. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc. Natl. Acad. Sci. USA 2008, 105, 13339–13344. [Google Scholar] [CrossRef] [Green Version]
- Gerson, S.L.; Kaplan, S.L.; Bruss, J.B.; Le, V.; Arellano, F.M.; Hafkin, B.; Kuter, D.J. Hematologic effects of linezolid: Summary of clinical experience. Antimicrob. Agent. Chem. 2002, 46, 2723–2726. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, A.; Kumar, M.; Kumar, A.; Kanchan, K. Comprehensive review on mechanism of action, resistance and evolution of antimycobacterial drugs. Life Sci. 2021, 274, 119301. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Lu, Y.; Sheng, L.; Yuan, Z.; Wang, B.; Wang, W.; Li, Y.; Ma, C.; Wang, X.; Zhang, D.; et al. Discovery of Fluorine-Containing Benzoxazinyl-oxazolidinones for the Treatment of Multidrug Resistant Tuberculosis. ACS Med. Chem. Lett. 2017, 8, 533–537. [Google Scholar] [CrossRef]
- Kong, Q.; Yang, Y. Recent advances in antibacterial agents. Bioorg. Med. Chem. Lett. 2021, 35, 127799. [Google Scholar] [CrossRef]
- Sellarès-Nadal, J.; Burgos, J.; Falcó, V.; Almirante, B. Investigational and Experimental Drugs for Community-Acquired Pneumonia: The Current Evidence. J. Exp. Pharmacol. 2020, 12, 529–538. [Google Scholar] [CrossRef]
- Cho, Y.L.; Jang, J. Development of Delpazolid for the Treatment of Tuberculosis. Appl. Sci. 2020, 10, 2211. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Wang, B.; Fu, L.; Li, G.; Lu, H.; Liu, Y.; Sheng, L.; Li, Y.; Zhang, B.; Lu, Y.; et al. Discovery of a Conformationally Constrained Oxazolidinone with Improved Safety and Efficacy Profiles for the Treatment of Multidrug-Resistant Tuberculosis. J. Med. Chem. 2020, 63. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, B.; Lu, H.; Zhao, H.; Yang, B.; Li, L.; Lu, Y.; Zhang, D.; Sun, N.; Huang, H. Identification of Novel Tricyclic Benzo[1, 3]oxazinyloxazolidinones as Potent Antibacterial Agents with Excellent Pharmacokinetic Profiles against Drug-Resistant Pathogens. J. Med. Chem. 2021, 64, 3234–3248. [Google Scholar] [CrossRef] [PubMed]
- Principi, N.; Argentiero, A.; Neglia, C.; Gramegna, A.; Esposito, S. New Antibiotics for the Treatment of Acute Bacterial Skin and Soft Tissue Infections in Pediatrics. Pharmacy 2020, 13, 333. [Google Scholar] [CrossRef]
- Jubeh, B.; Breijyeh, Z.; Karaman, R. Antibacterial Prodrugs to Overcome Bacterial Resistance. Molecules 2020, 25, 1543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dos Santos Fernandes, G.F.; Nunes Salgado, H.R.; dos Santos, J.L. A critical review of HPLC-based analytical methods for quantification of Linezolid. Crit. Rev. Anal. Chem. 2019, 50, 196–211. [Google Scholar] [CrossRef]
- Jiang, J.; Liu, Y.; Liu, X.; Zhang, D.; Huang, H.; Wang, B.; Sheng, L.; Li, Y. Simultaneous determination of a novel oxazolidinone anti-tuberculosis OTB-658 and its metabolites in monkey blood by LC-MS/MS. J. Chromatogr. B 2021, 1167, 122552. [Google Scholar] [CrossRef]
- Kalam, M.A.; Iqbal, M.; Alshememry, M.; Alkholief, M.; Alshamsan, A. UPLC-MS/MS assay of Tedizolid in rabbit aqueous humor: Application to ocular pharmacokinetic study. J. Chromatogr. B 2021, 1171, 122621. [Google Scholar] [CrossRef]
- Iqbal, M. A highly sensitive and efficient UPLC-MS/MS assay for rapid analysis of tedizolid (a novel oxazoli-dinone antibiotic) in plasma sample. Biomed. Chromatogr. 2016, 30, 1750–1756. [Google Scholar] [CrossRef]
- Belal, F.; Sharaf El-Din, M.K.; Eid, M.I.; El-Gamal, R.M. Spectrofluorimetric determination of terbinafine hydrochloride and linezolid in their dosage forms and human plasma. J. Fluoresc. 2013, 23, 1077–1087. [Google Scholar] [CrossRef] [PubMed]
- Moussa, B.A.; Mahrouse, M.A.; Hassan, M.A.; Fawzy, M.G. Spectrofluorimetric determination of gemifloxacin mesylate and linezolid in pharmaceutical formulations: Application of quinone-based fluorophores and enhanced native fluorescence. Acta Pharm. 2014, 64, 15–28. [Google Scholar] [CrossRef] [Green Version]
- Attiaa, A.K.; Al-Ghobashy, M.A.; El-Sayed, G.M.; Kamala, S.M. Voltammetric monitoring of linezolid, mero-penemand theophylline in plasma. Anal. Biochem. 2018, 545, 54–64. [Google Scholar] [CrossRef]
- Aydin, I.; Akgun, H.; Talay Pınar, P. Analytical determination of the oxazolidinone antibiotic linezolid at a pencil graphite and carbon paste electrodes. Chem. Select 2019, 4, 9966–9971. [Google Scholar] [CrossRef]
- Merli, D.; Pretali, L.; Fasani, E.; Albini, A.; Profumo, A. Analytical Determination and Electrochemical Characterization of the Oxazolidinone Antibiotic Linezolid. Electroanalysis 2011, 23, 2364–2372. [Google Scholar] [CrossRef]
- Mohammed, M.A.; Abdelwahab, N.S.; Banks, C.E. Electroanalytical sensing of the antimicrobial drug linezolid utilising an electrochemical sensing platform based upon a multiwalled carbon nanotubes/bromocresol green modified carbon paste electrode. Anal. Methods 2016, 8, 4345–4353. [Google Scholar] [CrossRef]
- Prashanth, S.; Teradal, N.; Seetharamappa, J.; Satpati, A.K.; Reddy, A. Fabrification of electroreduced graphene oxide–bentonite sodium composite modified electrode and its sensing application for linezolid. Electrochim. Acta 2014, 133, 49–56. [Google Scholar] [CrossRef]
- Leach, K.L.; Swaney, S.M.; Colca, J.R.; McDonald, W.G.; Blinn, J.R.; Thomasco, L.M.; Gadwood, R.C.; Shinabarger, D.; Xiong, L.; Mankin, A.S. The Site of Action of Oxazolidinone Antibiotics in Living Bacteria and in Human Mitochondria. Mol. Cell 2007, 26, 393–402. [Google Scholar] [CrossRef]
- Sadowy, E. Linezolid resistance genes and genetic elements enhancing their dissemination in enterococci and streptococci. Plasmid 2018, 99, 89–98. [Google Scholar] [CrossRef]
- Stefani, S.; Bongiorno, D.; Mongelli, G.; Campanile, F. Linezolid Resistance in Staphylococci. Pharmacy 2010, 3, 1988–2006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortún, J.; Martín-Dávila, P.; Navas, E.; Elias, M.J.P.; Cobo, J.; Tato, M.; De La Pedrosa, E.G.-G.; Gómez-Mampaso, E.; Moreno, S. Linezolid for the treatment of multidrug-resistant tuberculosis. J. Antimicrob. Chemother. 2005, 56, 180–185. [Google Scholar] [CrossRef] [Green Version]
- Falagas, M.E.; Siempos, I.I.; Papagelopoulos, P.J.; Vardakas, K.Z. Linezolid for the treatment of adults with bone and joint infections. Int. J. Antimicrob. Agents 2007, 29, 233–239. [Google Scholar] [CrossRef]
- Paczkowska-Walendowska, M.; Rosiak, N.; Tykarska, E.; Michalska, K.; Płazińska, A.; Płaziński, W.; Szymanowska, D.; Cielecka-Piontek, J. Tedizolid-Cyclodextrin System as Delayed-Release Drug Delivery with Antibacterial Activity. Int. J. Mol. Sci. 2020, 22, 115. [Google Scholar] [CrossRef]
- Paczkowska, M.; Cielecka-Piontek, J. Enhanced tedizolid solubility and permeability due to its interactions with hydrophilic biopolymers. Acta Pharm. Hung. 2018, 88, 195. [Google Scholar]
- Boncu, T.E.; Guclu, A.U.; Catma, M.F.; Savaser, A.; Gokce, A.; Ozdemir, N. In vitro and in vivo evaluation of linezolid loaded electrospun PLGA and PLGA/PCL fiber mats for prophylaxis and treatment of MRSA induced prosthetic infections. Int. J. Pharm. 2020, 573, 118758. [Google Scholar] [CrossRef]
- Boncu, T.E.; Ozdemir, N.; Guclu, A.U. Electrospinning of linezolid loaded PLGA nanofibers: Effect of solvents on its spinnability, drug delivery, mechanical properties, and antibacterial activities. Drug Dev. Ind. Pharm. 2020, 46, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Harjai, K.; Chhibber, S. Local delivery of linezolid from poly-d,l-lactide (PDLLA)–linezolid–coated or-thopaedic implants to prevent MRSA mediated post-arthroplasty infections. Diagn. Microbiol. Infect. Dis. 2014, 79, 387–392. [Google Scholar] [CrossRef]
- Guo, P.; Buttaro, B.A.; Xue, H.Y.; Tran, N.T.; Wong, H.L. Lipid-polymer hybrid nanoparticles carrying linezolid improve treatment of methicillin-resistant Staphylococcus aureus (MRSA) harbored inside bone cells and biofilms. Eur. J. Pharm. Biopharm. 2020, 151, 189–198. [Google Scholar] [CrossRef]
- Patil, V.; Bagade, S.B.; Bonde, S.C. In-vitro and ex-vivo characterization of novel mannosylated gelatin nano-particles of linezolid by quality-by-design approach. J. Drug Deliv. Sci. Technol. 2020, 60, 101976. [Google Scholar] [CrossRef]
- Boyce, J.H.; Dang, B.; Ary, B.; Edmondson, Q.; Craik, C.S.; DeGrado, W.F.; Seiple, I.B. Platform to Discover Protease-Activated Antibiotics and Application to Siderophore–Antibiotic Conjugates. J. Am. Chem. Soc. 2020, 142, 21310–21321. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Miller, P.A.; Vakulenko, S.B.; Stewart, N.K.; Boggess, W.C.; Miller, M.J. A Synthetic Dual Drug Sideromycin Induces Gram-Negative Bacteria To Commit Suicide with a Gram-Positive Antibiotic. J. Med. Chem. 2018, 61, 3845–3854. [Google Scholar] [CrossRef]
- Global Tuberculosis Report 2020. Released on 14 October 2020. Available online: https://www.who.int/publications/i/item/9789240013131 (accessed on 12 July 2021).
- Upadhyay, S.K.; Dan, S.; Girdhar, M.; Rastogi, K. Recent Advancement in SARS-CoV-2 Diagnosis, Treatment, and Vaccine Formulation: A New Paradigm of Nanotechnology in Strategic Combating of COVID-19 Pandemic. Curr. Pharmacol. Rep. 2021, 7, 1–14. [Google Scholar] [CrossRef]
- Micale, N.; Piperno, A.; Mahfoudh, N.; Schurigt, U.; Schultheis, M.; Mineo, P.G.; Schirmeister, T.; Scala, A.; Grassi, G. A hyaluronic acid–pentamidine bioconjugate as a macrophage mediated drug targeting delivery system for the treatment of leishmaniasis. RSC Adv. 2015, 5, 95545–95550. [Google Scholar] [CrossRef] [Green Version]
- Scala, A.; Piperno, A.; Micale, N.; Mineo, P.G.; Abbadessa, A.; Risoluti, R.; Castelli, G.; Bruno, F.; Vitale, F.; Cascio, A.; et al. “Click” on PLGA-PEG and hyaluronic acid: Gaining access to anti-leishmanial pentamidine bioconjugates. J. Biomed. Mater. Res. Part B Appl. Biomater. 2017, 106, 2778–2785. [Google Scholar] [CrossRef] [PubMed]
- Neri, F.; Scala, A.; Grimato, S.; Santoro, M.; Spadaro, S.; Barreca, F.; Cimino, F.; Speciale, A.; Saija, A.; Grassi, G.; et al. Biocompatible silver nanoparticles embedded in a PEG–PLA polymeric matrix for stimulated laser light drug release. J. Nanoparticle Res. 2016, 18. [Google Scholar] [CrossRef]
- Scala, A.; Piperno, A.; Grassi, G.; Scolaro, L.M.; Mazzaglia, A. Nanoconstructs Based on Cyclodextrins for Antimicrobial Applications. In Nano- and Microscale Drug Delivery Systems: Design and Fabrication; Elsevier: Amsterdam, The Netherlands, 2017; pp. 229–244. [Google Scholar]
- Sharma, S.; Tiwari, M.; Tiwari, V. Therapeutic strategies against autophagic escape by pathogenic bacteria. Drug Discov. Today 2021, 26, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Jiang, G.; Gao, R.; Chen, G.; Ren, Y.; Liu, J.; van der Mei, H.C.; Busscher, H.J. Circumventing antimicrobi-al-resistance and preventing its development in novel, bacterial infection-control strategies. Expert Opin. Drug Deliv. 2020, 17, 1151–1164. [Google Scholar] [CrossRef]
- Foti, C.; Giuffrè, O. Interaction of Ampicillin and Amoxicillin with Mn2+: A Speciation Study in Aqueous Solution. Molecules 2020, 25, 3110. [Google Scholar] [CrossRef]
- Giuffrè, O.; Angowska, S.; Foti, C.; Sammartano, S. Thermodynamic Study on the Interaction of Ampicillin and Amoxicillin with Ca2+ in Aqueous Solution at Different Ionic Strengths and Temperatures. J. Chem. Eng. Data 2019, 64, 800–809. [Google Scholar] [CrossRef]
- Cardiano, P.; Cigala, R.M.; Crea, F.; De Stefano, C.; Giuffrè, O.; Sammartano, S.; Vianelli, G. Potentiometric, UV and 1H NMR study on the interaction of penicillin derivatives with Zn(II) in aqueous solution. Biophys. Chem. 2017, 223, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cardiano, P.; Crea, F.; Foti, C.; Giuffrè, O.; Sammartano, S. Potentiometric, UV and 1H NMR study on the interaction of Cu2+ with ampicillin and amoxicillin in aqueous solution. Biophys. Chem. 2017, 224, 59–66. [Google Scholar] [CrossRef] [PubMed]
- De Barros, A.L.C.; Schmidt, F.F.; de Aquino, S.F.; Afonso, R.J.C.F. Determination of Nine Pharmaceutical Active Compounds in Surface Waters from Paraopeba River Basin in Brazil by LTPE-HPLC-ESI-MS/MS. Environ. Sci. Pollut. Res. Int. 2018, 25, 19962–19974. [Google Scholar] [CrossRef] [PubMed]

| Drug Name | Features | Ref. |
|---|---|---|
| Sutezolid (NU-100480)Pfizer | Phase II of clinical trials was completed. The results showed that the drug was well tolerated and safe. | [28,29] |
| Contezolid (MRX-1) | Phase III of clinical trials was completed in China. It is awaiting approval for the oral treatment of ABSSSI caused by GPB. | [30] |
| Radezolid (RX-1741) | Phase III clinical trials for the treatment of multidrug-resistant infections, including infections caused by LNZ-resistant strains. It is 11-times more active in comparison to LNZ. Currently, its safety profile has not been established and its advantages over LNZ and TDZ are not clear. | [14,31] |
| Delpazolid (LCB01-0371) | Phase I/Phase II of clinical trials are ongoing. The safety profile could be suitable for long-term therapies (i.e., TB). LegoChem Biosciences entered into a license agreement with RMX Biopharma for the development of Delpazolid in China. In addition, Delpazolid received an FDA orphan drug designation. | [32] |
| Posizolid (AZD2563/AZD5847) Astrazeneca | Phase II clinical trials discontinued. The results are not conclusive since the studies for the treatment of TB were discontinued. | [29] |
| TBI-223 | It is under phase I clinical trial (NCT03758612) with the aim to evaluate its safety, tolerability and pharmacokinetics. | [28,33,34] |
| Tedizolid (TZD) formerly Torezolid | TZD is approved for the treatment of acute bacterial skin and soft tissue infections by the FDA and EMA. Compared to LNZ, TZD is significantly less expensive. Oral and intravenous formulations are available. Tedizolid phosphate is an orally absorbed phosphate prodrug of TZD. Tedizolid phosphate was the second oxazolidinone drug approved by FDA for the treatment of MRSA skin infections in 2014. | [35,36] |
| Instrumental Techniques | Oxazolidinone | Matrices | LOD | Linear Concentration Range | Ref. |
|---|---|---|---|---|---|
| HPLC-UV | LNZ | Biological fluids | 0.007 ≤ LOD ≤ 0.5 μg/mL | [37] | |
| HPLC/UPLC-MS/MS | LNZ | Biological fluids | 0.05 ≤ LOD ≤ 0.1 μg/mL | [37] | |
| HPLC-MS/MS | OTB-658 | Monkey blood | 0.74 ng/mL | 10–2000 ng/mL | [38] |
| UPLC-MS/MS | TZD | Plasma | 10 ng/mL * | 0.74–1500 ng/mL | [40] |
| TZD | Humor of rabbit | 1.97 ng/mL | 4.98–1000 ng/mL | [39] | |
| Spectrofluorimetry | LNZ | Pharmaceutical formulations | 110 ng/mL | 0.5–50 μg/mL | [41] |
| LNZ | Pharmaceutical formulations | 4.28 ng/mL | 20–400 ng/mL | [42] | |
| Voltammetry | LNZ | Pharmaceutical formulations | 50 ng/mL | up to 200 μg/mL | [45] |
| LNZ | Pharmaceutical formulations | 11 ng/mL | 0.084–10.5 μg/mL | [47] | |
| LNZ | Plasma | 0.98 ng/mL | 0.0085–2.70 μg/mL | [43] | |
| LNZ | Pharmaceutical and biological samples | 0.47 ng/mL | 0.01–0.2 μg/mL | [44] | |
| LNZ | Cationic surfactant media | 50 ng/mL | 0.25–6.41 μg/mL | [21] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Foti, C.; Piperno, A.; Scala, A.; Giuffrè, O. Oxazolidinone Antibiotics: Chemical, Biological and Analytical Aspects. Molecules 2021, 26, 4280. https://doi.org/10.3390/molecules26144280
Foti C, Piperno A, Scala A, Giuffrè O. Oxazolidinone Antibiotics: Chemical, Biological and Analytical Aspects. Molecules. 2021; 26(14):4280. https://doi.org/10.3390/molecules26144280
Chicago/Turabian StyleFoti, Claudia, Anna Piperno, Angela Scala, and Ottavia Giuffrè. 2021. "Oxazolidinone Antibiotics: Chemical, Biological and Analytical Aspects" Molecules 26, no. 14: 4280. https://doi.org/10.3390/molecules26144280
APA StyleFoti, C., Piperno, A., Scala, A., & Giuffrè, O. (2021). Oxazolidinone Antibiotics: Chemical, Biological and Analytical Aspects. Molecules, 26(14), 4280. https://doi.org/10.3390/molecules26144280