Engineering of Streptoalloteichus tenebrarius 2444 for Sustainable Production of Tobramycin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and General Fermentation and Growth Conditions
2.2. Genome Sequencing, Analysis, and Cloning of the aprK Gene Inactivation Construct
2.3. Generation of the aprK Gene Inactivation Construct
2.4. Construction of the Tobramycin Cluster Overexpression Mutant
2.5. Production Assays
2.6. HPLC-MS Analysis
2.7. Statistics Using Student’s t-Test
3. Results and Discussion
3.1. Identification of the Tobramycin Biosynthetic Gene Cluster in Streptoalloteichus tenebrarius 2444
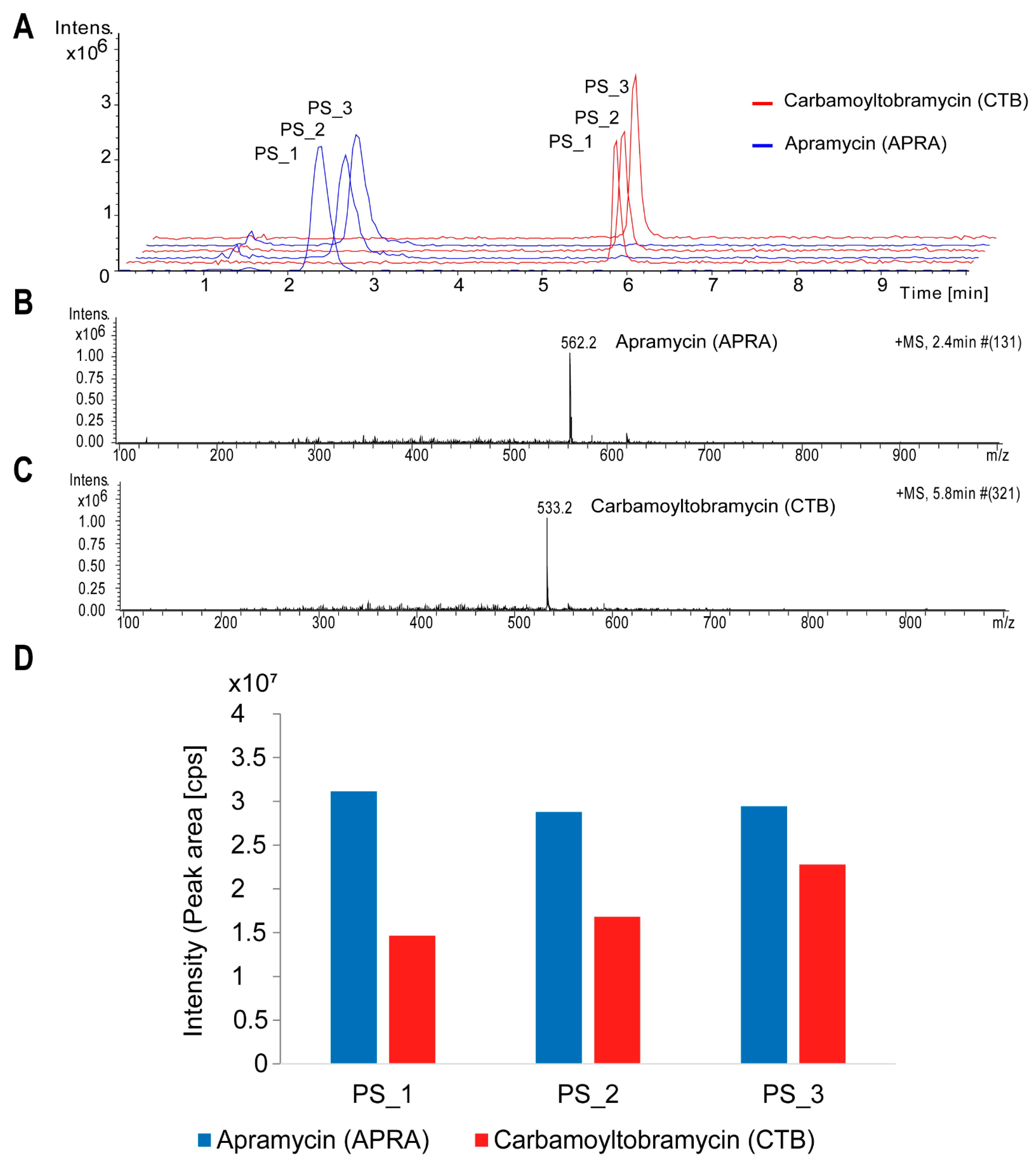
3.2. Aminoglycoside Production in Streptoalloteichus tenebrarius 2444 PS
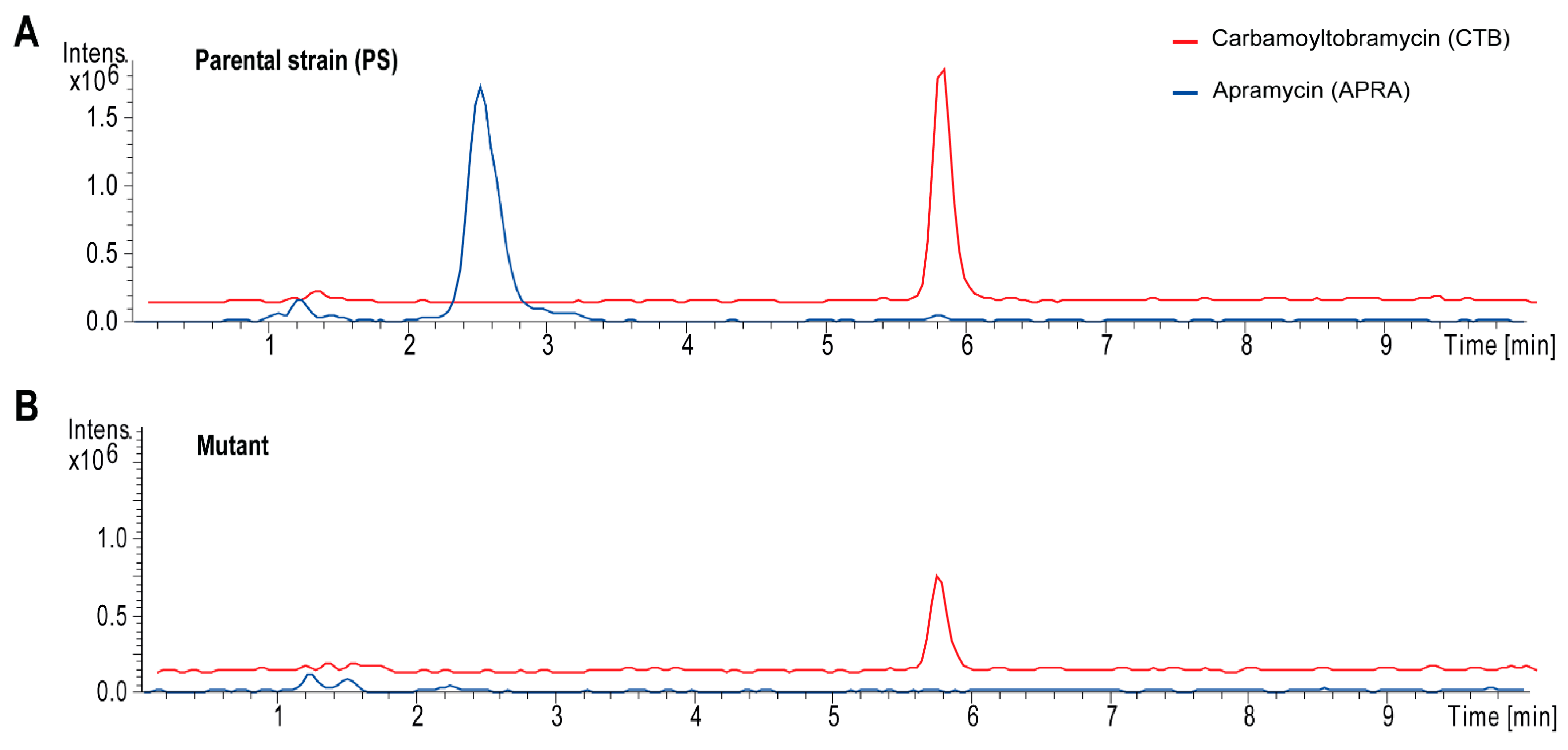
3.3. Elimination of Apramycin Production by Genetic Engineering
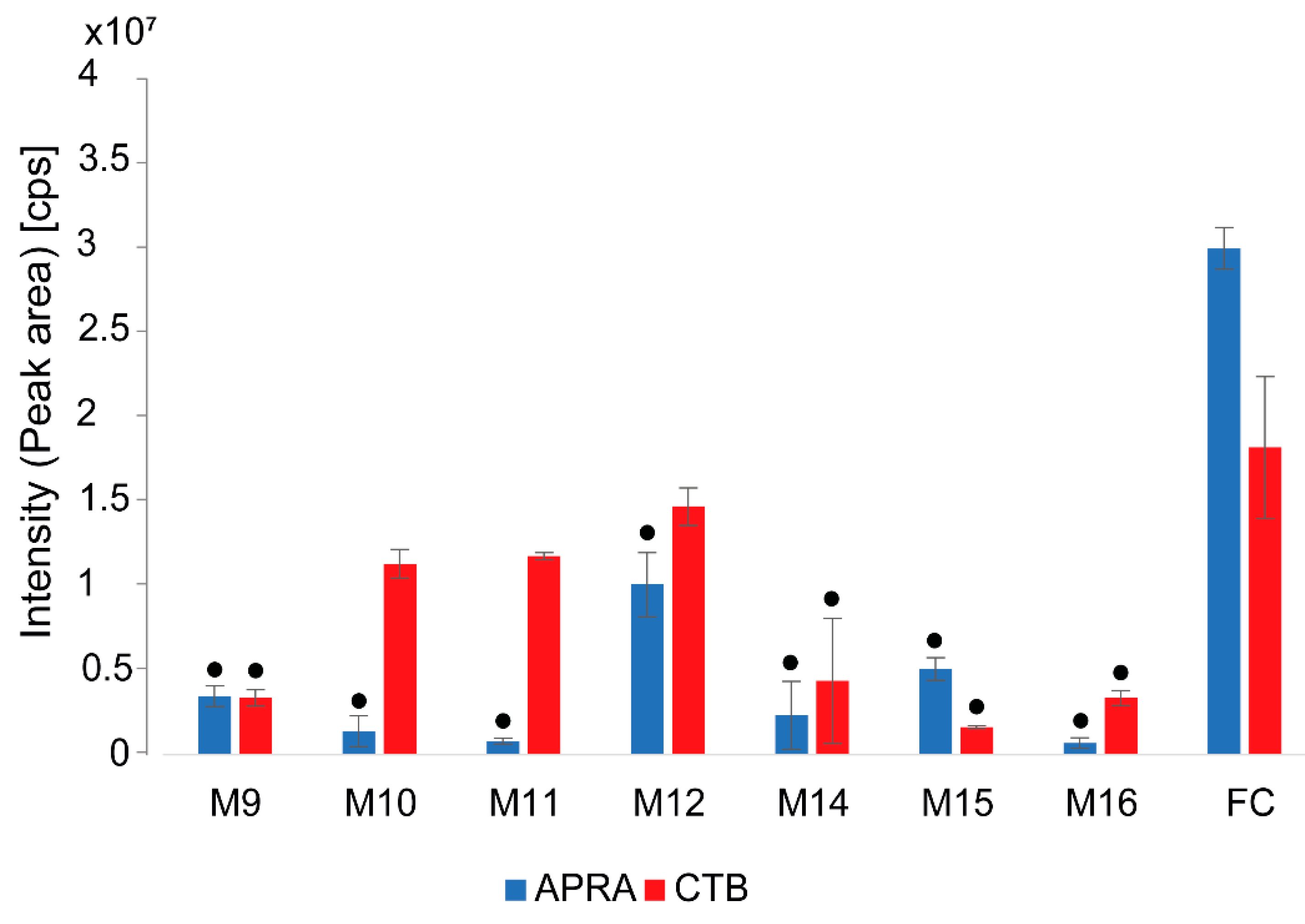
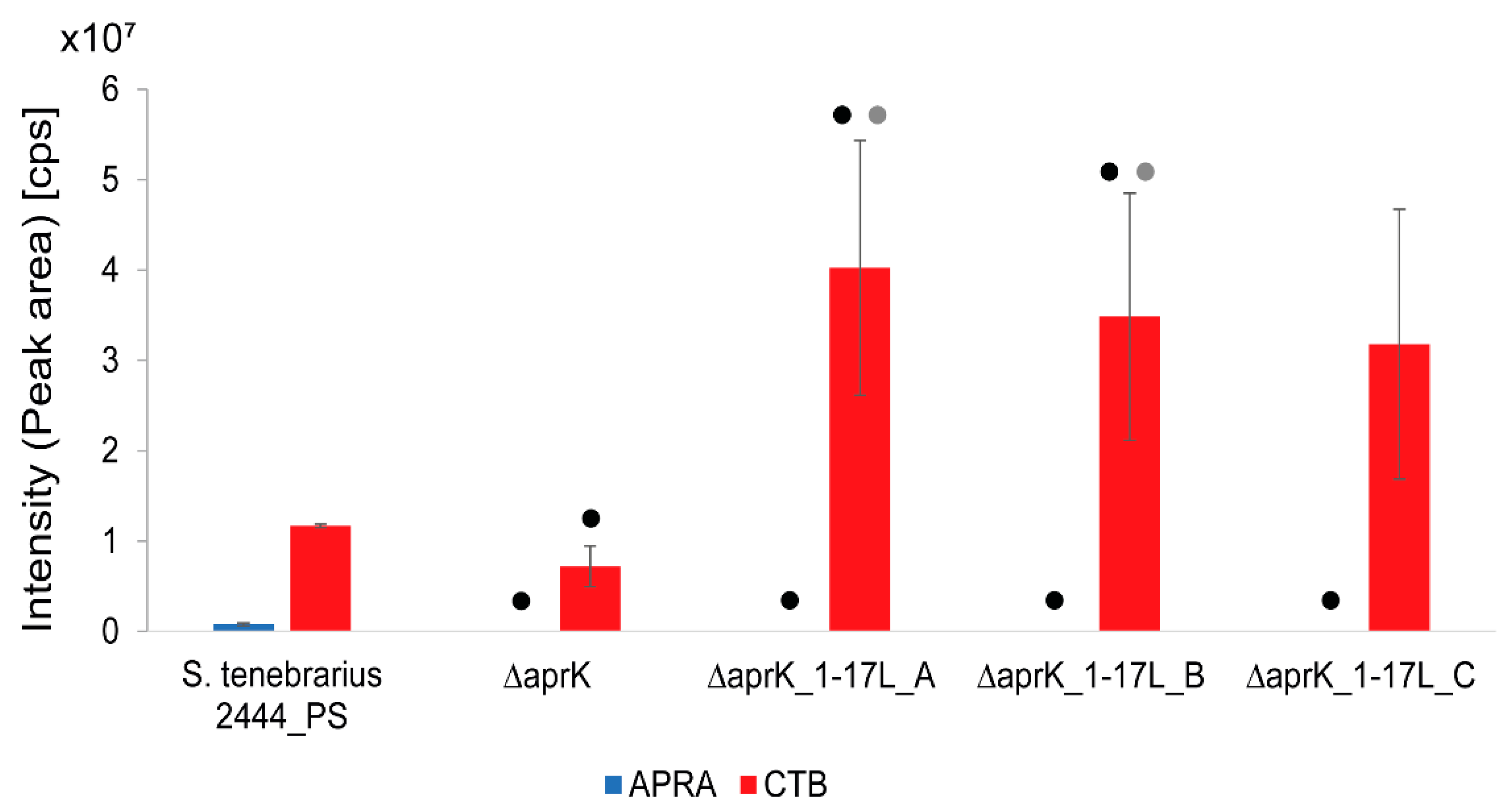
3.4. Overexpression of the Tobramycin Biosynthetic Gene Cluster
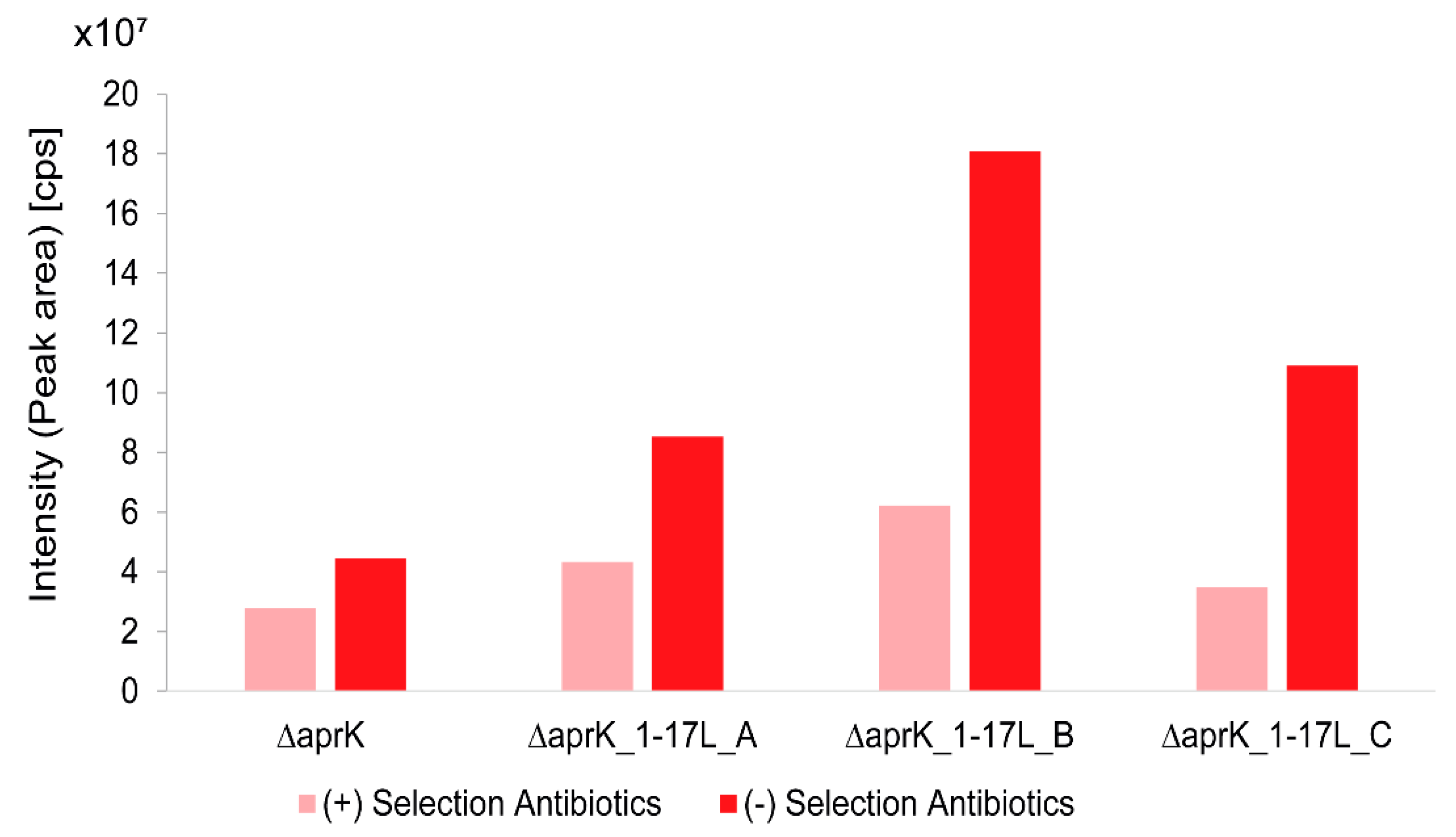
3.5. Effect of Selection Antibiotics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Fosso, M.Y.; Zhu, H.; Green, K.D.; Garneau-Tsodikova, S.; Fredrick, K. Tobramycin Variants with Enhanced Ribosome-Targeting Activity. ChemBioChem 2015, 16, 1565–1570. [Google Scholar] [CrossRef] [PubMed]
- Wasserman, M.R.; Pulk, A.; Zhou, Z.; Altman, R.B.; Zinder, J.C.; Green, K.D.; Garneau-Tsodikova, S.; Cate, J.H.; Blanchard, S.C. Chemically Related 4,5-Linked Aminoglycoside Antibiotics Drive Subunit Rotation in Opposite Directions. Nat. Commun. 2015, 6, 7896. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Pulk, A.; Wasserman, M.R.; Feldman, M.B.; Altman, R.B.; Cate, J.H.; Blanchard, S.C. Allosteric Control of the Ribosome by Small-Molecule Antibiotics. Nat. Struct. Mol. Biol. 2012, 19, 957–963. [Google Scholar] [CrossRef] [Green Version]
- Arya, D.P. Aminoglycoside Antibiotics: From Chemical Biology to Drug Discovery; John Wiley & Sons: Hoboken, NJ, USA, 2007; Volume 5. [Google Scholar]
- Benveniste, R.; Davies, J. Structure-Activity Relationships among the Aminoglycoside Antibiotics: Role of Hydroxyl and Amino Groups. Antimicrob. Agents Chemother. 1973, 4, 402–409. [Google Scholar] [CrossRef] [Green Version]
- Kudo, F.; Eguchi, T. Aminoglycoside Antibiotics: New Insights into the Biosynthetic Machinery of Old Drugs. Chem. Rec. 2016, 16, 4–18. [Google Scholar] [CrossRef]
- Llewellyn, N.M.; Spencer, J.B. Biosynthesis of 2-Deoxystreptamine-Containing Aminoglycoside Antibiotics. Nat. Prod. Rep. 2006, 23, 864–874. [Google Scholar] [CrossRef] [PubMed]
- Piepersberg, W.; Aboshanab, K.M.; Schmidt-Beißner, H.; Wehmeier, U.F. The Biochemistry and Genetics of Aminoglycoside Producers. In Aminoglycoside Antibiotics: From Chemical Biology to Drug Discovery; Arya, D.P., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2007; pp. 15–118. [Google Scholar]
- Lv, M.; Ji, X.; Zhao, J.; Li, Y.; Zhang, C.; Su, L.; Ding, W.; Deng, Z.; Yu, Y.; Zhang, Q. Characterization of a C3 Deoxygenation Pathway Reveals a Key Branch Point in Aminoglycoside Biosynthesis. J. Am. Chem. Soc. 2016, 138, 6427–6435. [Google Scholar] [CrossRef]
- Xiao, J.; Li, H.; Wen, S.; Hong, W. Concentrated Biosynthesis of Tobramycin by Genetically Engineered Streptomyces tenebrarius. J. Gen. Appl. Microbiol. 2014, 60, 256–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, K.; Davis, F.; Rhoades, J. Nebramycin: Separation of the Complex and Identification of Factors 4, 5, and 5′. J. Antibiot. 1973, 26, 745–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, T.; Ishida, Y.; Otoguro, M.; Hatano, K.; Suzuki, K.-I. Classification of ‘Streptomyces tenebrarius’ Higgins and Kastner as Streptoalloteichus tenebrarius nom. rev., comb. nov., and Emended Description of the Genus Streptoalloteichus. Int. J. Syst. Evol. Microbiol. 2008, 58, 688–691. [Google Scholar] [CrossRef] [Green Version]
- Kudo, F.; Eguchi, T. Biosynthetic Genes for Aminoglycoside Antibiotics. J. Antibiot. 2009, 62, 471–481. [Google Scholar] [CrossRef]
- Kharel, M.K.; Basnet, D.B.; Lee, H.C.; Liou, K.; Woo, J.S.; Kim, B.-G.; Sohng, J.K. Isolation and Characterization of the Tobramycin Biosynthetic Gene Cluster from Streptomyces tenebrarius. FEMS Microbiol. Lett. 2004, 230, 185–190. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Huang, F.; Huang, C.; Duan, X.; Jian, X.; Leeper, F.; Deng, Z.; Leadlay, P.F.; Sun, Y. Specificity and Promiscuity at the Branch Point in Gentamicin Biosynthesis. Chem. Biol. 2014, 21, 608–618. [Google Scholar] [CrossRef] [Green Version]
- Park, J.W.; Park, S.R.; Nepal, K.K.; Han, A.R.; Ban, Y.H.; Yoo, Y.J.; Kim, E.J.; Kim, E.M.; Kim, D.; Sohng, J.K.; et al. Discovery of Parallel Pathways of Kanamycin Biosynthesis Allows Antibiotic Manipulation. Nat. Chem. Biol. 2011, 7, 843–852. [Google Scholar] [CrossRef]
- Ni, X.; Li, D.; Yang, L.; Huang, T.; Li, H.; Xia, H. Construction of Kanamycin B Overproducing Strain by Genetic Engineering of Streptomyces tenebrarius. Appl. Microbiol. Biotechnol. 2011, 89, 723–731. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. AntiSMASH 3.0—A Comprehensive Resource for the Genome Mining of Biosynthetic Gene Clusters. Nucleic Acids Res. 2015, 43, 237–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Gish, W.; States, D.J. Identification of Protein Coding Regions by Database Similarity Search. Nat. Genet. 1993, 3, 266–272. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information (NCBI). Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 14 June 2021).
- Yu, Y.; Hou, X.; Ni, X.; Xia, H. Biosynthesis of 3′-Deoxy-Carbamoylkanamycin C in a Streptomyces tenebrarius Mutant Strain by tacB Gene Disruption. J. Antibiot. 2008, 61, 63–69. [Google Scholar] [CrossRef] [Green Version]
- Parthier, C.; Görlich, S.; Jaenecke, F.; Breithaupt, C.; Bräuer, U.; Fandrich, U.; Clausnitzer, D.; Wehmeier, U.F.; Böttcher, C.; Scheel, D.; et al. The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction. Angew. Chem. Int. Ed. Engl. 2012, 51, 4046–4052. [Google Scholar] [CrossRef]
- Kharel, M.K.; Subba, B.; Lee, H.C.; Liou, K.; Sohng, J.K. Characterization of L-Glutamine: 2-Deoxy-Scyllo-Inosose Aminotransferase (tbmB) from Streptomyces tenebrarius. Bioorg. Med. Chem. Lett. 2005, 15, 89–92. [Google Scholar] [CrossRef]
- Lin, Y.-S.; Hong, W.-R. Construction of Streptomyces tenebrarius Mutant with Knockout of tobS 1-C Genes. J. China Pharm. Univ. 2012, 43, 92–96. [Google Scholar]
- Pelzer, S.; Reichert, W.; Huppert, M.; Heckmann, D.; Wohlleben, W. Cloning and Analysis of a Peptide Synthetase Gene of the Balhimycin Producer Amycolatopsis mediterranei DSM5908 and Development of a Gene Disruption/Replacement System. J. Biotechnol. 1997, 56, 115–128. [Google Scholar] [CrossRef]
- Robertsen, H.L.; Musiol-Kroll, E.M.; Ding, L.; Laiple, K.J.; Hofeditz, T.; Wohlleben, W.; Lee, S.Y.; Grond, S.; Weber, T. Filling the Gaps in the Kirromycin Biosynthesis: Deciphering the Role of Genes Involved in Ethylmalonyl-CoA Supply and Tailoring Reactions. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; John Innes Foundation Norwich: Norwich, England, 2000; Volume 291. [Google Scholar]
- Musiol, E.M.; Greule, A.; Härtner, T.; Kulik, A.; Wohlleben, W.; Weber, T. The AT2 Domain of KirCI Loads Malonyl Extender Units to the ACPs of the Kirromycin PKS. ChemBioChem 2013, 14, 1343–1352. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinform 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Flatt, P.M.; Mahmud, T. Biosynthesis of Aminocyclitol-Aminoglycoside Antibiotics and Related Compounds. Nat. Prod. Rep. 2007, 24, 358–392. [Google Scholar] [CrossRef]
- Kharel, M.K.; Subba, B.; Lee, H.C.; Liou, K.; Woo, J.S.; Sohng, J.K. An Approach for Cloning Biosynthetic Genes of 2-Deoxystreptamine-Containing Aminocyclitol Antibiotics: Isolation of a Biosynthetic Gene Cluster of Tobramycin from Streptomyces tenebrarius. Biotechnol. Lett. 2003, 25, 2041–2047. [Google Scholar] [CrossRef] [PubMed]
- Borodina, I.; Schöller, C.; Eliasson, A.; Nielsen, J. Metabolic Network Analysis of Streptomyces tenebrarius, a Streptomyces Species with an Active Entner-Doudoroff Pathway. Appl. Environ. Microbiol. 2005, 71, 2294–2302. [Google Scholar] [CrossRef] [Green Version]
- Majumdar, M.K.; Majumdar, S. Utilization of Carbon-and Nitrogen-Containing Compounds for Neomycin Production by Streptomyces fradiae. Appl. Microbiol. 1967, 15, 744–749. [Google Scholar] [CrossRef]
- Yao, K.; Gao, S.; Wu, Y.; Zhao, Z.; Wang, W.; Mao, Q. Influence of Dextrins on the Production of Spiramycin and Impurity Components by Streptomyces ambofaciens. Folia Microbiol. 2018, 63, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Yanai, K.; Murakami, T.; Bibb, M. Amplification of the Entire Kanamycin Biosynthetic Gene Cluster during Empirical Strain Improvement of Streptomyces kanamyceticus. Proc. Natl. Acad. Sci. USA 2006, 103, 9661–9666. [Google Scholar] [CrossRef] [Green Version]
- Nah, H.-J.; Woo, M.-W.; Choi, S.-S.; Kim, E.-S. Precise Cloning and Tandem Integration of Large Polyketide Biosynthetic Gene Cluster Using Streptomyces Artificial Chromosome System. Microb. Cell Fact. 2015, 14, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.; Xia, L.; Ding, X.; Luo, Y.; Huang, F.; Jiang, Y. Duplication of Partial Spinosyn Biosynthetic Gene Cluster in Saccharopolyspora spinosa Enhances Spinosyn Production. FEMS Microbiol. Lett. 2011, 325, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Liao, G.; Li, J.; Li, L.; Yang, H.; Tian, Y.; Tan, H. Cloning, Reassembling and Integration of the Entire Nikkomycin Biosynthetic Gene Cluster into Streptomyces ansochromogenes Lead to an Improved Nikkomycin Production. Microb. Cell Fact. 2010, 9, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Thoma, L.; Sepulveda, E.; Latus, A.; Muth, G. The Stability Region of the Streptomyces lividans plasmid pIJ101 Encodes a DNA-Binding Protein Recognizing a Highly Conserved Short Palindromic Sequence Motif. Front. Microbiol. 2014, 5, 499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Gelder, L.; Ponciano, J.M.; Joyce, P.; Top, E.M. Stability of a Promiscuous Plasmid in Different Hosts: No Guarantee for a Long-Term Relationship. Microbiology 2007, 153, 452–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Gene | Protein | BlastP Hits (Accession) | Percent Identity (BlastP) | Putative Function | References for Functional Characterization |
|---|---|---|---|---|---|
| tobH1 (tacE) | TobH1 (tacE) | TobH1 (TacE) (CAH18547.1) | 100% | Unknown | - |
| YvrE (SHG81534.1) | 79.59% | Sugar lactone lactonase YvrE | - | ||
| tobX | TobX | TobX (CAH18549.1) | 100% | Unknown (dioxygenase) | - |
| tobE (tacD) | TobE (TacD) | TobE (Q2MF22.1) | 100% | 2-deoxy-scyllo-inosamine dehydrogenase (DOIA dehydrogenase) | - |
| tobT (tbmE) | TobT (TbmE) | TbmE (CAE22476.1) | 100% | Major facilitator superfamily protein | - |
| tobB (tacC) | TobB (TacC) | Hypothetical protein (CAE22475.1) | 100% | Unknown | - |
| Hypothetical protein (WP_073480773.1) | 86.84% | Aminotransferase class III-fold pyridoxal phosphate-dependent enzyme | - | ||
| tobQ (tacB) | TobQ (TacB) | Hypothetical protein (CAE22474.1) | 100% | 6′-dehydrogenase | [22] |
| Hypothetical protein (WP_073480774.1) | 89.37% | GMC family oxidoreductase | - | ||
| tobZ (tacA) | TobZ (TacA) | TacA (CAE22473.1) (Q70IY1.1) | 100% | O-carbamoyltransferase | [10,23] |
| tobS1 (tbmB) | TobS1 (TbmB) | TbmB (Q2MF17.1) | 100% | 2-deoxy-scyllo-inosose aminotransferase (l-glutamine:DOI aminotransferase) | [24,25] |
| tobC (tbmA) | TobC (TbmA) | TbmA (Q2MF16.1) | 100% | 2-deoxy-scyllo-inosose synthase (DOI synthase) | [14,25] |
| tobD2 (tbmC) | TobD2 (TbmC) | TbmC (CAE22470.1) | 100% | Dehydrogenase | - |
| tobM2 (tbmD) | TobM2 (TbmD) | TbmD (CAE22469.1) | 100% | 6-glycosyltransferase | [16] |
| tobN (tbmX) | TobN (TbmX) | TobN (CAH18559.1) | 100% | Amidase | - |
| tobS2 | TobS2 | TobS2 (Q2MF12.1) | 100% | l-glutamine:3-amino-2,3-dideoxy-scyllo-inosose aminotransferase (l-glutamine:amino-DOI aminotransferase) | - |
| tobH2 | TobH2 | Hypothetical protein (CAH18561.1) | 100% | Retrotransposon protein (possibly non-functional) | - |
| tobM1 | TobM1 | TobM1 (CAH18562.1) | 99.76% | Aminoglycoside 4-glucosaminyltransferase | [16] |
| tobH3 | TobH3 | No hits | - | Unknown | - |
| tobH4 | TobH4 | No hits | - | Unknown | - |
| tobL | TobL | TobL (CAH18563.1) | 100% | Carbamoyl-phosphate synthase | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitousis, L.; Maier, H.; Martinovic, L.; Kulik, A.; Stockert, S.; Wohlleben, W.; Stiefel, A.; Musiol-Kroll, E.M. Engineering of Streptoalloteichus tenebrarius 2444 for Sustainable Production of Tobramycin. Molecules 2021, 26, 4343. https://doi.org/10.3390/molecules26144343
Mitousis L, Maier H, Martinovic L, Kulik A, Stockert S, Wohlleben W, Stiefel A, Musiol-Kroll EM. Engineering of Streptoalloteichus tenebrarius 2444 for Sustainable Production of Tobramycin. Molecules. 2021; 26(14):4343. https://doi.org/10.3390/molecules26144343
Chicago/Turabian StyleMitousis, Lena, Hannes Maier, Luka Martinovic, Andreas Kulik, Sigrid Stockert, Wolfgang Wohlleben, Alfred Stiefel, and Ewa M. Musiol-Kroll. 2021. "Engineering of Streptoalloteichus tenebrarius 2444 for Sustainable Production of Tobramycin" Molecules 26, no. 14: 4343. https://doi.org/10.3390/molecules26144343
APA StyleMitousis, L., Maier, H., Martinovic, L., Kulik, A., Stockert, S., Wohlleben, W., Stiefel, A., & Musiol-Kroll, E. M. (2021). Engineering of Streptoalloteichus tenebrarius 2444 for Sustainable Production of Tobramycin. Molecules, 26(14), 4343. https://doi.org/10.3390/molecules26144343







