Effect of Hydrated Ionic Liquid on Photocycle and Dynamics of Photoactive Yellow Protein
Abstract
:1. Introduction
2. Results
2.1. CD Spectra of pG State in Hy[ch][dhp]
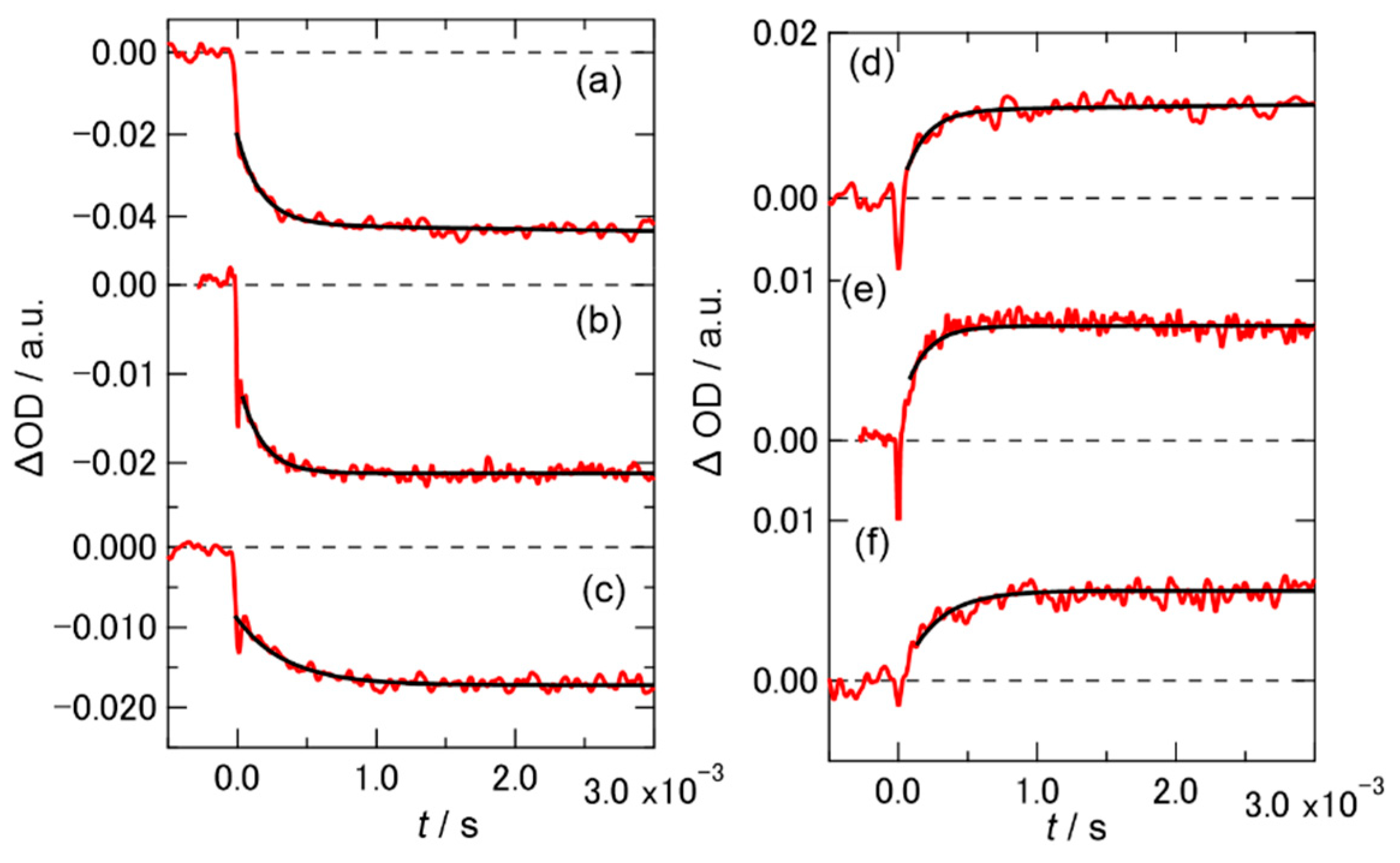
2.2. Time Profiles of the Transient Absorption of PYP in Hy[ch][dhp]
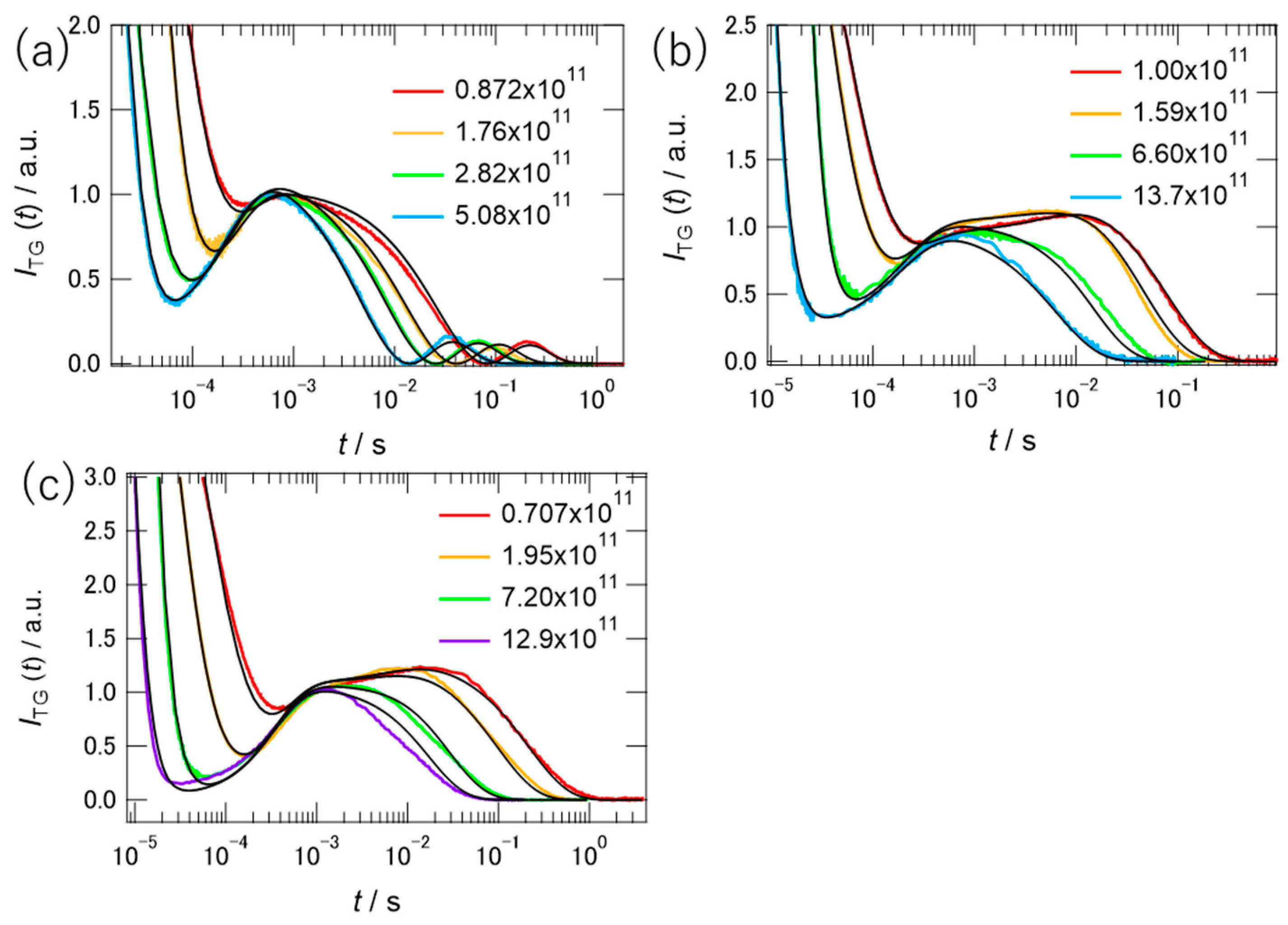
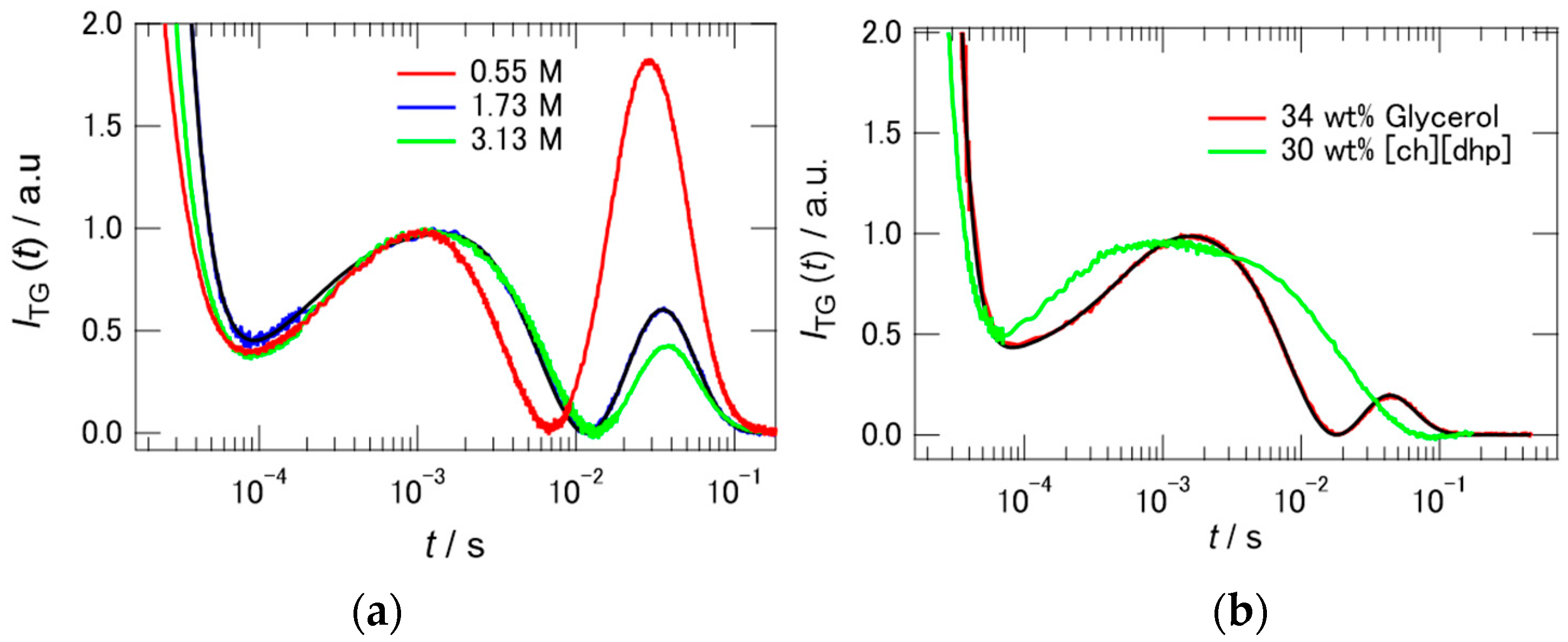
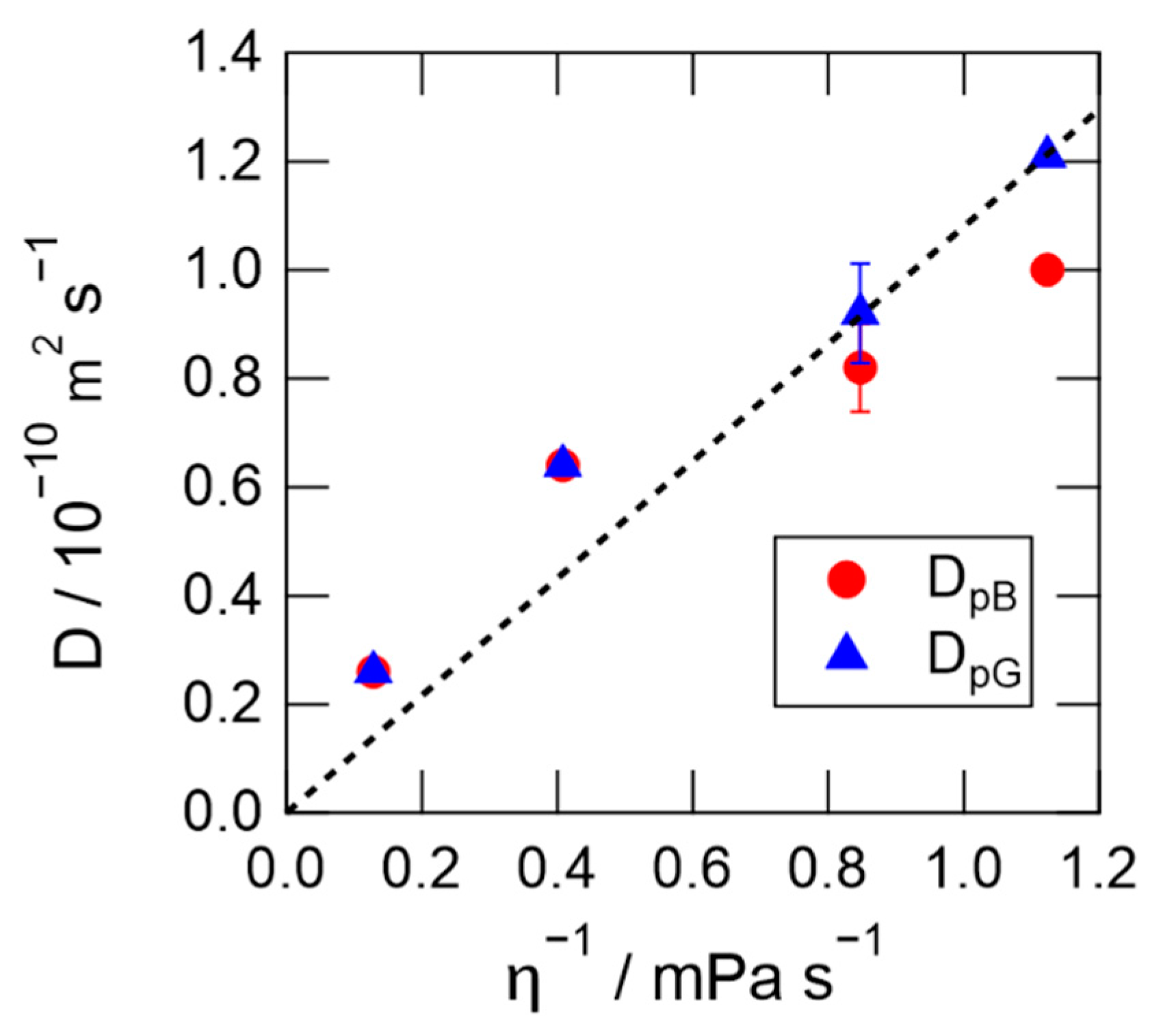
2.3. TG Signals of PYP in Hy[ch][dhp]
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Fujita, K. Ionic liquids as stabilization and refolding additives and solvents for proteins. Adv. Biochem. Eng. Biotechnol. 2021, 168, 215–226. [Google Scholar]
- Kumar, A.; Bhakuni, K.; Venkatesu, P. Strategic planning of proteins in ionic liquids: Future solvents for the enhanced stability of proteins against multiple stresses. Phys. Chem. Chem. Phys. PCCP 2019, 21, 23269–23282. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.Y.; Jonnalagadda, K.S.; Paradis, N.; Vaden, T.D.; Wu, C.; Caputo, G.A. Effects of ionic liquids on metalloproteins. Molecules 2021, 26, 514. [Google Scholar] [CrossRef] [PubMed]
- Sivapragasam, M.; Moniruzzaman, M.; Goto, M. Recent advances in exploiting ionic liquids for biomolecules: Solubility, stability and applications. Biotechnol. J. 2016, 11, 1000–1013. [Google Scholar] [CrossRef] [PubMed]
- Ohno, H.; Fujita, K.; Kohno, Y. Is seven the minimum number of water molecules per ion pair for assured biological activity in ionic liquid-water mixtures? Phys. Chem. Chem. Phys. 2015, 17, 14454–14460. [Google Scholar] [CrossRef]
- Weingartner, H.; Cabrele, C.; Herrmann, C. How ionic liquids can help to stabilize native proteins. Phys. Chem. Chem. Phys. 2012, 14, 415–426. [Google Scholar] [CrossRef] [Green Version]
- Fujita, K.; MacFarlane, D.R.; Forsyth, M. Protein solubilising and stabilising ionic liquids. Chem. Comm. 2005, 38, 4804–4806. [Google Scholar] [CrossRef]
- Fujita, K.; MacFarlane, D.R.; Forsyth, M.; Yoshizawa-Fujita, M.; Murata, K.; Nakamura, N.; Ohno, H. Solubility and stability of cytochrome c in hydrated ionic liquids: Effect of oxo acid residues and kosmotropicity. Biomacromolecules 2007, 8, 2080–2086. [Google Scholar] [CrossRef]
- Sasmal, D.K.; Mondal, T.; Sen Mojumdar, S.; Choudhury, A.; Banerjee, R.; Bhattacharyya, K. An FCS study of unfolding and refolding of CPM-labeled human serum albumin: Role of ionic liquid. J. Phys. Chem. B 2011, 115, 13075–13083. [Google Scholar] [CrossRef]
- Fiebig, O.C.; Mancini, E.; Caputo, G.; Vaden, T.D. Quantitative evaluation of myoglobin unfolding in the presence of guanidinium hydrochloride and ionic liquids in solution. J. Phys. Chem. B 2014, 118, 406–412. [Google Scholar] [CrossRef]
- Singh, U.K.; Patel, R. Dynamics of ionic liquid-assisted refolding of denatured cytochrome c: A study of preferential interactions toward renaturation. Mol. Pharm. 2018, 15, 2684–2697. [Google Scholar] [CrossRef] [PubMed]
- Pabbathi, A.; Samanta, A. On the stability and conformational dynamics of cytochrome c in ammonium ionic liquids. J. Phys. Chem. B 2020, 124, 8132–8140. [Google Scholar] [CrossRef] [PubMed]
- Takekiyo, T.; Miyazaki, K.; Watanabe, Y.; Uesugi, Y.; Tanaka, S.; Ishikawa, Y.; Yoshimura, Y. Solubilization and recovery of heat-aggregated cytochrome c using alkylammonium nitrate. J. Mol. Liq. 2019, 291, 111239. [Google Scholar] [CrossRef]
- Fujita, K.; Kajiyama, M.; Liu, Y.; Nakamura, N.; Ohno, H. Hydrated ionic liquids as a liquid chaperon for refolding of aggregated recombinant protein expressed in Escherichia coli. Chem. Commun. 2016, 52, 13491–13494. [Google Scholar] [CrossRef] [PubMed]
- Meyer, T.E. Isolation and characterization of soluble cytochromes, ferredoxins and other chromophoric proteins from the halophilic phototrophic bacterium Ectothiorhodospira halophila. Biochim. Biophys. Acta Bioenerg. 1985, 806, 175–183. [Google Scholar] [CrossRef]
- Meyer, T.E.; Tollin, G.; Hazzard, J.H.; Cusanovich, M.A. Photoactive yellow protein from the purple phototrophic bacterium, Ectothiorhodospira halophila. Quantum yield of photobleaching and effects of temperature, alcohols, glycerol, and sucrose on kinetics of photobleaching and recovery. Biophys. J. 1989, 56, 559–564. [Google Scholar] [CrossRef] [Green Version]
- Hoff, W.D.; van Stokkum, I.H.; van Ramesdonk, H.J.; van Brederode, M.E.; Brouwer, A.M.; Fitch, J.C.; Meyer, T.E.; van Grondelle, R.; Hellingwerf, K.J. Measurement and global analysis of the absorbance changes in the photocycle of the photoactive yellow protein from Ectothiorhodospira halophila. Biophys. J. 1994, 67, 1691–1705. [Google Scholar] [CrossRef] [Green Version]
- Hellingwerf, K.J.; Hendriks, J.; Gensch, T. Photoactive yellow protein, a new type of photoreceptor protein: Will this “Yellow Lab” bring us where we want to go? J. Phys. Chem. A 2003, 107, 1082–1094. [Google Scholar] [CrossRef] [Green Version]
- Baca, M.; Borgstahl, G.E.O.; Boissinot, M.; Burke, P.M.; Williams, D.R.; Slater, K.A.; Getzoff, E.D. Complete chemical structure of photoactive yellow protein: Novel thioester-linked 4-hydroxycinnamyl chromophore and photocycle chemistry. Biochemistry 1994, 33, 14369–14377. [Google Scholar] [CrossRef]
- Kort, R.; Vonk, H.; Xu, X.; Hoff, W.D.; Crielaard, W.; Hellingwerf, K.J. Evidence for trans-cis isomerization of the p-coumaric acid chromophore as the photochemical basis of the photocycle of photoactive yellow protein. FEBS Lett. 1996, 382, 73–78. [Google Scholar] [CrossRef] [Green Version]
- Mix, L.T.; Carroll, E.C.; Morozov, D.; Pan, J.; Gordon, W.R.; Philip, A.; Fuzell, J.; Kumauchi, M.; van Stokkum, I.; Groenhof, G.; et al. Excitation-wavelength-dependent photocycle initiation dynamics resolve heterogeneity in the photoactive yellow protein from Halorhodospira halophila. Biochemistry 2018, 57, 1733–1747. [Google Scholar] [CrossRef] [Green Version]
- Chen, E.; Gensch, T.; Gross, A.B.; Hendriks, J.; Hellingwerf, K.J.; Kliger, D.S. Dynamics of protein and chromophore structural changes in the photocycle of photoactive yellow protein monitored by time-resolved optical rotatory dispersion. Biochemistry 2003, 42, 2062–2071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Wilderen, L.J.G.W.; van der Horst, M.A.; van Stokkum, I.H.M.; Hellingwerf, K.J.; van Grondelle, R.; Groot, M.L. Ultrafast infrared spectroscopy reveals a key step for successful entry into the photocycle for photoactive yellow protein. Proc. Natl. Acad. Sci. USA 2006, 103, 15050–15055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schotte, F.; Cho, H.S.; Kaila, V.R.; Kamikubo, H.; Dashdorj, N.; Henry, E.R.; Graber, T.J.; Henning, R.; Wulff, M.; Hummer, G.; et al. Watching a signaling protein function in real time via 100-ps time-resolved Laue crystallography. Proc. Natl. Acad. Sci. USA 2012, 109, 19256–19261. [Google Scholar] [CrossRef] [Green Version]
- Jung, Y.O.; Lee, J.H.; Kim, J.; Schmidt, M.; Moffat, K.; Šrajer, V.; Ihee, H. Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography. Nat. Chem. 2013, 5, 212–220. [Google Scholar] [CrossRef] [Green Version]
- Yeremenko, S.; van Stokkum, I.H.; Moffat, K.; Hellingwerf, K.J. Influence of the crystalline state on photoinduced dynamics of photoactive yellow protein studied by ultraviolet-visible transient absorption spectroscopy. Biophys. J. 2006, 90, 4224–4235. [Google Scholar] [CrossRef] [Green Version]
- Takeshita, K.; Imamoto, Y.; Kataoka, M.; Mihara, K.I.; Tokunaga, F.; Terazima, M. Structural change of site-directed mutants of PYP: New dynamics during pR state. Biophys. J. 2002, 83, 1567–1577. [Google Scholar] [CrossRef] [Green Version]
- Takeshita, K.; Imamoto, Y.; Kataoka, M.; Tokunaga, F.; Terazima, M. Themodynamic and transport properties of intermediate states of the photocyclic reaction of photoactive yellow protein. Biochemistry 2002, 41, 3037–3048. [Google Scholar] [CrossRef]
- Khan, J.S.; Imamoto, Y.; Harigai, M.; Kataoka, M.; Terazima, M. Conformational changes of PYP monitored by diffusion coefficient: Effect of N-terminal alpha-helices. Biophys. J. 2006, 90, 3686–3693. [Google Scholar] [CrossRef] [Green Version]
- Hoshihara, Y.; Imamoto, Y.; Kataoka, M.; Tokunaga, F.; Terazima, M. Conformational changes in the N-terminal region of photoactive yellow protein: A time-resolved diffusion study. Biophys. J. 2008, 94, 2187–2193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, A.; Kelemen, L.; Hendriks, J.; White, B.J.; Hellingwerf, K.J.; Hoff, W.D. Formation of a new buried charge drives a large-amplitude protein quake in photoreceptor activation. Biochemistry 2001, 40, 1510–1517. [Google Scholar] [CrossRef] [PubMed]
- Changenet-Barret, P.; Plaza, P.; Martin, M.M.; Chosrowjan, H.; Taniguchi, S.; Mataga, N.; Imamoto, Y.; Kataoka, M. Structural effects on the ultrafast photoisomerization of photoactive yellow protein. Transient absorption spectroscopy of two point mutants. J. Phys. Chem. C 2009, 113, 11605–11613. [Google Scholar] [CrossRef]
- Harigai, M.; Imamoto, Y.; Kamikubo, H.; Yamazaki, Y.; Kataoka, M. Role of an N-terminal loop in the secondary structural change of photoactive yellow protein. Biochemistry 2003, 42, 13893–13900. [Google Scholar] [CrossRef]
- Borucki, B.; Otto, H.; Meyer, T.E.; Cusanovich, M.A.; Heyn, M.P. Sensitive circular dichroism marker for the chromophore environment of photoactive yellow protein: Assignment of the 307 and 318 nm Bands to the n→π* transition of the carbonyl. J. Phys. Chem. B 2005, 109, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Borucki, B.; Kyndt, J.A.; Joshi, C.P.; Otto, H.; Meyer, T.E.; Cusanovich, M.A.; Heyn, M.P. Effect of salt and pH on the activation of photoactive yellow protein and gateway mutants Y98Q and Y98F. Biochemistry 2005, 44, 13650–13663. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, Y.; Terazima, M.; Kimura, Y. Charge effect on the diffusion coefficient and the bimolecular reaction rate of diiodide anion radical in room temperature ionic liquids. J. Phys. Chem. B 2009, 113, 5188–5193. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Umezaki, U.; Hatakenaka, M.; Onodera, K.; Mizutani, H.; Kim, S.; Nakasone, Y.; Terazima, M.; Kimura, Y. Effect of Hydrated Ionic Liquid on Photocycle and Dynamics of Photoactive Yellow Protein. Molecules 2021, 26, 4554. https://doi.org/10.3390/molecules26154554
Umezaki U, Hatakenaka M, Onodera K, Mizutani H, Kim S, Nakasone Y, Terazima M, Kimura Y. Effect of Hydrated Ionic Liquid on Photocycle and Dynamics of Photoactive Yellow Protein. Molecules. 2021; 26(15):4554. https://doi.org/10.3390/molecules26154554
Chicago/Turabian StyleUmezaki, Utana, Miu Hatakenaka, Kana Onodera, Hiroto Mizutani, Suhyang Kim, Yusuke Nakasone, Masahide Terazima, and Yoshifumi Kimura. 2021. "Effect of Hydrated Ionic Liquid on Photocycle and Dynamics of Photoactive Yellow Protein" Molecules 26, no. 15: 4554. https://doi.org/10.3390/molecules26154554





