Spectroscopic Studies of Quinobenzothiazine Derivative in Terms of the In Vitro Interaction with Selected Human Plasma Proteins. Part 1
Abstract
:1. Introduction
2. Results and Discussion
2.1. Salt1–HSA, Salt1–AGP, Salt1–HGG and Salt1–CNS Interaction
2.2. Salt1–Protein Binding Site Assessment
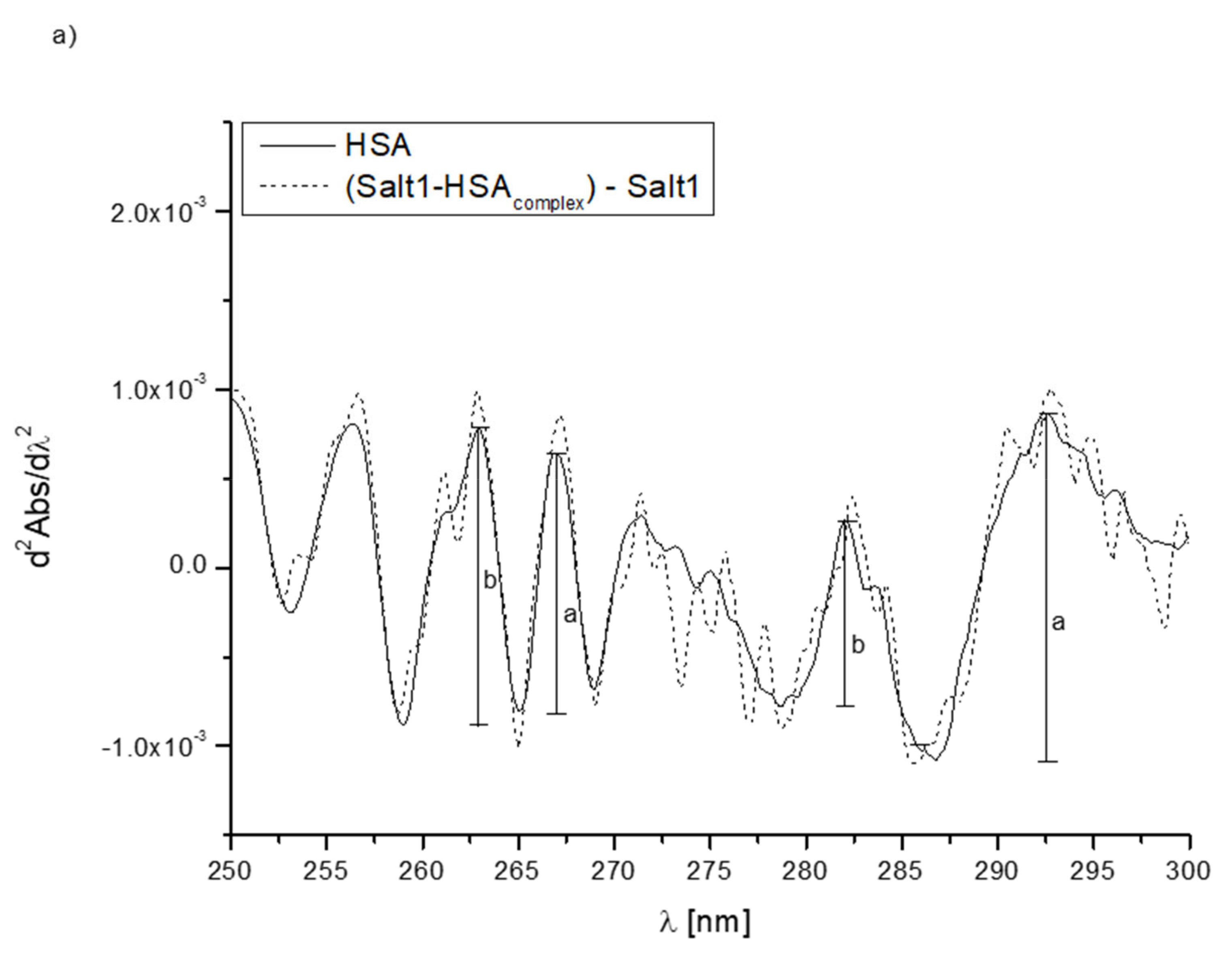
2.2.1. HSA Binding Site Assessment
2.2.2. AGP Binding Site Assessment
3. Materials and Methods
3.1. Chemicals
3.2. Methods
3.2.1. Sample Preparation
3.2.2. Emission and Absorption Spectra Measurements
3.2.3. Circular Dichroism (CD) Measurements
3.3. Statistics
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gerhard, M.; William, S. Principles and Medical Biochemistry, 4th ed.; Elsevier: Philadelphia, PA, USA, 2012; pp. 232–260. [Google Scholar]
- Bteich, M. An overview of albumin and alpha-1-acid glycoprotein main characteristics: Highlighting the roles of amino acids in binding kinetics and molecular interactions. Heliyon 2019, 5, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Carter, D.; He, X.M. Atomic structure and chemistry of human serum albumin. Nature 1992, 358, 209–215. [Google Scholar]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, NY, USA, 2006; pp. 1–535. [Google Scholar]
- Lu, J.; Stewart, A.J.; Sadler, P.; Pinheiro, T.J.T.; Blindauer, C.A. Albumin as a zinc carrier, properties of its high-affinity zinc-binding site. Biochem. Soc. Trans. 2008, 36, 1317–1324. [Google Scholar] [CrossRef] [Green Version]
- Dockal, M.; Carter, C.D.; Ruler, F. The Three Recombinant Domains of Human Serum Albumin. J. Biol. Chem. 1999, 274, 29303–29310. [Google Scholar] [CrossRef] [Green Version]
- Maciążek-Jurczyk, M.; Janas, K.; Pożycka, J.; Szkudlarek, A.; Rogóż, W.; Owczarzy, A.; Kulig, K. Human Serum Albumin Aggregation/Fibrillation and its Abilities to Drugs Binding. Molecules 2020, 25, 618. [Google Scholar] [CrossRef] [Green Version]
- Ghuman, J.; Zunszain, P.A.; Petitpas, I.; Bhattacharya, A.A.; Otagiri, M.; Curry, S. Structural basis of the drug-binding specificity of human serum albumin. J. Mol. Biol. 2005, 353, 38–52. [Google Scholar] [CrossRef]
- Yang, F.; Yue, J.; Ma, L.; Ma, Z.; Li, M.; Wu, X.; Liang, H. Interactive associations of drug–drug and drug–drug–drug with IIA subdomain of human serum albumin. Mol. Pharm. 2012, 9, 3259–3265. [Google Scholar] [CrossRef] [PubMed]
- Curry, S. Lessons from the crystallographic analysis of small molecule binding to human serum albumin. Drug Metab. Pharmacokinet. 2009, 24, 342–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curry, S.; Mandelkow, H.; Brick, P.; Franks, N. Crystal structure of human serum albumin complexes with fatty acid reveals an asymmetric distribution of binding site. Nat. Struct. Biol. 1998, 5, 827–835. [Google Scholar] [CrossRef]
- Dente, L.; Rüther, U.; Tripodi, M.; Wagner, E.F.; Cortese, R. Expression of human α1-acid glycoprotein genes m cultured cells and in transgenic mice. Genes Dev. 2015, 2, 259–266. [Google Scholar] [CrossRef] [Green Version]
- Azad, M.A.K.; Huang, J.X.; Cooper, M.A.; Roberts, K.D.; Thompson, P.E.; Nation, R.L.; Li, J.; Velkov, T. Structure activity relationships for the binding of polymyxins with human α–1 acid glycoproteins. Biochem. Pharmacol. 2012, 84, 278–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albani, J.R. Relation between the secondary structure of carbohydrate residues of 1-acid glycoprotein (orosomucoid) and the fluorescence of the protein. Carbohydr Res. 2003, 338, 1097–1101. [Google Scholar] [CrossRef]
- Olins, D.E.; Edelman, M.D. The antigenic structure of the polypeptide chains of human ɣ-globulin. J. Exp. Med. 1962, 116, 635–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merler, E.; Rosen, F.S. The gamma globulins–structure and synthesis of the immunoglobulins. N. Engl. J. Med. 1966, 275, 480–486. [Google Scholar] [CrossRef]
- Sędek, Ł.; Mazur, B. The application of monoclonal and polyclonal antibodies in flow cytometry. PBKom. 2008, 35, 17–24. [Google Scholar]
- Crumpton, M.J.; Wilkinson, J.M. Amino Acid Compositions of Human and Rabbit ɣ-Globulins and of the Fragments Produced by Reduction. Biochem. J. 1963, 88, 228. [Google Scholar] [CrossRef] [Green Version]
- Valeur, B.; Berberan-Santos, M.N. Molecular fluorescence, Principles and Applications. J. Biomed. Opt. 2009, 34–123. [Google Scholar]
- Eftink, C.A.; Ghiron, C.A. Exposure of Tryptophanyl Residues in Proteins. Quantitative Determination by Fluorescence Quenching Studies. Biochemistry 1976, 15, 672–680. [Google Scholar] [CrossRef]
- Turoverov, K.K.; Haitlina, S.Y.; Pinaev, G.P. Ultra-violet fluorescence of actin. Determination of native actin content in actin preparations. FEBS Lett. 1976, 62, 4–6. [Google Scholar] [CrossRef] [Green Version]
- Maciążek-Jurczyk, M.; Morak–Młodawska, B.; Jeleń, M.; Kopeć, W.; Szkudlarek, A.; Owczarzy, A.; Kulig, K.; Rogóż, W.; Pożycka, J. The Influence of Oxidative Stress on Serum Albumin Structure as a Carrier of Selected Diazaphenothiazine with Potential Anticancer Activity. Pharmaceuticals 2021, 14, 285. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, T.; Terada, H. Estimation of State amount of Phenylalanine residues in proteins by second derivative spectrophotometry. BBA 1979, 580, 120–128. [Google Scholar] [CrossRef]
- Imre, T.; Schlosser, G.; Pocsfalvi, G.; Siciliano, R.; Molnár–Szöllösi, É.; Kremmer, T.; Malorni, A.; Vèkey, K. Glycosylation site analysis of human alpha-1-acid glycoprotein (AGP) by capillary liquid chromatography–electrospray mass spectrometry. J. Mass Spectrom. 2005, 40, 1483–1742. [Google Scholar] [CrossRef]
- Sierra, T.; Moreno, B.; Crevillen, A.G.; González, M.C.; Escapra, A. Total α1- acid glycoprotein determination in serum samples using disposable screen-printed electrodes and Osmium (VI) as electrochemical tag. Talanta 2018, 180, 206–210. [Google Scholar] [CrossRef]
- Fournier, T.; Medjoubi, N.N.; Porquet, D. Alpha-1-acid glycoprotein. Biochim. Biophys. Acta 2000, 1482, 157–171. [Google Scholar] [CrossRef]
- Hochepied, T.; Berger, F.G.; Baumann, H.; Libert, C. α-Acid glycoprotein: An acute phase protein with inflammatory and immunomodulating properties. Cytokine Growth Factor Rev. 2003, 14, 25–34. [Google Scholar] [CrossRef]
- Smith, S.A.; Waters, N. Pharmacokinetic and Pharmacodynamic Considerations for Drugs Binding to Alpha-1-Acid Glycoprotein. Pharm Res. 2019, 36, 1–19. [Google Scholar] [CrossRef]
- Sochacka, J.; Pawełczak, P. Characterization of 6-Mercaptopurine binding site on human α1-acid glycoprotein (orosomucoid) using molecular docking. Acta Pol. Pharm. Drug Res. 2012, 69, 161–166. [Google Scholar]
- Pitekova, B.; Uhlikova, E.; Kupcova, V.; Durfinova, M.; Mojto, V.; Turecky, L. Can alpha-1-acid glycoprotein affect the outcome of treatment in a cancer patient? Bratisl Med. J. 2019, 120, 9–14. [Google Scholar] [CrossRef]
- Bailey, D.N.; Briggs, J.R. The Binding of Selected Therapeutic Drugs to Human Serum α1 Acid Glycoprotein and to Human Serum Albumin In Vitro. Drug Monit. 2004, 26, 40–43. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.K.; Devenny, J.J.; Bitensky, M.W. Micropinocytic ingestion of glycosylated albumin by isolated microvessels: Possible role in pathogenesis of diabetic microangiopathy. Proc. Natl. Acad. Sci. USA 1981, 78, 2393–2397. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Kai, G.; Yang, F.; Chunxi, L.; Xiaochen, X.; Koichiro, Y. Molecular structure-affinity relationship of natural polyphenols for bovine c-globulin. Mol. Nutr. Food Res. 2011, 55, 86–92. [Google Scholar] [CrossRef]
- Ying, L.; Chao, W.; Guanghua, L. Interaction of Jatrorrhizine with Human Gamma Globulin in membrane mimetic environments: Probing of the binding mechanism and binding site by spectroscopic and molecular modeling methods. J. Mol. Struct. 2010, 980, 108–113. [Google Scholar] [CrossRef]
- Hein, K.L.; Kragh-Hansen, U.; Morth, J.P.; Jeppesen, M.D.; Otzen, D.; Møller, J.V.; Nissen, P. Crystallographic analysis reveals a unique lidocaine binding site on human serum albumin. J. Struct. Biol. 2010, 171, 353–360. [Google Scholar] [CrossRef]
- Mao, H.; Hajduk, P.J.; Craig, R.; Bell, R.; Borre, T.; Fesik, S.W. Rational design of diflunisal analogues with reduced affinity for human serum albumin. J. Am. Chem. Soc. 2001, 123, 10429–10435. [Google Scholar] [CrossRef]
- Gokara, M.; Sudhamalla, B.; Amooru, D.G.; Subramanyam, R. Molecular interaction studies of trimethoxy flavone with human serum albumin. PLoS ONE 2010, 1, 8834. [Google Scholar] [CrossRef] [Green Version]
- Oltersdorf, T.; Elmore, S.W.; Shoemaker, A.R.; Armstrong, R.C.; Augeri, D.J.; Belli, B.A.; Bruncko, M.; Deckwerth, T.L.; Dinges, J.; Hajduk, P.J. An inhibitor of Bcl-2 family proteins induces regression of solid tumors. Nature 2005, 435, 677–681. [Google Scholar] [CrossRef]
- Vallianatou, T.; Lambrinidis, G.; Tsantili-Kakoulidou, A. In silico prediction of human serum albumin binding for drug leads. Expert. Opin. Drug Discov. 2013, 8, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Peyrin, E.; Guillaume, Y.C.; Morin, N.; Guinchard, C. Sucrose dependence of solute retention on human serum albumin stationary chase: Hydrophobic effect and surface tension considerations. Anal. Chem. 1998, 70, 2812–2818. [Google Scholar] [CrossRef] [PubMed]
- Taira, Z.; Terada, H. Specific and nonspecific ligand binding to serum albumin. Biochem. Pharmacol. 1998, 34, 1999–2005. [Google Scholar]
- Kibbey, C.E.; Poole, S.K.; Robinson, B.; Jackson, J.D.; Durham, D. An integrated process for measuring the physicochemical properties of drug candidates in a preclinical discovery environment. J. Pharm. Sci. 2001, 90, 1164–1175. [Google Scholar] [CrossRef] [PubMed]
- Cancer Today. Available online: https://gco.iarc.fr/today (accessed on 14 February 2021).
- Zięba, A.; Sochanik, A.; Szurkoc, A.; Rams, M.; Mrożek, A.; Cmoch, P. Synthesis and in vitro antiproliferative activity of 5-alkyl-12(H)-quino[3,4-b] [1,4]benzothiazinium salts. Eur. J. Med. Chem. 2010, 45, 4733–4739. [Google Scholar] [CrossRef] [PubMed]
- Sułkowska, A.; Maciążek-Jurczyk, M.; Bojko, B.; Równicka-Zubik, J.; Skupień, I.; Temba, E.; Pentak, D.; Sułkowski, W. Competitive binding of phenylbutazone and colchicine to serum albumin in multidrug therapy: A spectroscopic study. J. Mol. Struct. 2008, 881, 97–106. [Google Scholar] [CrossRef]
- Zięba, A.; Bober, K. Lipophilicity analysis of newly synthetized quinobenzothiaines by use of TLC. J. Liq. Chromatogr. Relat. Technol. 2016, 39, 104–109. [Google Scholar] [CrossRef]
- Zsila, F. Subdomain Ib Is the Third major Drug Binding Region of Human Serum Albumin: Toward the Tree–Sites Models. Mol. Pharmaceutics 2013, 10, 1668–1682. [Google Scholar] [CrossRef]
- Balestrieri, C.; Colonna, G.; Giovane, A.; Irace, G.; Servillo, L. Second–derivative Spectroscopy of Proteins. A Method for the Quantitative Determination of Aroma Acids in Proteins. Eur. J. Biochem. 1978, 90, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Terada, H.; Inoue, Y.; Ichikawa, T. Second derivative spectral properties of tryptophan and tyrosine residues in Proteins. Effects of Guanidine Hydrochloride and Dodecyl Sulfate in the Residues in Lysozyme, Ribonuclease and Serum Albumin. Chem. Pharm. Bull. 1984, 32, 585–590. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Jia, B.; Wand, H.; Li, N.; Chen, G.; Lin, Y.; Gao, W. The interaction of 2- mercaptobenzimidazole with human serum albumin as determined by spectroscopy, atomic force microscopy and molecular modeling. Colloids Surf. 2013, 104, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef]
- Miles, A.J.; Wallace, B.A. Circular dichroism spectroscopy of membrane proteins. Chem. Soc. Rev. 2016, 45, 4859–4872. [Google Scholar] [CrossRef] [Green Version]
- StatPearls. Available online: https://www.ncbi.nlm.nih.gov/books/NBK470235 (accessed on 18 January 2021).
- Matsumoto, K.; Sukimoto, K.; Nishi, K.; Maruyama, T.; Suenaga, A.; Otagiri, M. Characterization of Ligand Binding Sites on the a1-Acid Glycoprotein in Humans, Bovines and Dogs. Drug Metab. Pharmacokin. 2002, 17, 300–306. [Google Scholar] [CrossRef]
- Bohnert, T.; Gan, L.S. Plasma Protein Binding: From Discovery to Development. J. Pharm Sci. 2013, 102, 2953–2994. [Google Scholar] [CrossRef] [PubMed]
- Sudlow, G.; Birkett, D.J.; Wade, D.N. Further Characterization of Specific Drug Binding Sites on Human Serum Albumin. Mol. Pharmacol. 1976, 12, 1052–1061. [Google Scholar]
- Patela, S.; Sharma, K.K.; Datta, A. Competitive binding of Chlorinp6and Dansyl-L-Proline to Sudlow’s site II of human serum albumin. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2015, 138, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Klotz, I.M.; Hunston, D.L. Properties of graphical representations of multiple classes of binding sites. Biochemistry 1971, 10, 3065–3069. [Google Scholar] [CrossRef]
- Ryan, A.J.; Ghuman, J.; Zunszain, P.A.; Chung, C.; Curry, S. Structural basis of binding of fluorescent, site-specific dansylated amino acids to human serum albumin. J. Struct. Biol. 2011, 174, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Schönfeld, D.L.; Ravelli, R.B.; Mueller, U. Skerra, A. The 1.8-Å Crystal Structure of α1-Acid Glycoprotein (Orosomucoid) Solved by UV RIP Reveals the Broad Drug-Binding Activity of This Human Plasma Lipocalin. J. Mol. Biol. 2008, 384, 393–405. [Google Scholar] [CrossRef]
- Otagiri, M.; Miyoshi, T.; Yamamichi, R.; Maruyama, T.; Perrin, J.H. Effects of tricyclic drug on induced circular dichroism spectra of dicumarol bound to α1-acid glycoprotein. Biochem. Phamacol. 1991, 42, 729–733. [Google Scholar] [CrossRef]
- Israeli, Z.H.; Dayton, P.G. Human alpha-1-Glycoprotein And Its Interactions With Drugs. Drug Metab. Rev. 2005, 20, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Nishi, K.; Fukunaga, N.; Otagiri, M. Construction of expression system for human alpha–1–acid glycoproteins in Pichia pastoris and evaluation of this drug-binding properties. Drug Metab. Dispos. 2004, 32, 1069–1074. [Google Scholar] [CrossRef] [Green Version]









| Salt1–Protein Complex | λex 275 nm | λex 295 nm |
|---|---|---|
| Percentage of Fluorescence Quenching (%) | ||
| Salt1–HSAcomplex | 32.04 | 34.51 |
| Salt1–AGPcomplex | 54.34 | 59.00 |
| Salt1–HGGcomplex | 25.20 | 28.99 |
| Salt1–CNScomplex | 25.47 | 25.43 |
| Protein | λex 275 nm | λex 295 nm | ||||
|---|---|---|---|---|---|---|
| Δλmax (nm) | ΔA | ΔFWHM (nm) | Δλ(nm) | ΔA | ΔFWHM (nm) | |
| HSA | 6 | 0.09 | 0.37 | 5 | 0.14 | 1.41 |
| AGP | 2 | 0.08 | 2.42 | 6 | 0.13 | 3.62 |
| HGG | 2 | 0.06 | 1.59 | 6 | 0.07 | 5.17 |
| CNS | 2 | 0.08 | 1.87 | 3 | 0.11 | 4.26 |
| λ 250–270 nm | λ > 270 nm |  | |
| HSA | 0.87 | 1.86 | |
| Salt1–HSAcomplex | 1.03 | 1.97 | |
| AGP | 1.19 | 3.93 | |
| Salt1–AGPcomplex | 0.67 | 4.10 | |
| HGG | 0.54 | 0.80 | |
| Salt1–HGGcomplex | 1.32 | 0.34 | |
| CNS | 0.91 | 0.84 | |
| Salt1–CNScomplex | 1.36 | 0.89 |
| [θ]mrw at 209.8 nm (mdeg·cm2·dmol−1) | [θ]mrw at 219.4 nm (mdeg·cm2·dmol−1) | % α-Helix | % β-Sheet | % Turn | % Random | |
|---|---|---|---|---|---|---|
| HAS a | −17063.3333 | −16147.26496 | 37.7 | 10.8 | 21.0 | 30.4 |
| Salt1–HSAcomplex a | −16991.3675 | −16028.88890 | 37.3 | 10.1 | 21.6 | 30.0 |
| AGP b | - | −7892.39560 | 17.1 | 82.9 | - | - |
| Salt1–AGPcomplex b | - | −7257.86810 | 14.9 | 85.1 | - | - |
| λex 275 nm | λex 295 nm | |||||
|---|---|---|---|---|---|---|
| Pearson’s | 104) ±SD * (mol·L−1) | 1014) ±SD * (mol−1·L·s−1) | Pearson’s | 104) ±SD * (mol·L−1) | 1014) ±SD * (mol−1·L·s−1) | |
| Salt1–HSAcomplex | 0.997 | 2.70 ± 0.10 | 0.43 ± 0.10 | 0.998 | 2.84 ± 0.08 | 0.46 ± 0.08 |
| Salt1–AGPcomplex | 0.999 | 5.40 ± 0.10 | 2.40 ± 0.10 | 0.998 | 6.72 ± 0.20 | 2.94 ± 0.02 |
| Salt1–HGGcomplex | 0.998 | 1.60 ± 0.03 | 0.81 ± 0.03 | 0.996 | 1.14 ± 0.04 | 0.11 ± 0.04 |
| Salt1–CNScomplex | 0.990 | 1.28 ± 0.10 | 0.14 ± 0.10 | 0.999 | 1.38 ± 0.02 | 0.14 ± 0.02 |
| λex 275 nm | λex 295 nm | |||
|---|---|---|---|---|
| 104) ±SD * (mol·L−1) | n ± SD * | 104) ±SD * (mol·L−1) | n ± SD * | |
| Salt1–HSAcomplex | 2.45 ± 0.01 | 1.62 ± 0.10 | 2.71 ± 0.01 | 1.38 ± 0.10 |
| Salt1–AGPcomplex | 7.35 ± 0.01 | 1.15 ± 0.02 | 7.86 ± 0.03 | 1.17 ± 0.01 |
| Salt1–HGGcomplex | 1.41 ± 0.02 | 1.01 ± 0.20 | 1.33 ± 0.02 | 0.94 ± 0.20 |
| Salt1–CNScomplex | 6.35 ± 0.01 | 0.95 ± 0.10 | 2.36 ± 0.02 | 0.82 ± 0.20 |
| CSalt1 (mol·L−1) | [HSA]:[dGlu] | [HSA]:[dGly] | [HSA]:[dPhe] | [HSA]:[dPro] |
|---|---|---|---|---|
| 1:1 | ||||
| percentage of displacement (%) | ||||
| 0 | - | - | - | - |
| 3·10−6 | 7.43 | 5.19 | 2.99 | 7.53 |
| 6·106 | 12.35 | 9.68 | 5.75 | 11.75 |
| 9·10−6 | 15.85 | 13.59 | 8.98 | 15.53 |
| 12·10−6 | 20.72 | 18.80 | 11.78 | 20.98 |
| 15·10−6 | 24.13 | 22.87 | 14.84 | 24.11 |
| 18·10−6 | 27.78 | 25.68 | 18.30 | 29.09 |
| 21·10−6 | 31.27 | 29.79 | 21.29 | 31.61 |
| 24·10−6 | 34.20 | 32.80 | 23.88 | 36.83 |
| 27·10−6 | 36.34 | 35.61 | 25.81 | 39.78 |
| 30·10−6 | 38.75 | 37.76 | 27.79 | 41.66 |
| 33·10−6 | 40.99 | 41.35 | 30.31 | 44.82 |
| CSalt1 (mol·L−1) | [AGP]:[QR] 1:0.5 | [AGP]:[QR] 1:1 |
|---|---|---|
| percentage of displacement (%) | ||
| 0 | - | - |
| 3·10−6 | 9.2 | 10.7 |
| 6·10−6 | 14.8 | 18.9 |
| 12·10−6 | 26.8 | 26.4 |
| 18·10−6 | 32.7 | 34.8 |
| 24·10−6 | 41.2 | 39.8 |
| 30·10−6 | 46.2 | 46.1 |
| 36·10−6 | 49.3 | 50.3 |
| 42·10−6 | 51.8 | 52.6 |
| 48·10−6 | 58.5 | 57.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Owczarzy, A.; Zięba, A.; Pożycka, J.; Kulig, K.; Rogóż, W.; Szkudlarek, A.; Maciążek-Jurczyk, M. Spectroscopic Studies of Quinobenzothiazine Derivative in Terms of the In Vitro Interaction with Selected Human Plasma Proteins. Part 1. Molecules 2021, 26, 4776. https://doi.org/10.3390/molecules26164776
Owczarzy A, Zięba A, Pożycka J, Kulig K, Rogóż W, Szkudlarek A, Maciążek-Jurczyk M. Spectroscopic Studies of Quinobenzothiazine Derivative in Terms of the In Vitro Interaction with Selected Human Plasma Proteins. Part 1. Molecules. 2021; 26(16):4776. https://doi.org/10.3390/molecules26164776
Chicago/Turabian StyleOwczarzy, Aleksandra, Andrzej Zięba, Jadwiga Pożycka, Karolina Kulig, Wojciech Rogóż, Agnieszka Szkudlarek, and Małgorzata Maciążek-Jurczyk. 2021. "Spectroscopic Studies of Quinobenzothiazine Derivative in Terms of the In Vitro Interaction with Selected Human Plasma Proteins. Part 1" Molecules 26, no. 16: 4776. https://doi.org/10.3390/molecules26164776






