Magnesium-Free Immobilization of DNA Origami Nanostructures at Mica Surfaces for Atomic Force Microscopy
Abstract
:1. Introduction
2. Results and Discussion
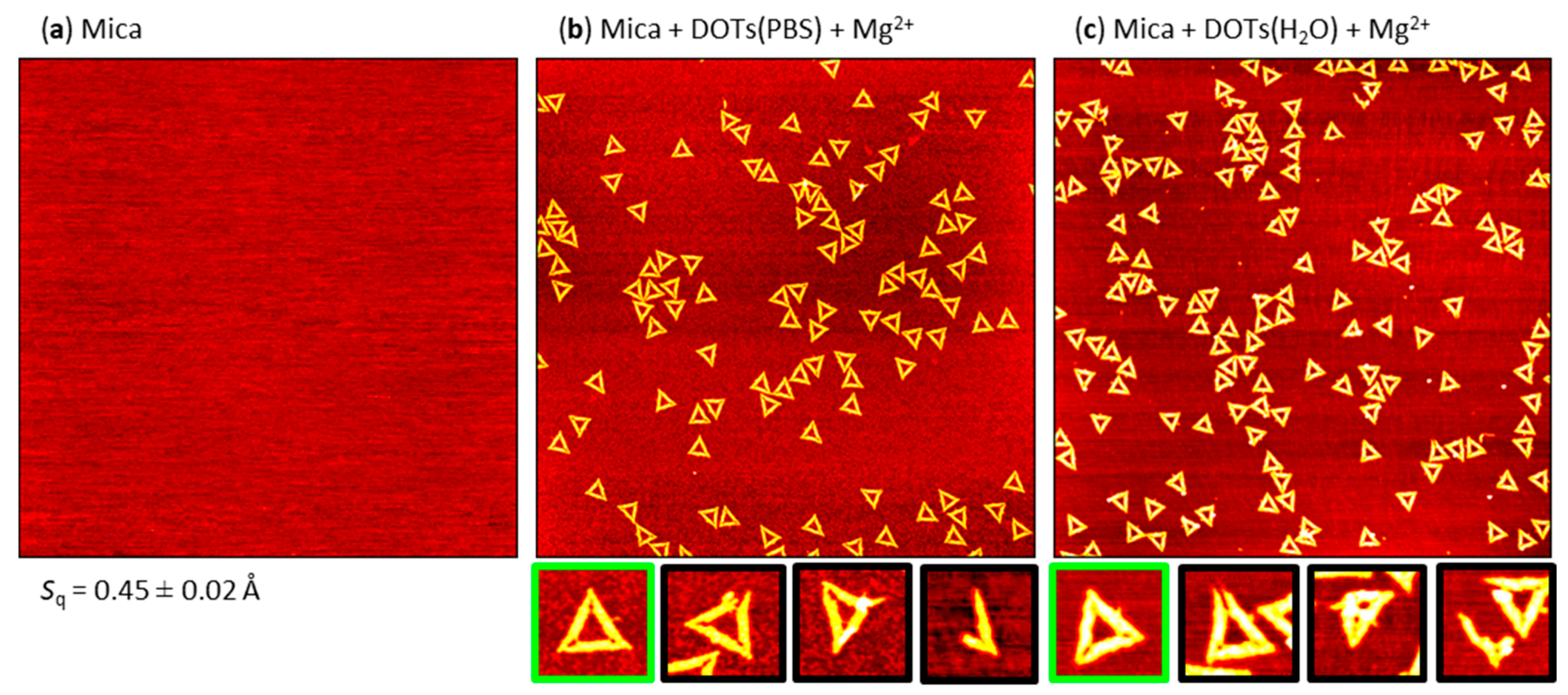
2.1. Mg2+-Mediated Adsorption
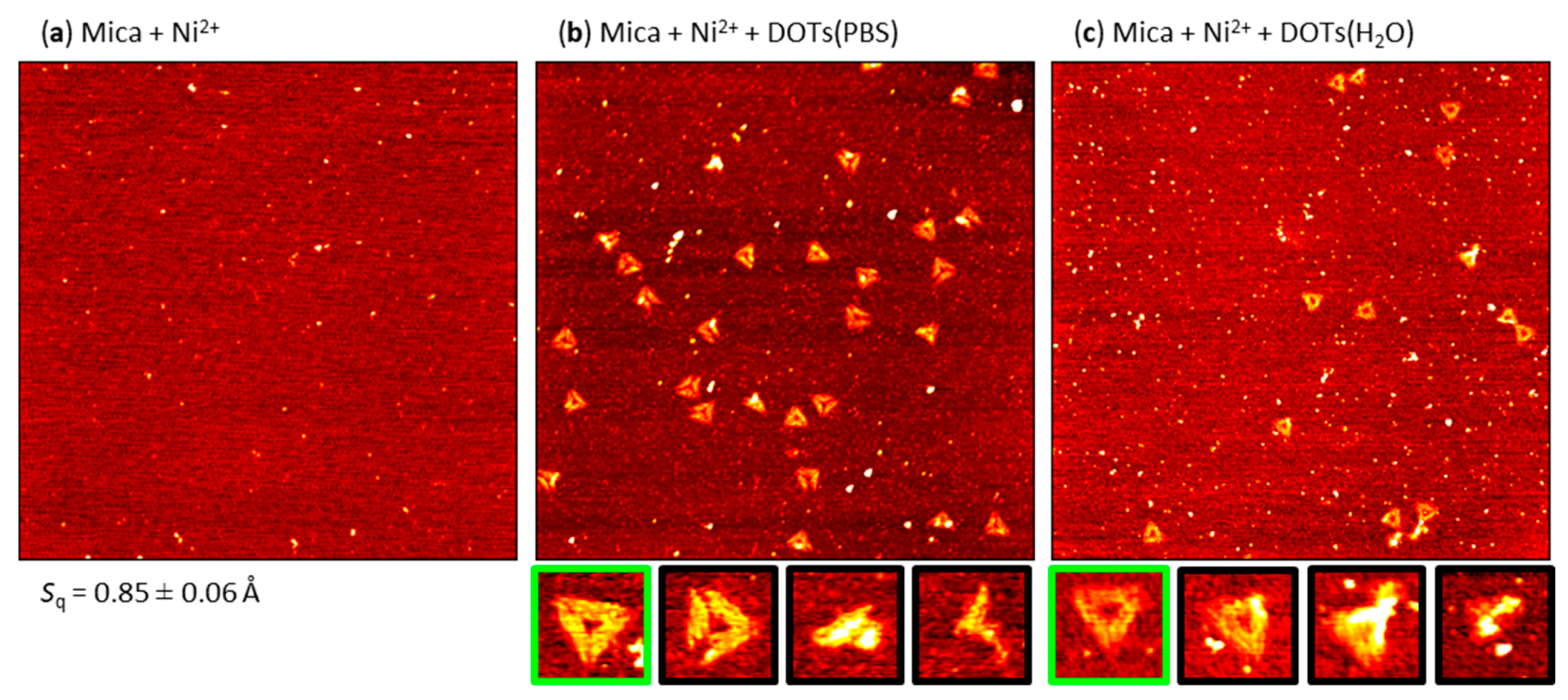
2.2. Pre-Adsorption of Ni2+
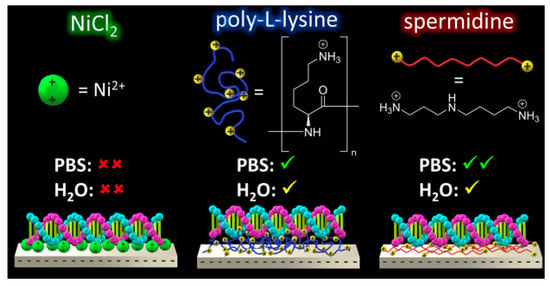
2.3. Pre-Adsorption of Poly-l-Lysine (PLL)
2.4. Pre-Adsorption of Spermidine (Spdn)
2.5. Effect of PBS and H2O Exposure on the Pre-Adsorbed Polyelectrolyte Films
2.6. Effect of DON Shape
3. Materials and Methods
3.1. DON Assembly and Buffer Exchange
3.2. Mica Surface Modification
- (1)
- NiCl2 pretreatment: 10 mM NiCl2 aqueous solution was deposited onto a freshly cleaved mica surface and incubated for 1 h. An incubation time of 1 h was chosen based on our previous work [45]. It should be noted, however, that equivalent results as reported here were also obtained with shorter incubation times, i.e., 1 min to 30 min. The mica substrate was then rinsed with HPLC-grade water to remove excess NiCl2.
- (2)
- PLL pretreatment: PLL was dissolved in HPLC-grade water to yield a 0.1% w/v PLL solution. The PLL solution was deposited onto a freshly cleaved mica surface and incubated for 1 h. An incubation time of 1 h was chosen based on literature to ensure maximum surface coverage [64]. The mica substrate was then rinsed with HPLC-grade water to remove excess PLL.
- (3)
- Spdn pretreatment: Spdn was dissolved in HPLC-grade water to yield a 5 mg/mL Spdn solution and then deposited onto a freshly cleaved mica surface. After incubation for 5 min the mica substrate was rinsed with HPLC-grade water. An incubation time of 5 min was chosen based on literature [58].
3.3. DON Immobilization and AFM Imaging
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Högberg, B.; Graf, F.; Shih, W.M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. [Google Scholar] [CrossRef]
- Keller, A.; Linko, V. Challenges and Perspectives of DNA Nanostructures in Biomedicine. Angew. Chem. Int. Ed. Engl. 2020, 59, 15818–15833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, D.M.; Keller, A. DNA Nanostructures in the Fight Against Infectious Diseases. Adv. NanoBiomed Res. 2021, 1, 2000049. [Google Scholar] [CrossRef] [PubMed]
- Bald, I.; Keller, A. Molecular processes studied at a single-molecule level using DNA origami nanostructures and atomic force microscopy. Molecules 2014, 19, 13803–13823. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.J.; Wälti, C. DNA nanostructures: A versatile lab-bench for interrogating biological reactions. Comput. Struct. Biotechnol. J. 2019, 17, 832–842. [Google Scholar] [CrossRef]
- Endo, M.; Sugiyama, H. Single-molecule imaging of dynamic motions of biomolecules in DNA origami nanostructures using high-speed atomic force microscopy. Acc. Chem. Res. 2014, 47, 1645–1653. [Google Scholar] [CrossRef] [PubMed]
- Engelen, W.; Dietz, H. Advancing Biophysics Using DNA Origami. Annu. Rev. Biophys. 2021, 50, 469–492. [Google Scholar] [CrossRef]
- Wamhoff, E.-C.; Banal, J.L.; Bricker, W.P.; Shepherd, T.R.; Parsons, M.F.; Veneziano, R.; Stone, M.B.; Jun, H.; Wang, X.; Bathe, M. Programming Structured DNA Assemblies to Probe Biophysical Processes. Annu. Rev. Biophys. 2019, 48, 395–419. [Google Scholar] [CrossRef]
- Kogikoski, S.; Paschoalino, W.J.; Kubota, L.T. Supramolecular DNA origami nanostructures for use in bioanalytical applications. Trends Anal. Chem. 2018, 108, 88–97. [Google Scholar] [CrossRef]
- Wang, S.; Zhou, Z.; Ma, N.; Yang, S.; Li, K.; Teng, C.; Ke, Y.; Tian, Y. DNA Origami-Enabled Biosensors. Sensors 2020, 20, 6899. [Google Scholar] [CrossRef]
- Schoenit, A.; Cavalcanti-Adam, E.A.; Göpfrich, K. Functionalization of Cellular Membranes with DNA Nanotechnology. Trends Biotechnol. 2021. [Google Scholar] [CrossRef]
- Zhao, N.; Chen, Y.; Chen, G.; Xiao, Z. Artificial Cells Based on DNA Nanotechnology. ACS Appl. Bio Mater. 2020, 3, 3928–3934. [Google Scholar] [CrossRef]
- Funke, J.J.; Dietz, H. Placing molecules with Bohr radius resolution using DNA origami. Nat. Nanotechnol. 2016, 11, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Suma, A.; Cui, M.; Grundmeier, G.; Carnevale, V.; Zhang, Y.; Kielar, C.; Keller, A. Arranging Small Molecules with Subnanometer Precision on DNA Origami Substrates for the Single-Molecule Investigation of Protein–Ligand Interactions. Small Struct. 2020, 1, 2000038. [Google Scholar] [CrossRef]
- Kielar, C.; Reddavide, F.V.; Tubbenhauer, S.; Cui, M.; Xu, X.; Grundmeier, G.; Zhang, Y.; Keller, A. Pharmacophore Nanoarrays on DNA Origami Substrates as a Single-Molecule Assay for Fragment-Based Drug Discovery. Angew. Chem. Int. Ed. Engl. 2018, 57, 14873–14877. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Ijäs, H.; Linko, V.; Keller, A. Structural stability of DNA origami nanostructures under application-specific conditions. Comput. Struct. Biotechnol. J. 2018, 16, 342–349. [Google Scholar] [CrossRef]
- Ijäs, H.; Shen, B.; Heuer-Jungemann, A.; Keller, A.; Kostiainen, M.A.; Liedl, T.; Ihalainen, J.A.; Linko, V. Unraveling the interaction between doxorubicin and DNA origami nanostructures for customizable chemotherapeutic drug release. Nucleic Acids Res. 2021, 49, 3048–3062. [Google Scholar] [CrossRef]
- Kollmann, F.; Ramakrishnan, S.; Shen, B.; Grundmeier, G.; Kostiainen, M.A.; Linko, V.; Keller, A. Superstructure-Dependent Loading of DNA Origami Nanostructures with a Groove-Binding Drug. ACS Omega 2018, 3, 9441–9448. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Li, N.; Liu, S.; Wang, L.; Wang, Z.-G.; Zhang, Z.; Ding, B. Site-Specific Synthesis of Silica Nanostructures on DNA Origami Templates. Adv. Mater. 2020, 32, e2000294. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.; Döblinger, M.; Liedl, T.; Heuer-Jungemann, A. DNA-Origami-Templated Silica Growth by Sol-Gel Chemistry. Angew. Chem. Int. Ed. Engl. 2019, 58, 912–916. [Google Scholar] [CrossRef] [PubMed]
- Ijäs, H.; Hakaste, I.; Shen, B.; Kostiainen, M.A.; Linko, V. Reconfigurable DNA Origami Nanocapsule for pH-Controlled Encapsulation and Display of Cargo. ACS Nano 2019, 13, 5959–5967. [Google Scholar] [CrossRef] [Green Version]
- Kroener, F.; Traxler, L.; Heerwig, A.; Rant, U.; Mertig, M. Magnesium-Dependent Electrical Actuation and Stability of DNA Origami Rods. ACS Appl. Mater. Interfaces 2019, 11, 2295–2301. [Google Scholar] [CrossRef]
- Kielar, C.; Xin, Y.; Shen, B.; Kostiainen, M.A.; Grundmeier, G.; Linko, V.; Keller, A. On the Stability of DNA Origami Nanostructures in Low-Magnesium Buffers. Angew. Chem. Int. Ed. Engl. 2018, 57, 9470–9474. [Google Scholar] [CrossRef]
- Teschome, B.; Facsko, S.; Gothelf, K.V.; Keller, A. Alignment of Gold Nanoparticle-Decorated DNA Origami Nanotubes: Substrate Prepatterning versus Molecular Combing. Langmuir 2015, 31, 12823–12829. [Google Scholar] [CrossRef] [PubMed]
- Kopielski, A.; Csaki, A.; Fritzsche, W. Surface Mobility and Ordered Rearrangement of Immobilized DNA Origami. Langmuir 2015, 31, 12106–12110. [Google Scholar] [CrossRef] [PubMed]
- Brassat, K.; Ramakrishnan, S.; Bürger, J.; Hanke, M.; Doostdar, M.; Lindner, J.K.N.; Grundmeier, G.; Keller, A. On the Adsorption of DNA Origami Nanostructures in Nanohole Arrays. Langmuir 2018, 34, 14757–14765. [Google Scholar] [CrossRef] [PubMed]
- Kershner, R.J.; Bozano, L.D.; Micheel, C.M.; Hung, A.M.; Fornof, A.R.; Cha, J.N.; Rettner, C.T.; Bersani, M.; Frommer, J.; Rothemund, P.W.K.; et al. Placement and orientation of individual DNA shapes on lithographically patterned surfaces. Nat. Nanotechnol. 2009, 4, 557–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakrishnan, S.; Krainer, G.; Grundmeier, G.; Schlierf, M.; Keller, A. Cation-Induced Stabilization and Denaturation of DNA Origami Nanostructures in Urea and Guanidinium Chloride. Small 2017, 13, 1702100. [Google Scholar] [CrossRef] [PubMed]
- Sala, L.; Zerolová, A.; Rodriguez, A.; Reimitz, D.; Davídková, M.; Ebel, K.; Bald, I.; Kočišek, J. Folding DNA into origami nanostructures enhances resistance to ionizing radiation. Nanoscale 2021, 13, 11197–11203. [Google Scholar] [CrossRef]
- Kielar, C.; Zhu, S.; Grundmeier, G.; Keller, A. Quantitative Assessment of Tip Effects in Single-Molecule High-Speed Atomic Force Microscopy Using DNA Origami Substrates. Angew. Chem. Int. Ed. Engl. 2020, 59, 14336–14341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liu, X.; Liu, P.; Wang, F.; Ariyama, H.; Ando, T.; Lin, J.; Wang, L.; Hu, J.; Li, B.; et al. Capturing transient antibody conformations with DNA origami epitopes. Nat. Commun. 2020, 11, 3114. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, S.; Shen, B.; Kostiainen, M.A.; Grundmeier, G.; Keller, A.; Linko, V. Real-Time Observation of Superstructure-Dependent DNA Origami Digestion by DNase I Using High-Speed Atomic Force Microscopy. ChemBioChem 2019, 20, 2818–2823. [Google Scholar] [CrossRef]
- Suzuki, Y.; Endo, M.; Cañas, C.; Ayora, S.; Alonso, J.C.; Sugiyama, H.; Takeyasu, K. Direct analysis of Holliday junction resolving enzyme in a DNA origami nanostructure. Nucleic Acids Res. 2014, 42, 7421–7428. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, A.; Endo, M.; Hidaka, K.; Sugiyama, H. Direct and real-time observation of rotary movement of a DNA nanomechanical device. J. Am. Chem. Soc. 2013, 135, 1117–1123. [Google Scholar] [CrossRef]
- Willner, E.M.; Kamada, Y.; Suzuki, Y.; Emura, T.; Hidaka, K.; Dietz, H.; Sugiyama, H.; Endo, M. Single-Molecule Observation of the Photoregulated Conformational Dynamics of DNA Origami Nanoscissors. Angew. Chem. Int. Ed. Engl. 2017, 56, 15324–15328. [Google Scholar] [CrossRef] [PubMed]
- Kielar, C.; Xin, Y.; Xu, X.; Zhu, S.; Gorin, N.; Grundmeier, G.; Möser, C.; Smith, D.M.; Keller, A. Effect of Staple Age on DNA Origami Nanostructure Assembly and Stability. Molecules 2019, 24, 2577. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishnan, S.; Krainer, G.; Grundmeier, G.; Schlierf, M.; Keller, A. Structural stability of DNA origami nanostructures in the presence of chaotropic agents. Nanoscale 2016, 8, 10398–10405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakrishnan, S.; Schärfen, L.; Hunold, K.; Fricke, S.; Grundmeier, G.; Schlierf, M.; Keller, A.; Krainer, G. Enhancing the stability of DNA origami nanostructures: Staple strand redesign versus enzymatic ligation. Nanoscale 2019, 11, 16270–16276. [Google Scholar] [CrossRef]
- Xin, Y.; Kielar, C.; Zhu, S.; Sikeler, C.; Xu, X.; Möser, C.; Grundmeier, G.; Liedl, T.; Heuer-Jungemann, A.; Smith, D.M.; et al. Cryopreservation of DNA Origami Nanostructures. Small 2020, 16, 1905959. [Google Scholar] [CrossRef] [Green Version]
- Pastré, D.; Piétrement, O.; Fusil, S.; Landousy, F.; Jeusset, J.; David, M.-O.; Hamon, L.; Le Cam, E.; Zozime, A. Adsorption of DNA to Mica Mediated by Divalent Counterions: A Theoretical and Experimental Study. Biophys. J. 2003, 85, 2507–2518. [Google Scholar] [CrossRef] [Green Version]
- Piétrement, O.; Pastré, D.; Fusil, S.; Jeusset, J.; David, M.-O.; Landousy, F.; Hamon, L.; Zozime, A.; Le Cam, E. Reversible Binding of DNA on NiCl 2 -Treated Mica by Varying the Ionic Strength. Langmuir 2003, 19, 2536–2539. [Google Scholar] [CrossRef]
- Hansma, H.G.; Laney, D.E. DNA binding to mica correlates with cationic radius: Assay by atomic force microscopy. Biophys. J. 1996, 70, 1933–1939. [Google Scholar] [CrossRef] [Green Version]
- Bezanilla, M.; Drake, B.; Nudler, E.; Kashlev, M.; Hansma, P.K.; Hansma, H.G. Motion and enzymatic degradation of DNA in the atomic force microscope. Biophys. J. 1994, 67, 2454–2459. [Google Scholar] [CrossRef] [Green Version]
- Xin, Y.; Shen, B.; Kostiainen, M.A.; Grundmeier, G.; Castro, M.; Linko, V.; Keller, A. Scaling Up DNA Origami Lattice Assembly. Chem. Eur. J. 2021, 27, 8564–8571. [Google Scholar] [CrossRef]
- Woo, S.; Rothemund, P.W.K. Self-assembly of two-dimensional DNA origami lattices using cation-controlled surface diffusion. Nat. Commun. 2014, 5, 4889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duguid, J.; Bloomfield, V.A.; Benevides, J.; Thomas, G.J. Raman spectroscopy of DNA-metal complexes. I. Interactions and conformational effects of the divalent cations: Mg, Ca, Sr, Ba, Mn, Co, Ni, Cu, Pd, and Cd. Biophys. J. 1993, 65, 1916–1928. [Google Scholar] [CrossRef] [Green Version]
- Fan, S.; Wang, D.; Cheng, J.; Liu, Y.; Luo, T.; Cui, D.; Ke, Y.; Song, J. Information Coding in a Reconfigurable DNA Origami Domino Array. Angew. Chem. Int. Ed. Engl. 2020, 59, 12991–12997. [Google Scholar] [CrossRef] [PubMed]
- Ido, S.; Kimura, K.; Oyabu, N.; Kobayashi, K.; Tsukada, M.; Matsushige, K.; Yamada, H. Beyond the helix pitch: Direct visualization of native DNA in aqueous solution. ACS Nano 2013, 7, 1817–1822. [Google Scholar] [CrossRef] [PubMed]
- Pyne, A.; Thompson, R.; Leung, C.; Roy, D.; Hoogenboom, B.W. Single-molecule reconstruction of oligonucleotide secondary structure by atomic force microscopy. Small 2014, 10, 3257–3261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sushko, M.L.; Shluger, A.L.; Rivetti, C. Simple model for DNA adsorption onto a mica surface in 1:1 and 2:1 electrolyte solutions. Langmuir 2006, 22, 7678–7688. [Google Scholar] [CrossRef]
- Lee, A.J.; Szymonik, M.; Hobbs, J.K.; Wälti, C. Tuning the translational freedom of DNA for high speed AFM. Nano Res. 2015, 8, 1811–1821. [Google Scholar] [CrossRef] [Green Version]
- Heenan, P.R.; Perkins, T.T. Imaging DNA Equilibrated onto Mica in Liquid Using Biochemically Relevant Deposition Conditions. ACS Nano 2019, 13, 4220–4229. [Google Scholar] [CrossRef]
- Akpinar, B.; Haynes, P.J.; Bell, N.A.W.; Brunner, K.; Pyne, A.L.B.; Hoogenboom, B.W. PEGylated surfaces for the study of DNA-protein interactions by atomic force microscopy. Nanoscale 2019, 11, 20072–20080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Zheng, M.; Li, Z.; Li, Q.; Mao, C. Patterning Nanoparticles with DNA Molds. ACS Appl. Mater. Interfaces 2019, 11, 13853–13858. [Google Scholar] [CrossRef]
- Franquelim, H.G.; Dietz, H.; Schwille, P. Reversible membrane deformations by straight DNA origami filaments. Soft Matter 2021, 17, 276–287. [Google Scholar] [CrossRef]
- Nakazawa, K.; El Fakih, F.; Jallet, V.; Rossi-Gendron, C.; Mariconti, M.; Chocron, L.; Hishida, M.; Saito, K.; Morel, M.; Rudiuk, S.; et al. Reversible Supra-Folding of User-Programmed Functional DNA Nanostructures on Fuzzy Cationic Substrates. Angew. Chem. Int. Ed. Engl. 2021, 60, 15214–15219. [Google Scholar] [CrossRef] [PubMed]
- Tanigawa, M.; Okada, T. Atomic force microscopy of supercoiled DNA structure on mica. Anal. Chim. Acta 1998, 365, 19–25. [Google Scholar] [CrossRef]
- Chu, C.-H.; Sarangadharan, I.; Regmi, A.; Chen, Y.-W.; Hsu, C.-P.; Chang, W.-H.; Lee, G.-Y.; Chyi, J.-I.; Chen, C.-C.; Shiesh, S.-C.; et al. Beyond the Debye length in high ionic strength solution: Direct protein detection with field-effect transistors (FETs) in human serum. Sci. Rep. 2017, 7, 5256. [Google Scholar] [CrossRef] [PubMed]
- Kielar, C.; Ramakrishnan, S.; Fricke, S.; Grundmeier, G.; Keller, A. Dynamics of DNA Origami Lattice Formation at Solid-Liquid Interfaces. ACS Appl. Mater. Interfaces 2018, 10, 44844–44853. [Google Scholar] [CrossRef]
- Xin, Y.; Martinez Rivadeneira, S.; Grundmeier, G.; Castro, M.; Keller, A. Self-assembly of highly ordered DNA origami lattices at solid-liquid interfaces by controlling cation binding and exchange. Nano Res. 2020, 13, 3142–3150. [Google Scholar] [CrossRef]
- Bui, H.; Onodera, C.; Kidwell, C.; Tan, Y.; Graugnard, E.; Kuang, W.; Lee, J.; Knowlton, W.B.; Yurke, B.; Hughes, W.L. Programmable periodicity of quantum dot arrays with DNA origami nanotubes. Nano Lett. 2010, 10, 3367–3372. [Google Scholar] [CrossRef]
- Opherden, L.; Oertel, J.; Barkleit, A.; Fahmy, K.; Keller, A. Paramagnetic decoration of DNA origami nanostructures by Eu3+ coordination. Langmuir 2014, 30, 8152–8159. [Google Scholar] [CrossRef] [PubMed]
- Morga, M.; Adamczyk, Z.; Kosior, D.; Kujda-Kruk, M. Kinetics of Poly-l-lysine Adsorption on Mica and Stability of Formed Monolayers: Theoretical and Experimental Studies. Langmuir 2019, 35, 12042–12052. [Google Scholar] [CrossRef] [PubMed]







| Strategy | Solution | Total | Intact | Damaged | Percentage Intact |
|---|---|---|---|---|---|
| Mg2+ | PBS | 654 | 614 | 40 | 93.9 |
| H2O | 808 | 717 | 91 | 88.7 | |
| Ni2+ | PBS | 315 | 38 | 277 | 12.1 |
| H2O | 92 | 34 | 58 | 37.0 | |
| PLL | PBS | 1557 | 932 | 625 | 59.9 |
| H2O | 179 | 49 | 130 | 27.4 | |
| Spdn | PBS | 3635 | 2657 | 978 | 73.1 |
| H2O | 161 | 110 | 51 | 68.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xin, Y.; Zargariantabrizi, A.A.; Grundmeier, G.; Keller, A. Magnesium-Free Immobilization of DNA Origami Nanostructures at Mica Surfaces for Atomic Force Microscopy. Molecules 2021, 26, 4798. https://doi.org/10.3390/molecules26164798
Xin Y, Zargariantabrizi AA, Grundmeier G, Keller A. Magnesium-Free Immobilization of DNA Origami Nanostructures at Mica Surfaces for Atomic Force Microscopy. Molecules. 2021; 26(16):4798. https://doi.org/10.3390/molecules26164798
Chicago/Turabian StyleXin, Yang, Amir Ardalan Zargariantabrizi, Guido Grundmeier, and Adrian Keller. 2021. "Magnesium-Free Immobilization of DNA Origami Nanostructures at Mica Surfaces for Atomic Force Microscopy" Molecules 26, no. 16: 4798. https://doi.org/10.3390/molecules26164798
APA StyleXin, Y., Zargariantabrizi, A. A., Grundmeier, G., & Keller, A. (2021). Magnesium-Free Immobilization of DNA Origami Nanostructures at Mica Surfaces for Atomic Force Microscopy. Molecules, 26(16), 4798. https://doi.org/10.3390/molecules26164798