Saccharpiscinols A–C: Flavans with Potential Anti-Inflammatory Activities from One Actinobacteria Saccharomonospora piscinae
Abstract
:1. Introduction
2. Results
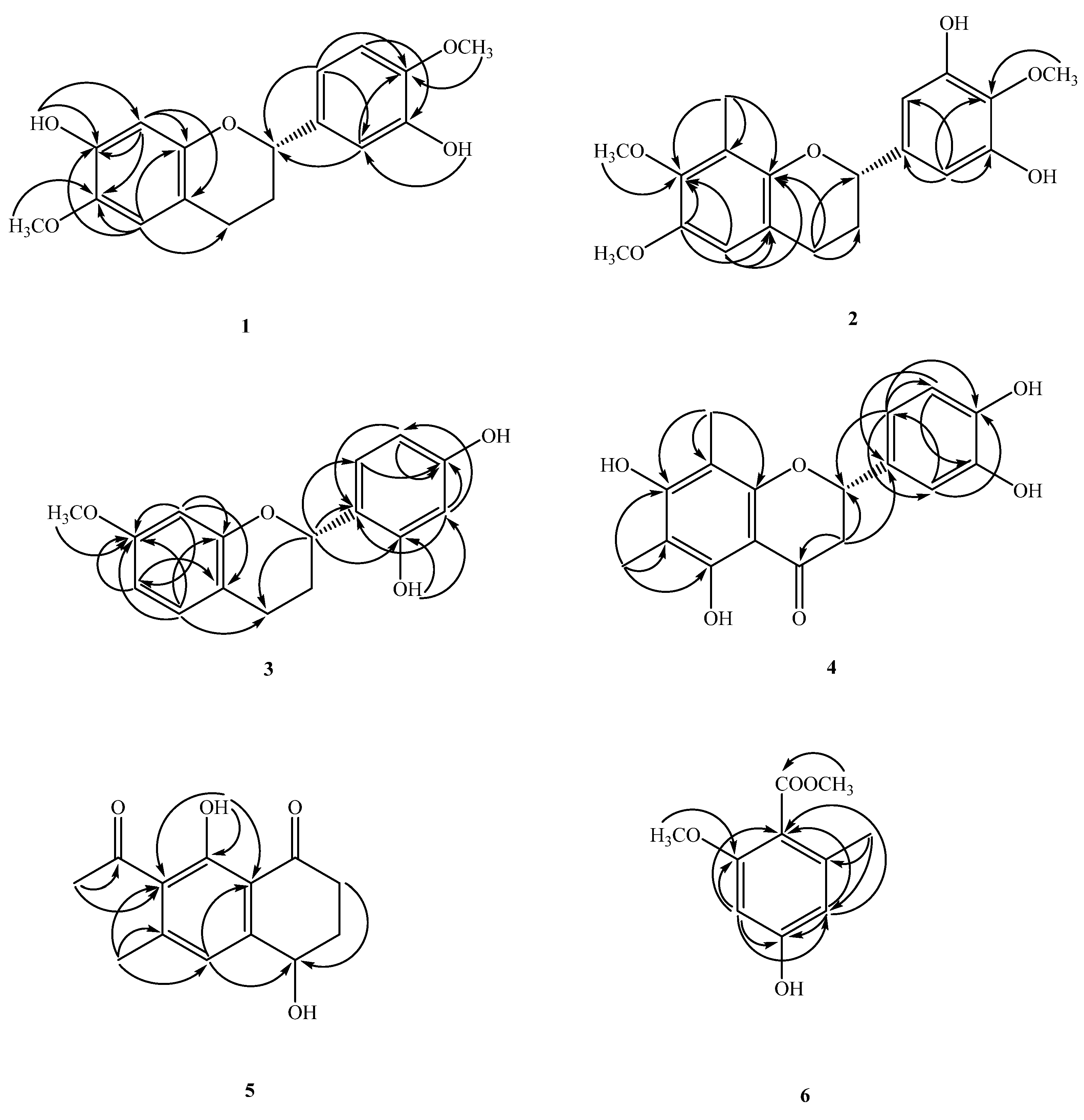
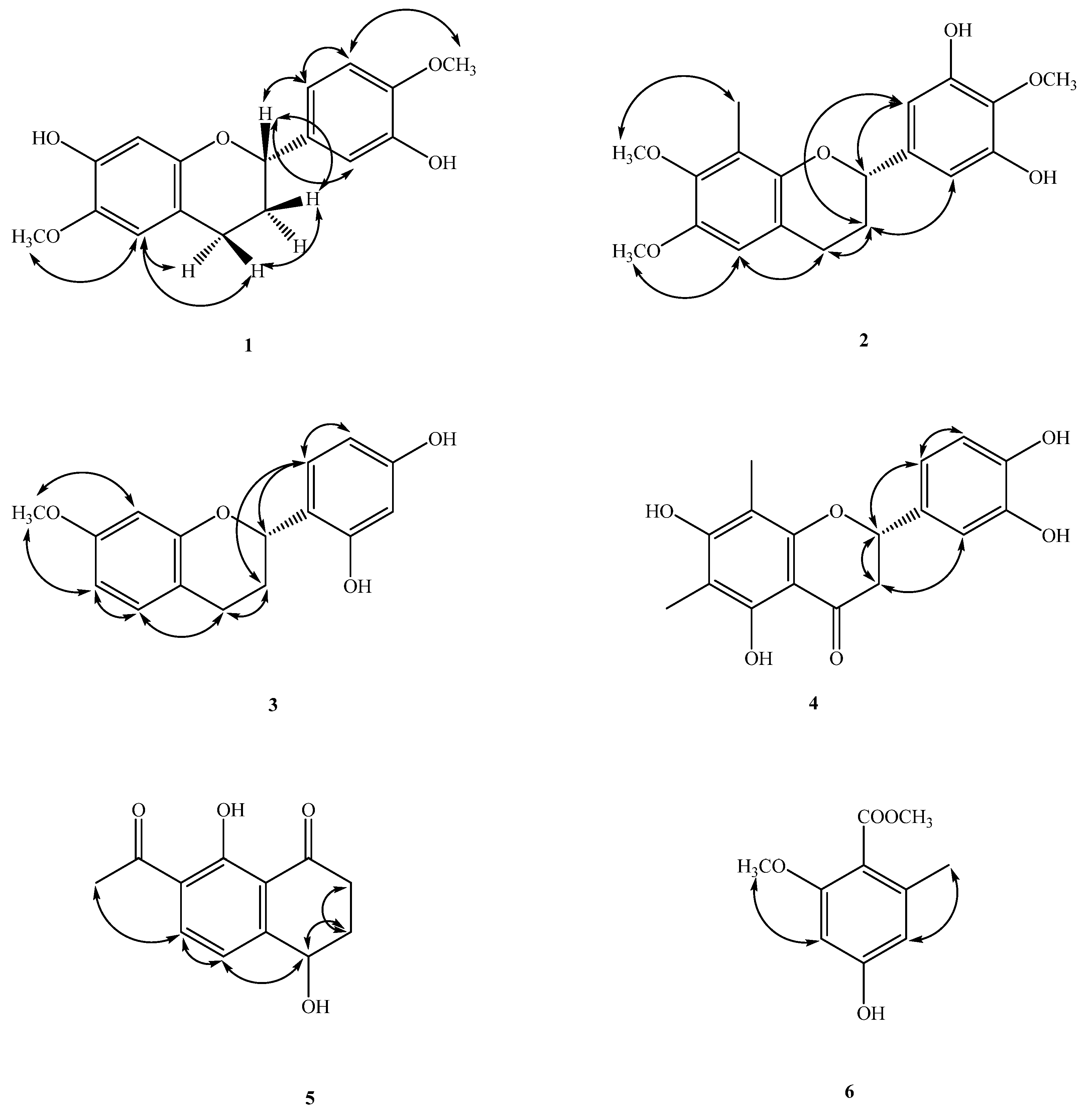
2.1. Structure Elucidation of Compounds
Analysis EtOAc Crude Extracts of Compounds by HPLC
2.2. Biological Studies
3. Materials and Methods
3.1. General Experimental Procedures
3.1.1. Microorganism
3.1.2. Cultivation and Preparation of the Fungal Strain
3.1.3. Isolation and Characterization of Secondary Metabolites
3.1.4. Determination of NO Production and Cell Viability Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Osada, H. Fascinating bioactive compounds from actinomycetes. Actinomycetologica 1995, 9, 254–262. [Google Scholar] [CrossRef]
- Mythili, B.; Ayyappa Das, M.P. Studies on antimicrobial activity of Streptomyces spp. isolates from tea plantation soil. Res. J. Agric. Sci. 2011, 2, 104–106. [Google Scholar]
- Kuster, E. The Actinomycetes. In Soil Biology; Burges, A., Raw, F., Eds.; Academic Press: London, UK, 1968; pp. 111–124. [Google Scholar]
- Tseng, M.; Chiang, W.P.; Liao, H.C.; Hsieh, S.Y.; Yuan, G.F. Saccharomonospora piscinae sp. nov., a novel actinobacterium from fishpond sediment in Taiwan. Int. J. Syst. Evol. Microbiol. 2018, 68, 1418–1422. [Google Scholar] [CrossRef]
- Ali, A.; Makboul, M.A.; Attia, A.A.; Ali, D.T. Chromones and flavans from Pancratium maritimum. Phytochemistry 1990, 29, 625–627. [Google Scholar] [CrossRef]
- Rezende, L.C.; Juck, D.B.F.; David, J.M.; David, J.P.; Lima, M.V.B.; Lima, L.S.; Alves, C.Q. New flavans isolated from the leaves and stems of Cratylia mollis (Leguminosae). Phytochem. Lett. 2015, 14, 165–169. [Google Scholar] [CrossRef]
- Achenbach, H.; Stoecker, M.; Constenla, M.A. Flavonoid and other constituents of Bauhinia manca. Phytochemistry 1988, 27, 1835–1842. [Google Scholar]
- Loset, J.D.; Marston, A.; Gupta, M.P.; Hostettmann, K. A methylflavan with free radical scavenging properties from Pancratium littorale. Fitoterapia 2001, 72, 35–39. [Google Scholar]
- Masaoud, M.; Ripperger, H. Flavonoids of dragon’s blood from Dracaena cinnabari. Phytochemistry 1995, 38, 745–749. [Google Scholar] [CrossRef]
- Abd El-Hafiz, M.A.; Ramadan, M.A.; Anton, R. Minor Phenolic Constituents of Crinum augustum. J. Nat. Prod. 1990, 53, 1349–1352. [Google Scholar] [CrossRef]
- Tan, B.Q.; Huang, S.S.; Liang, Y.E.; Sun, J.B.; Ma., Y.; Zeng, B.; Lee, S.M.Y.; Lu, J.L. Two new flavans from the roots of Dianella ensifolia (L.) DC. Nat. Prod. Res. 2017, 31, 1561–1565. [Google Scholar]
- Youssef, D.T.A.; Ramadan, M.A.; Khalifa, A.A. Acetophenones, a chalcone, a chromone and flavonoids from Pancratium maritimum. Phytochemistry 1998, 49, 2579–2583. [Google Scholar] [CrossRef]
- Kishimoto, Y. Pharmaceutical studies on ferns. XI. flavonoids of Cyrtomium species (3). Constitution of cyrtominetin and cyrtopterinetin. Chem. Pharm. Bull. 1956, 4, 24–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nhung, L.T.H.; Linh, N.T.T.; Cham, B.T.; Thuy, T.T.; Tam, N.T.; Thien, D.D.; Huong, P.T.M.; Tan, V.M.; Tai, B.H.; Anh, N.T.H. New phenolics from Dianella ensifolia. Nat. Prod. Res. 2019, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Elix, J.A.; Lajide, L. Three new tridepsides from the lichen Pseudocyphellaria faveolata. Aust. J. Chem. 1981, 34, 2005–2011. [Google Scholar] [CrossRef]
- Gieni, R.S.; Li, Y.; Hay Glass, K.T. Comparison of [3H]thymidine incorporation with MTT- and MTS-based bioassays for human and murine IL-2 and IL-4 analysis. Tetrazolium assays provide markedly enhanced sensitivity. J. Immunol. Methods 1995, 70, 85–93. [Google Scholar] [CrossRef]
- Johansson, M.; Kopcke, B.; Anke, H.; Sterner, O. Biologically active secondary metabolites from the ascomycete A111–95. 2. structure elucidation. J. Antibiot. 2002, 55, 104–106. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.J.; Wu, M.D.; Chan, H.Y.; Chang, H.S.; Wu, H.C.; Chen, J.J.; Yuan, G.F.; Weng, J.R.; Chang, C.T.; Lin, H.C. A new azaphilone derivative from the Monascus kaoliang fermented rice. Chem. Nat. Compd. 2019, 55, 79–81. [Google Scholar] [CrossRef]
- Wu, H.C.; Cheng, M.J.; Wu, M.D.; Chen, J.J.; Chen, Y.L.; Chang, H.S. Three new constituents from the fungus of monascus purpureus and their anti-inflammatory activity. Phytochem. Lett. 2019, 31, 242–248. [Google Scholar] [CrossRef]
- Cheng, M.J.; Wu, M.D.; Su, Y.S.; Chan, H.Y.; Hsieh, S.Y.; Chen, Y.L.; Chen, J.J.; Chou, Y.T.; Hsiao, C.Y.; Wu, H.S. Additional chemical constituents of an endophytic fungus Xylaria papulis. Chem. Nat. Compd. 2019, 55, 340–342. [Google Scholar] [CrossRef]
- Feng, K.J.; Cheng, M.J.; Yang, S.H.; Wu, M.D.; Hsieh, S.Y.; Chan, H.Y.; Su, Y.S.; Chou, Y.T.; Chang, H.S. Chemical constituents of the endophytic fungus Ophiocordyceps sobolifera. Chem. Nat. Compd. 2019, 55, 309–312. [Google Scholar] [CrossRef]
- Cheng, M.J.; Wu, M.D.; Wu, H.C.; Chan, H.Y.; Chen, Y.L.; Chang, H.S.; Chen, J.J.; Kuo, Y.H. Benzenoid derivatives and amide constituents of the Monascus sp.-fermented rice. Chem. Nat. Compd. 2019, 55, 787–789. [Google Scholar] [CrossRef]
- Wu, H.C.; Chen, J.J.; Wu, M.D.; Cheng, M.J.; Chang, H.S. Identification of new pigments produced by the fermented rice of the fungus Monascus pilosus and their anti-inflammatoryactivity. Phytochem. Lett. 2020, 40, 181–187. [Google Scholar] [CrossRef]


| Compound | IC50 | Cell Viability |
|---|---|---|
| Nitrite (μM) | (% Control) | |
| 1 | 12.5 ± 0.2 * | 91.1 ± 4.2 |
| 2 | 18.0 ± 0.3 * | 82.2 ± 3.2 # |
| 3 | 21.8 ± 0.5 * | 76.3 ± 5.4 # |
| 4 | 20.0 ± 1.5 * | 68.2 ± 3.3 # |
| Quercetin | 35.45 ± 3.78 * | 95.9 ± 1.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, Y.-S.; Chen, J.-J.; Cheng, M.-J.; Chai, C.-Y.; Kwan, A.-L.; Huang, J.-C.; Kuo, Y.-H. Saccharpiscinols A–C: Flavans with Potential Anti-Inflammatory Activities from One Actinobacteria Saccharomonospora piscinae. Molecules 2021, 26, 4909. https://doi.org/10.3390/molecules26164909
Su Y-S, Chen J-J, Cheng M-J, Chai C-Y, Kwan A-L, Huang J-C, Kuo Y-H. Saccharpiscinols A–C: Flavans with Potential Anti-Inflammatory Activities from One Actinobacteria Saccharomonospora piscinae. Molecules. 2021; 26(16):4909. https://doi.org/10.3390/molecules26164909
Chicago/Turabian StyleSu, Yung-Shun, Jih-Jung Chen, Ming-Jen Cheng, Chee-Yin Chai, Aij-Lie Kwan, Jheng-Cian Huang, and Yueh-Hsiung Kuo. 2021. "Saccharpiscinols A–C: Flavans with Potential Anti-Inflammatory Activities from One Actinobacteria Saccharomonospora piscinae" Molecules 26, no. 16: 4909. https://doi.org/10.3390/molecules26164909
APA StyleSu, Y.-S., Chen, J.-J., Cheng, M.-J., Chai, C.-Y., Kwan, A.-L., Huang, J.-C., & Kuo, Y.-H. (2021). Saccharpiscinols A–C: Flavans with Potential Anti-Inflammatory Activities from One Actinobacteria Saccharomonospora piscinae. Molecules, 26(16), 4909. https://doi.org/10.3390/molecules26164909








