Interaction between Curcumin and β-Casein: Multi-Spectroscopic and Molecular Dynamics Simulation Methods
Abstract
:1. Introduction
2. Results
2.1. Fluorescence Spectra
2.2. Ultraviolet-Visible Spectra
2.3. Homology Modeling and Evaluation of Three-Dimensional Structure of β-Casein
2.4. Molecular Docking
2.5. MD Simulation
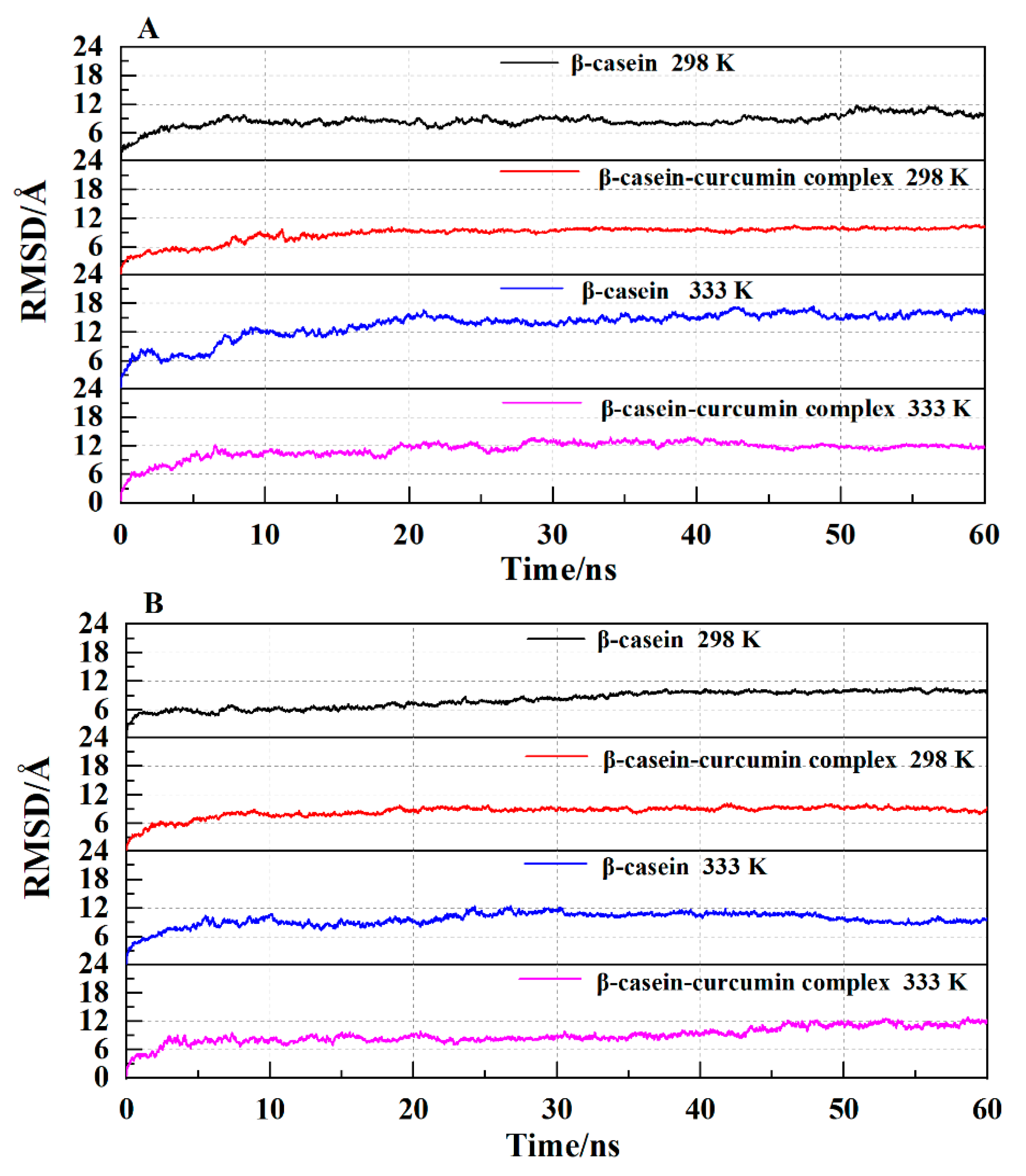
2.5.1. Analysis of the RMSD Value
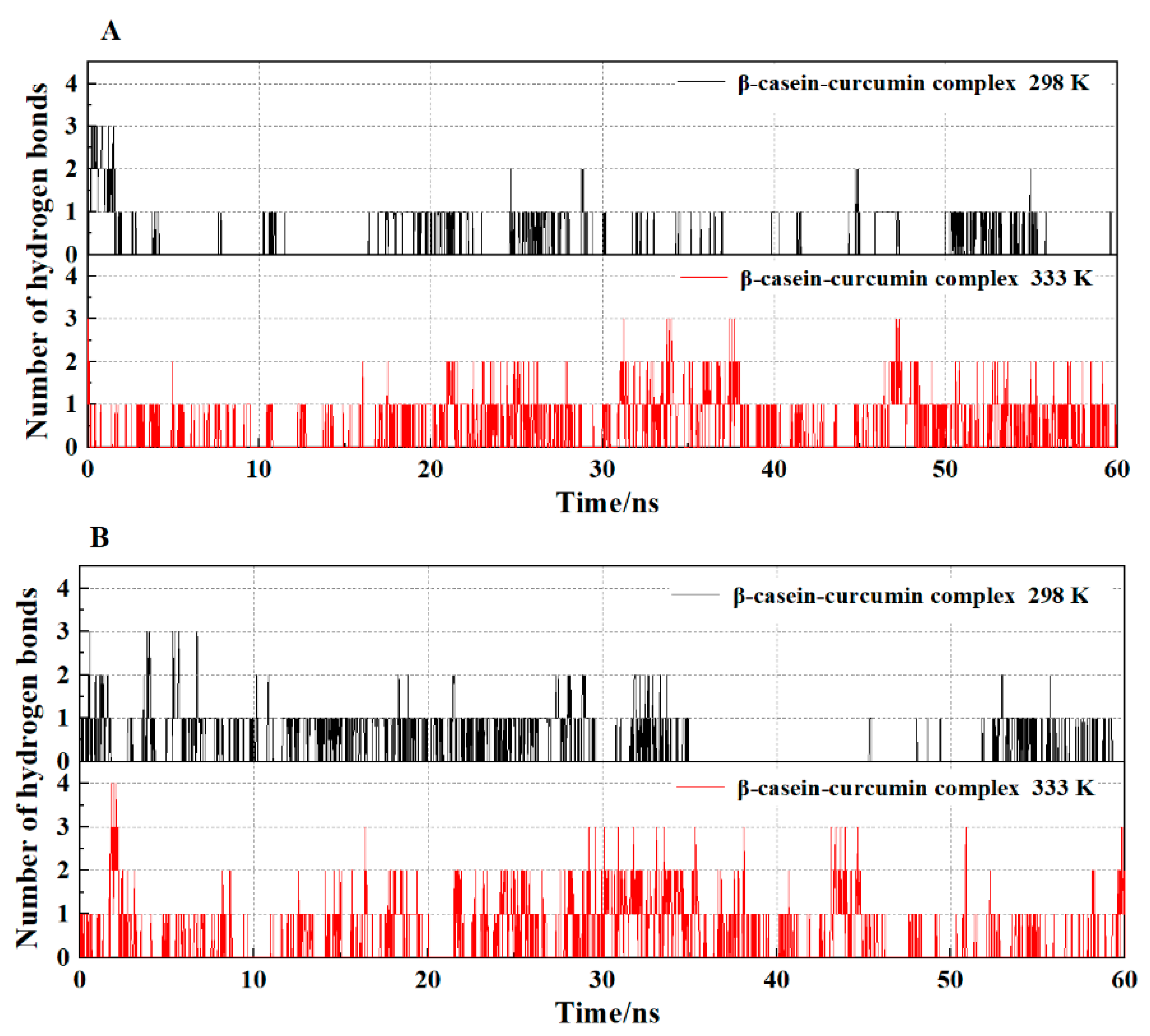
2.5.2. Hydrogen Bond Analysis
2.5.3. System Energy Analysis
2.5.4. Analysis of Secondary Structure Content
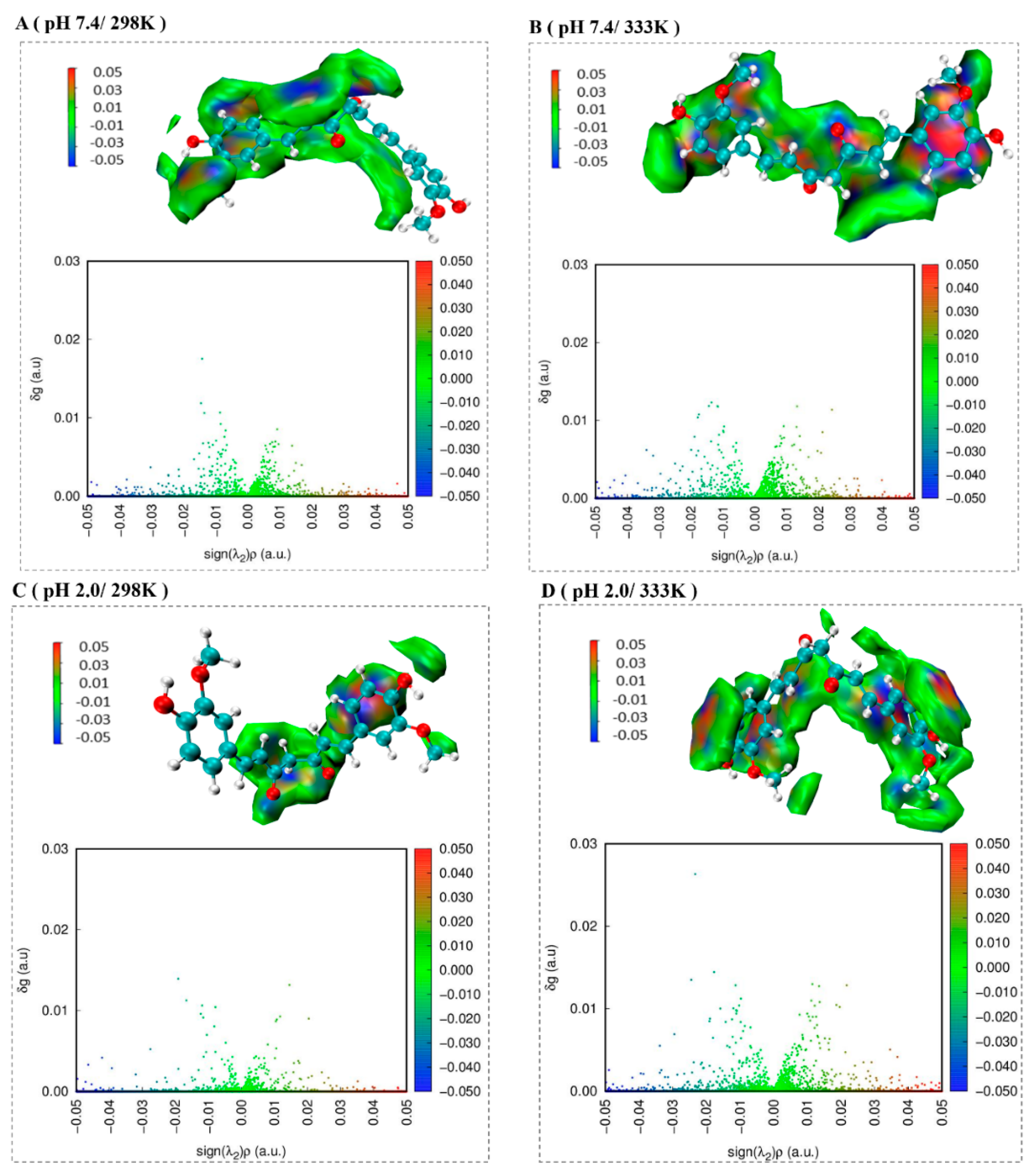
2.6. The Independent Gradient Model Analysis
3. Materials and Methods
3.1. Materials
3.2. Methods
3.2.1. Solution Preparation
3.2.2. Fluorescence Spectroscopy
3.2.3. Ultraviolet-Visible Spectroscopy
3.2.4. β-Casein Homology Modeling
3.2.5. Molecular Docking
3.2.6. MD Simulation
3.2.7. Weak Interaction Analysis
3.2.8. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Fu, Y.S.; Chen, T.H.; Weng, L.B.; Huang, L.Y.; Lai, D.; Weng, C.F. Pharmacological Properties and Underlying Mechanisms of Curcumin and Prospects in Medicinal Potential. Biomed. Pharmacother. 2021, 141, 111888. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.C.; Prasad, S.; Kim, J.H.; Patchva, S.; Webb, L.J.; Priyadarsini, I.K.; Aggarwal, B.B. Multitargeting by Curcumin as Revealed by Molecular Interaction Studies. Cheminform 2011, 28, 1937–1955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, J.; Wang, T.; Wang, M. Investigation of the Anti-Cataractogenic Mechanisms of Curcumin through in vivo and in vitro Studies. BMC Ophthalmol. 2018, 18, 48. [Google Scholar] [CrossRef] [Green Version]
- Ashry, A.M.; Hassan, A.M.; Habiba, M.M.; Elzayat, A.; Elsharnouby, M.E.; Sewilam, H.; Dawood, M.A.O. The Impact of Dietary Curcumin on the Growth Performance, Intestinal Antibacterial Capacity, and Haemato-Biochemical Parameters of Gilthead Seabream (Sparus aurata). Animals 2021, 11, 1779. [Google Scholar] [CrossRef] [PubMed]
- Sandhiutami, N.M.D.; Arozal, W.; Louisa, M.; Rahmat, D.; Wuyung, P.E. Curcumin Nanoparticle Enhances the Anticancer Effect of Cisplatin by Inhibiting PI3K/AKT and JAK/STAT3 Pathway in Rat Ovarian Carcinoma Induced by DMBA. Front. Pharmacol. 2021, 11, 2199. [Google Scholar] [CrossRef] [PubMed]
- Iova, G.M.; Calniceanu, H.; Popa, A.; Szuhanek, C.A.; Marcu, O.; Ciavoi, G.; Scrobota, I. The Antioxidant Effect of Curcumin and Rutin on Oxidative Stress Biomarkers in Experimentally Induced Periodontitis in Hyperglycemic Wistar Rats. Molecules 2021, 26, 1332. [Google Scholar] [CrossRef]
- Ma, Z.W.; Wang, N.; He, H.B.; Tang, X. Pharmaceutical Strategies of Improving Oral Systemic Bioavailability of Curcumin for Clinical Application. J. Control. Release 2019, 316, 359–380. [Google Scholar] [CrossRef]
- Michele, D.C.; Riccardo, G. Dietary Curcumin: Correlation between Bioavailability and Health Potential. Nutrients 2019, 11, 2147. [Google Scholar] [CrossRef] [Green Version]
- Esmaili, M.; Ghaffari, S.M.; Moosavi-Movahedi, Z.; Atri, M.S.; Sharifizadeh, A.; Farhadi, M.; Yousefi, R.; Chobert, J.M.; Haertlé, T.; Moosavi-Movahedi, A.A. Beta Casein-Micelle as a Nano Vehicle for Solubility Enhancement of Curcumin; Food Industry Application. LWT-Food Sci. Technol. 2011, 44, 2166–2172. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, P.; Yang, Z.Y.; Du, F.F.; Li, Z.; Wu, C.L.; Fang, A.H.; Xu, X.L.; Zhou, G.H. Molecular Dynamics Simulation Exploration of the Interaction between Curcumin and Myosin Combined with the Results of Spectroscopy Techniques. Food Hydrocoll. 2020, 101, 105455. [Google Scholar] [CrossRef]
- Rajabi, M.; Farhadian, S.; Shareghi, B.; Asgharzadeh, S.; Momeni, L. Noncovalent Interactions of Bovine Trypsin with Curcumin and Effect on Stability, Structure, and Function. Colloids Surf. B 2019, 183, 110287. [Google Scholar] [CrossRef]
- Wang, R.J.; Wei, Y.Y.; Wang, M.Q. Interaction of Natural Compounds in Licorice and Turmeric with HIV-NCp7 Zinc Finger Domain: Potential Relevance to the Mechanism of Antiviral Activity. Molecules 2021, 26, 3563. [Google Scholar] [CrossRef]
- Allahdad, Z.; Varidi, M.; Zadmard, R.; Saboury, A.A.; Haertlé, T. Binding of β-Carotene to Whey Proteins: Multi-Spectroscopic Techniques and Docking Studies. Food Chem. 2018, 277, 96–106. [Google Scholar] [CrossRef]
- Saleh, T.; Soudi, T.; Shojaosadati, S.A. Redox Responsive Curcumin-Loaded Human Serum Albumin Nanoparticles: Preparation, Characterization and in vitro Evaluation. Int. J. Biol. Macromol. 2018, 114, 759–766. [Google Scholar] [CrossRef]
- Safavi, M.S.; Shojaosadati, S.A.; Yang, H.G.; Kim, Y.J. Reducing Agent-Free Synthesis of Curcumin-Loaded Albumin Nanoparticles by Self-Assembly at Room Temperature. Int. J. Pharm. 2017, 529, 303–309. [Google Scholar] [CrossRef]
- Mirpoor, S.F.; Hosseini, S.M.H.; Yousefi, G.H. Mixed Biopolymer Nanocomplexes Conferred Physicochemical Stability and Sustained Release Behavior to Introduced Curcumin. Food Hydrocoll. 2017, 71, 216–224. [Google Scholar] [CrossRef]
- Chen, F.P.; Li, B.S.; Tang, C.H. Nanocomplexation between Curcumin and Soy Protein Isolate: Influence on Curcumin Stability/Bioaccessibility and in vitro Protein Digestibility. J. Agric. Food Chem. 2015, 63, 3559–3569. [Google Scholar] [CrossRef]
- Wusigale; Liang, L.; Luo, Y.C. Casein and Pectin: Structures, Interactions, and Applications. Trends Food Sci. Technol. 2020, 97, 391–403. [Google Scholar] [CrossRef]
- Zhang, M.; Lai, T.T.; Yao, M.K.; Zhang, M.; Yang, Z.N. Interaction of the Exopolysaccharide from Lactobacillus Plantarum YW11 with Casein and Bioactivities of the Polymer Complex. Foods 2021, 10, 1153. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.J.; Yang, M.; Wang, Y.C.; Wa, W.Q.; Zheng, J. Interaction between Caffeic Acid/Caffeic Acid Phenethyl Ester and Micellar Casein. Food Chem. 2021, 349, 129154. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.Y.; He, Y.L.; Kong, Y.C.; Mei, X.Y.; Huo, Y.P.; He, Y.; Liu, J.L. Elucidating the Interaction Mechanism of Eriocitrin with β-Casein by Multi-Spectroscopic and Molecular Simulation Methods. Food Hydrocoll. 2019, 94, 63–70. [Google Scholar] [CrossRef]
- Valenti, L.; Riso, P.; Mazzocchi, A.; Porrini, M.; Fargion, S.; Agostoni, C.; Angeloni, C. Dietary Anthocyanins As Nutritional Therapy for Nonalcoholic Fatty Liver Disease. Oxid. Med. Cell. Longev. 2013, 2013, 145421–145428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, J.; Xu, D.X.; Yang, J.; Zhang, X.; Mu, T.; Wang, Q.Y. Analysis of the Interaction Mechanism of Anthocyanins (Aronia Melanocarpa Elliot) with β-Casein. Food Hydrocoll. 2018, 84, 276–281. [Google Scholar] [CrossRef]
- Ross, P.D.; Subramanian, S. Thermodynamics of Protein Association Reactions: Forces Contributing to Stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef]
- Sun, Q.; He, J.; Yang, H.; Li, S.; Zhao, L.; Li, H. Analysis of Binding Properties and Interaction of Thiabendazole and Its Metabolite with Humanserum Albumin via Multiple Spectroscopic Methods. Food Chem. 2017, 233, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Brennan, J.D. Using Intrinsic Fluorescence to Investigate Proteins Entrapped in Sol–Gel Derived Materials. Appl. Spectrosc. 1999, 53, 106A–121A. [Google Scholar] [CrossRef]
- Wieduwilt, E.K.; Boisson, J.C.; Terraneo, G.; Hénon, E.; Genoni, A. A Step toward the Quantification of Noncovalent Interactions in Large Biological Systems: The Independent Gradient Model-Extremely Localized Molecular Orbital Approach. J. Chem. Inf. Model. 2021, 61, 795–809. [Google Scholar] [CrossRef] [PubMed]
- He, C.Y.; Liu, X.L.; Jiang, Z.J.; Geng, S.; Ma, H.J.; Liu, B.J. Interaction Mechanism of Flavonoids and α-Glucosidase: Experimental and Molecular Modelling Studies. Foods 2019, 8, 355. [Google Scholar] [CrossRef] [Green Version]
- Chung, C.; Rojanasasithara, T.; Mutilangi, W.; McClements, D.J. Enhancement of Colour Stability of Anthocyanins in Model Beverages by Gum Arabic Addition. Food Chem. 2016, 201, 14–22. [Google Scholar] [CrossRef]
- Roy, S.; Nandi, R.K.; Ganai, S.; Majumdar, K.C.; Tapan, K.D. Binding Interaction of Phosphorus Heterocycles with Bovine Serum Albumin: A Biochemical Study. J. Pharm. Anal. 2017, 7, 19–26. [Google Scholar] [CrossRef]
- Jang, J.C.; Liu, H.; Chen, W.; Zou, G.L. Binding of Mitomycin C to Blood Proteins: A Spectroscopic Analysis and Molecular Docking. J. Mol. Struct. 2009, 928, 72–77. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Rullmann, J.A.C.; Macarthur, M.W.; Kaptein, R.; Thornton, J.M. QUA and PROCHECK-NMR: Programs for Checking the Quality of Protein Structures Solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Elmar, K.; Gert, V. YASARA View-Molecular Graphics for All Devices-from Smartphones to Workstations. Bioinformatics 2014, 20, 2981–2982. [Google Scholar] [CrossRef] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, Efficient Generation of High-quality Atomic Charges. AM1-BCC Model: II. Parameterization and Validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef]
- Corentin, L.; Gaëtan, R.; Hassan, K.; Jean-Charles, B.; Julia, C.G.; Eric, H. Accurately Extracting the Signature of Intermolecular Interactions Present in the NCI Plot of the Reduced Density Gradient versus Electron Density. Phys. Chem. Chem. Phys. 2017, 19, 17928–17936. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F.W. Multiwfn: A Multifunctional Wavefunction Analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
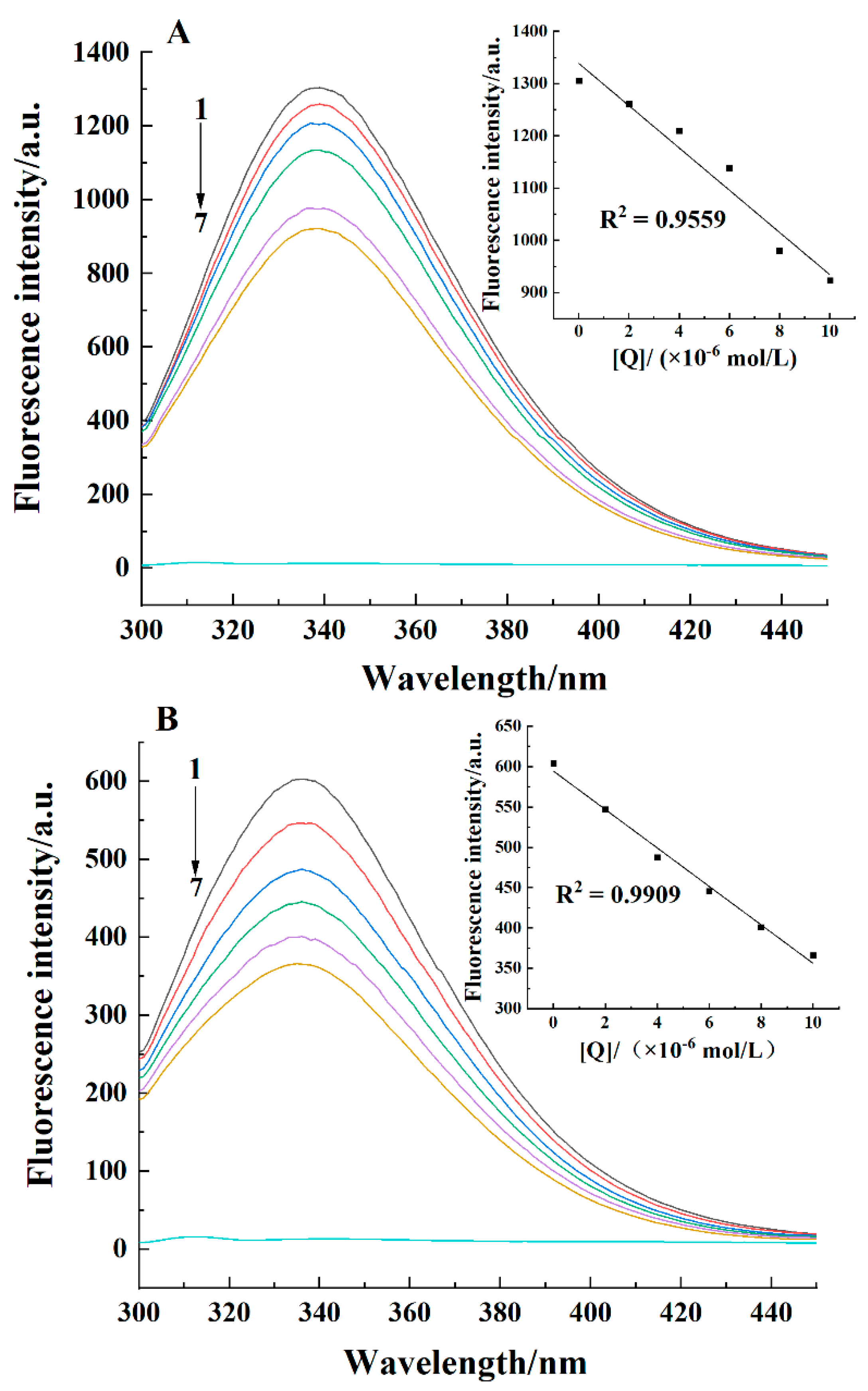
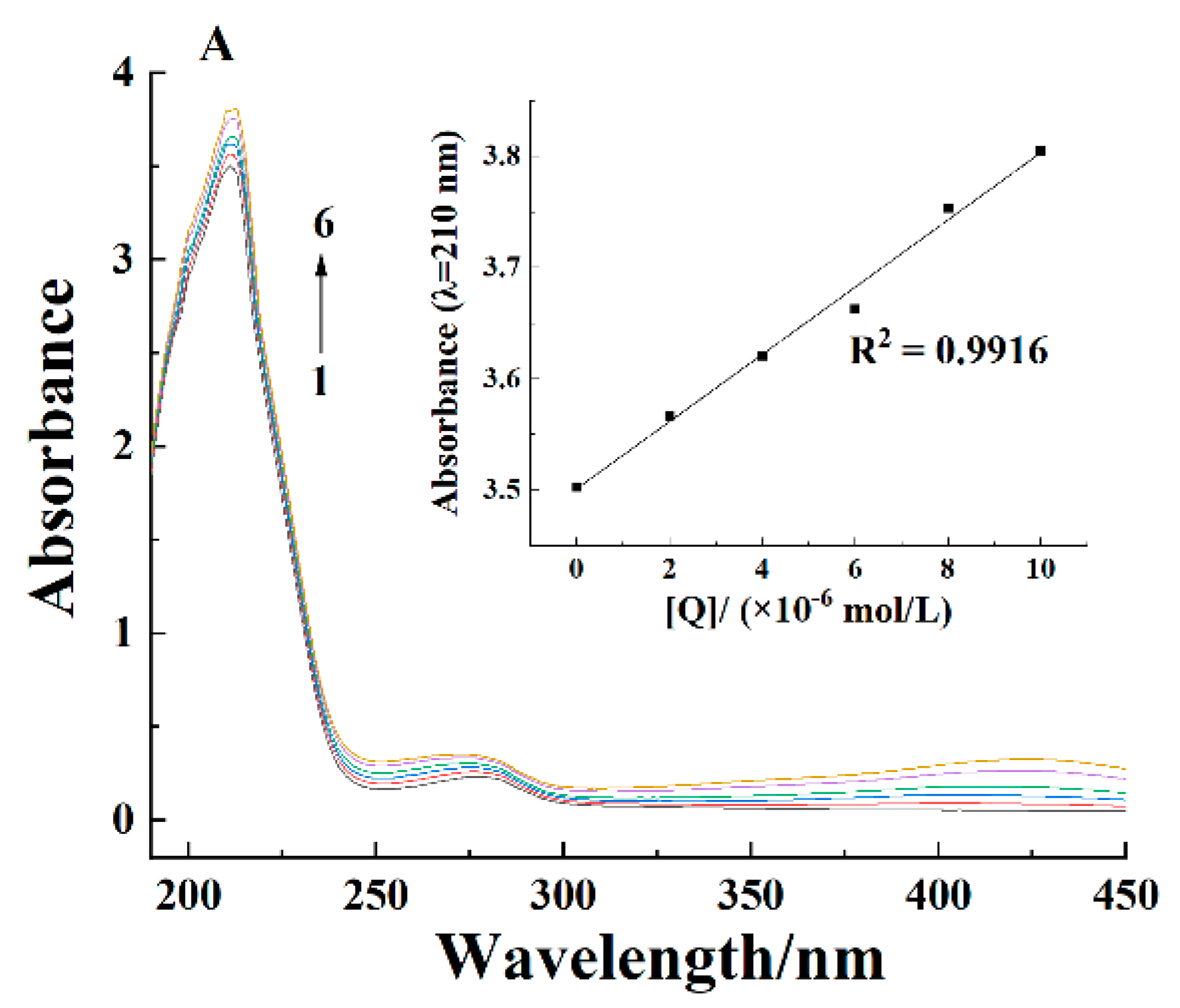
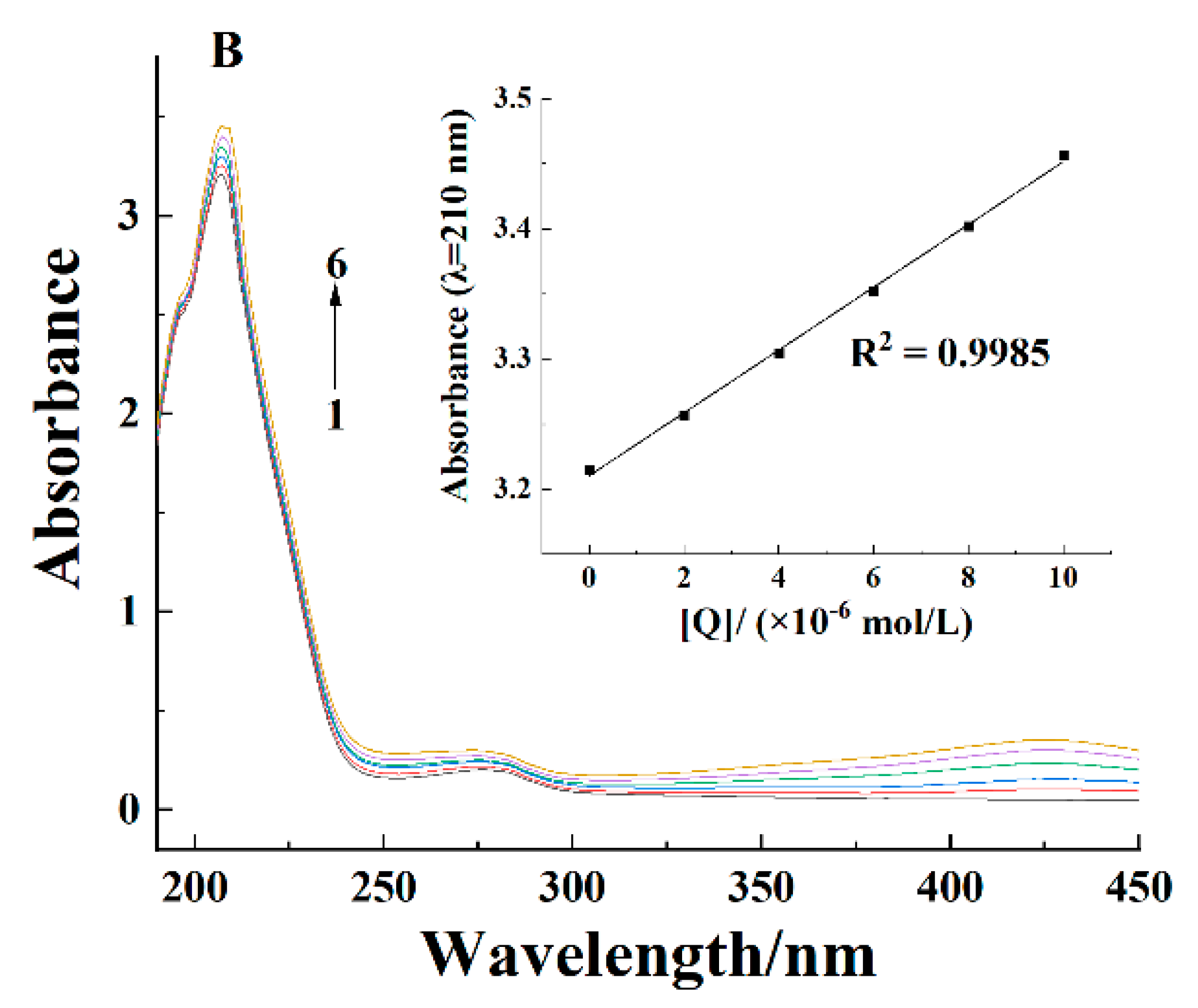

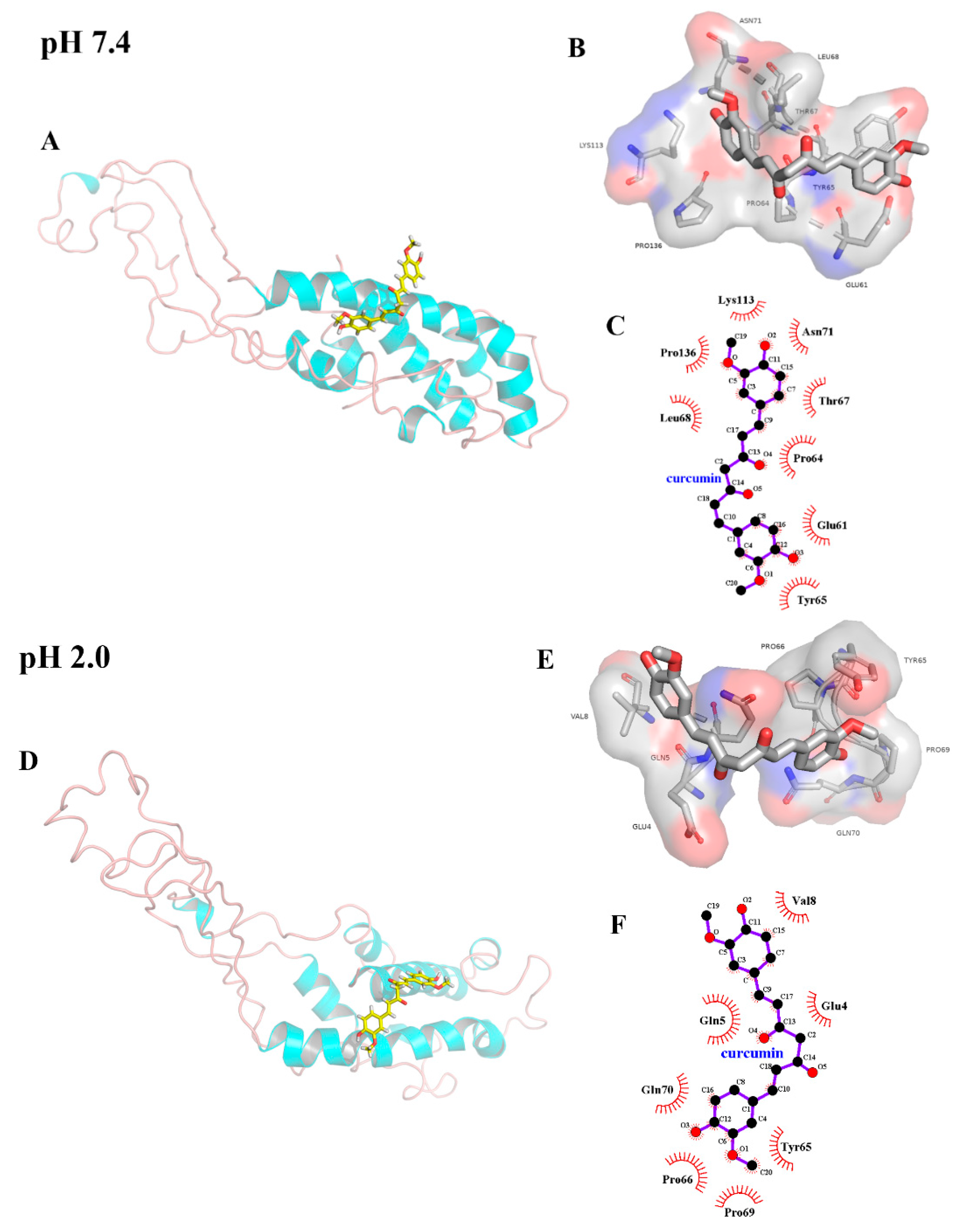
| pH | T/K | Ksv × 104/(L/mol) | Kq × 1012/(L/(mol·s)) | n | Kb × 104/(L/mol) | ΔH/(kJ/mol) | ΔS/(J/(mol·K)) | ΔG/(kJ/mol) |
|---|---|---|---|---|---|---|---|---|
| 7.4 | 298 | 3.67 ± 0.19 | 3.67 ± 0.19 | 1.59 ± 0.14 | 3.98 ± 0.26 | −8.43 ± 0.10 | 59.93 ± 0.28 | −17.87 ± 0.09 |
| 305 | 3.49 ± 0.07 | 3.49 ± 0.07 | 1.35 ± 0.08 | 3.90 ± 0.15 | −8.43 ± 0.10 | 59.93 ± 0.28 | −18.29 ± 0.10 | |
| 312 | 3.42 ± 0.07 | 3.42 ± 0.07 | 1.01 ± 0.01 | 3.41 ± 0.06 | −8.43 ± 0.10 | 59.93 ± 0.28 | −18.71 ± 0.09 | |
| 2.0 | 298 | 6.28 ± 0.05 | 6.28 ± 0.05 | 1.13 ± 0.02 | 6.48 ± 0.03 | −6.84 ± 0.11 | 69.21 ± 0.40 | −20.63 ± 0.12 |
| 305 | 5.98 ± 0.09 | 5.98 ± 0.09 | 1.17 ± 0.05 | 6.17 ± 0.05 | −6.84 ± 0.11 | 69.21 ± 0.40 | −21.12 ± 0.12 | |
| 312 | 5.74 ± 0.09 | 5.74 ± 0.09 | 1.07 ± 0.07 | 5.73 ± 0.03 | −6.84 ± 0.11 | 69.21 ± 0.40 | −21.60 ± 0.12 |
| pH | Conformation | Eb/(kcal/mol) | Kd/(× 10−7 mol/L) | Present Interacting Receptor Residues |
|---|---|---|---|---|
| 7.4 | 1 | −7.01 | 0.73 | 10 |
| 2 | −6.42 | 1.90 | 11 | |
| 3 | −6.38 | 2.10 | 10 | |
| 4 | −6.34 | 2.20 | 11 | |
| 5 | −6.15 | 3.10 | 6 | |
| 6 | −6.08 | 3.50 | 7 | |
| 7 | −5.67 | 7.10 | 10 | |
| 8 | −5.58 | 8.20 | 9 | |
| 9 | −5.54 | 8.70 | 9 | |
| 2.0 | 1 | −7.53 | 0.30 | 11 |
| 2 | −7.17 | 0.55 | 9 | |
| 3 | −6.79 | 1.00 | 10 | |
| 4 | −6.62 | 1.40 | 12 | |
| 5 | −6.10 | 3.40 | 7 | |
| 6 | −5.94 | 4.50 | 8 | |
| 7 | −5.89 | 4.90 | 8 | |
| 8 | −5.72 | 6.40 | 9 |
| pH | T/K | ΔEvdw/ (kJ/mol) | ΔEelec/ (kJ/mol) | ΔEinter/(kJ/mol) | ΔE/ (kJ/mol) | Ebind/ (kJ/mol) | |||
|---|---|---|---|---|---|---|---|---|---|
| ΔEbond | ΔEangle | ΔEdihedral | ΔEplanarity | ||||||
| 7.4 | 298 | 2.14 × 105 | −1.61 × 106 | 1.04 × 105 | 5.69 × 104 | 5.22 × 104 | 397.52 | −1.18 × 106 | −88.85 |
| 333 | 2.33 × 105 | −1.78 × 106 | 1.29 × 105 | 6.97 × 104 | 5.29 × 104 | 440.38 | −1.30 × 106 | −35.70 | |
| 2.0 | 298 | 3.36 × 105 | −2.44 × 106 | 1.59 × 105 | 8.40 × 104 | 5.08 × 104 | 398.18 | −1.81 × 106 | −131.07 |
| 333 | 2.58 × 105 | −1.95 × 106 | 1.42 × 105 | 7.63 × 104 | 5.11 × 104 | 445.60 | −1.42 × 106 | −79.28 | |
| pH | T/K | α-Helix Content/% | β-Sheet Content/% | Turn Content/% | Random Coil Content/% | |
|---|---|---|---|---|---|---|
| β-Casein | 7.4 | 298 | 21.47 | 4.10 | 20.11 | 50.72 |
| 333 | 21.93 | 5.57 | 21.56 | 47.18 | ||
| 2.0 | 298 | 26.27 | 1.63 | 17.75 | 51.89 | |
| 333 | 27.29 | 1.83 | 21.74 | 46.81 | ||
| Complex | 7.4 | 298 | 15.56 | 3.71 | 20.24 | 55.50 |
| 333 | 19.22 | 5.90 | 20.73 | 50.36 | ||
| 2.0 | 298 | 19.69 | 0.84 | 18.64 | 57.16 | |
| 333 | 24.18 | 2.63 | 21.05 | 47.87 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, R.; Qin, X.; Zhong, J. Interaction between Curcumin and β-Casein: Multi-Spectroscopic and Molecular Dynamics Simulation Methods. Molecules 2021, 26, 5092. https://doi.org/10.3390/molecules26165092
Zhao R, Qin X, Zhong J. Interaction between Curcumin and β-Casein: Multi-Spectroscopic and Molecular Dynamics Simulation Methods. Molecules. 2021; 26(16):5092. https://doi.org/10.3390/molecules26165092
Chicago/Turabian StyleZhao, Ruichen, Xiaoli Qin, and Jinfeng Zhong. 2021. "Interaction between Curcumin and β-Casein: Multi-Spectroscopic and Molecular Dynamics Simulation Methods" Molecules 26, no. 16: 5092. https://doi.org/10.3390/molecules26165092
APA StyleZhao, R., Qin, X., & Zhong, J. (2021). Interaction between Curcumin and β-Casein: Multi-Spectroscopic and Molecular Dynamics Simulation Methods. Molecules, 26(16), 5092. https://doi.org/10.3390/molecules26165092








