NMR-Based Metabolomic Analysis on the Protective Effects of Apolipoprotein A-I Mimetic Peptide against Contrast Media-Induced Endothelial Dysfunction
Abstract
:1. Introduction
2. Results
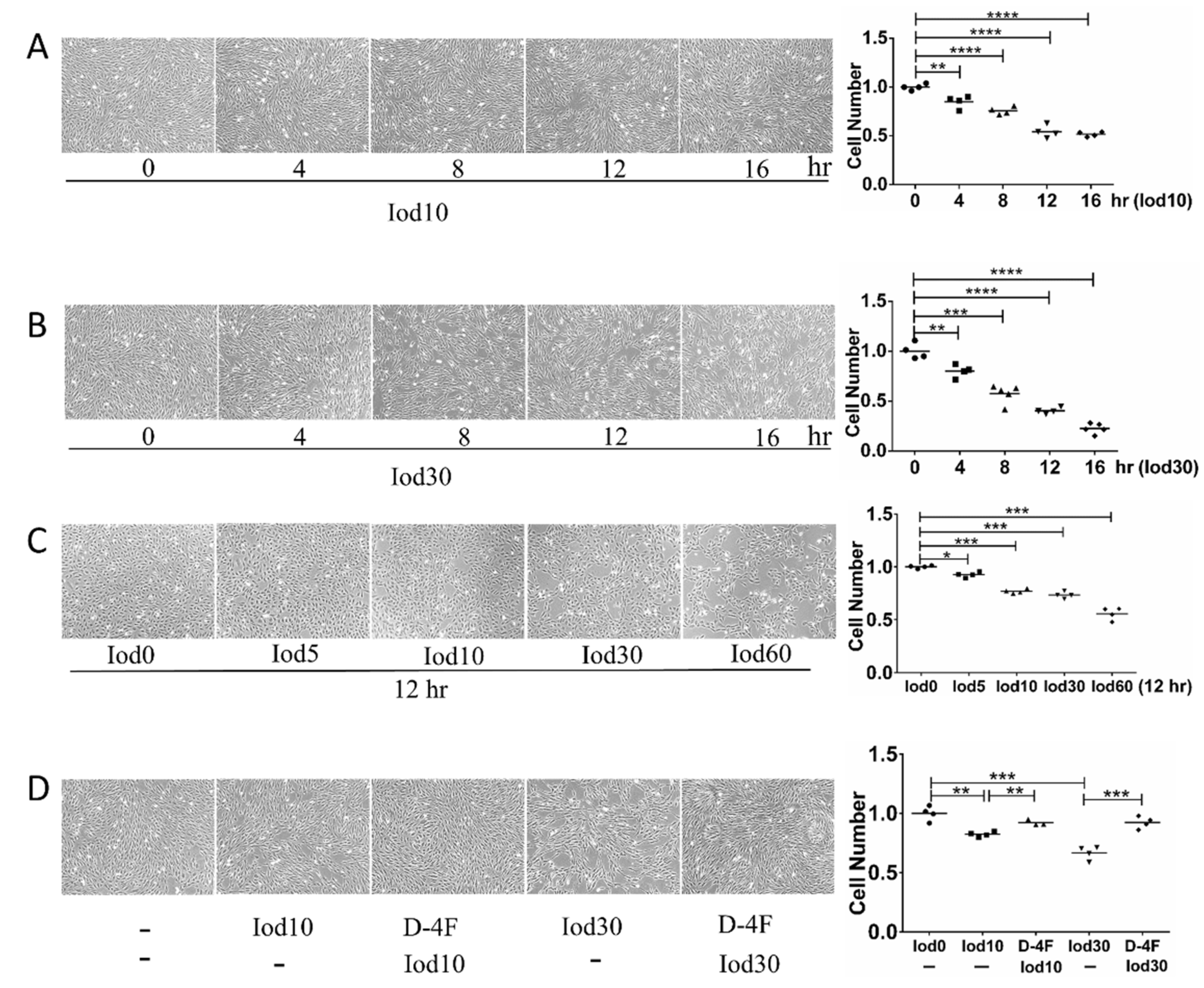
2.1. Protective Effects of D-4F on HUVEC Vitality Impaired by Iodixanol
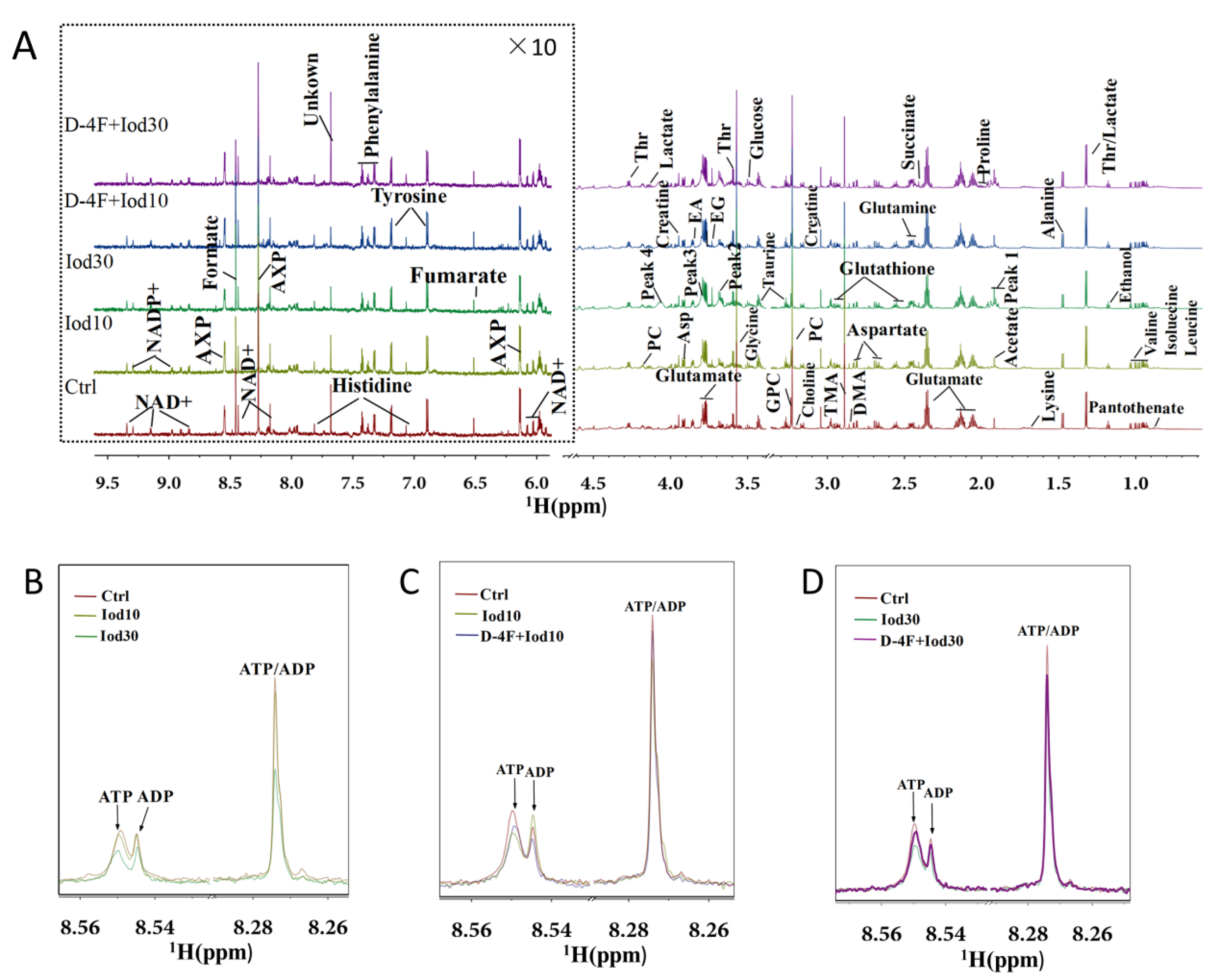
2.2. 1H NMR Spectra of Intracellular Metabolites
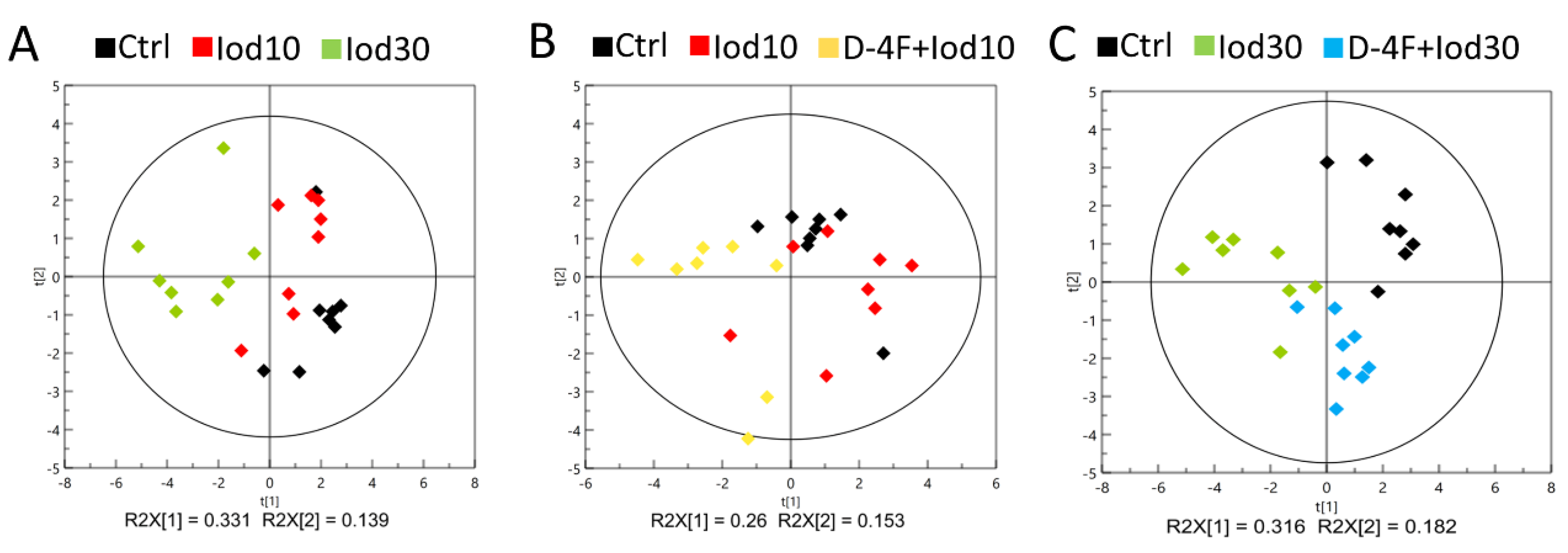
2.3. Metabolic Profiles of HUVECs
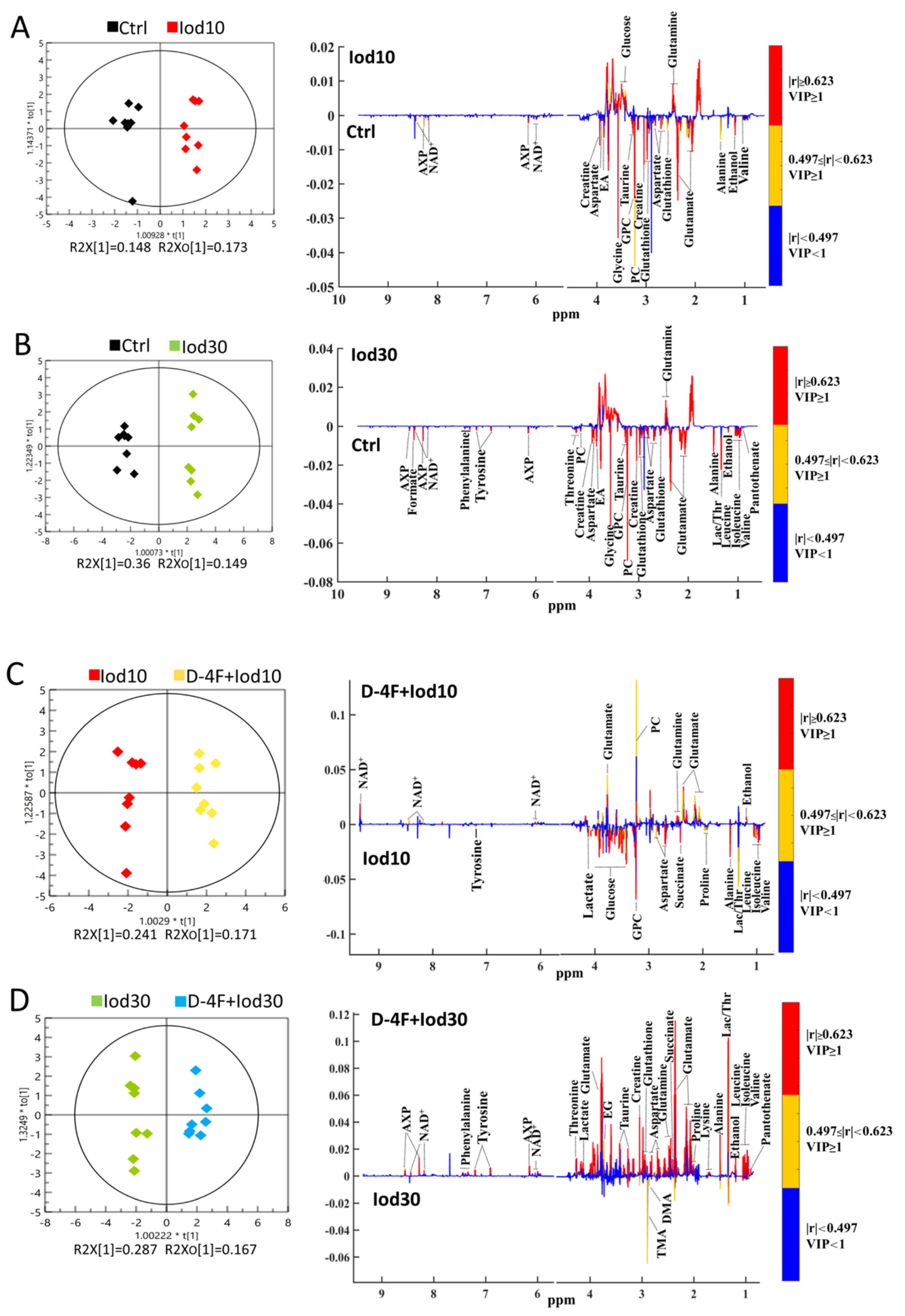
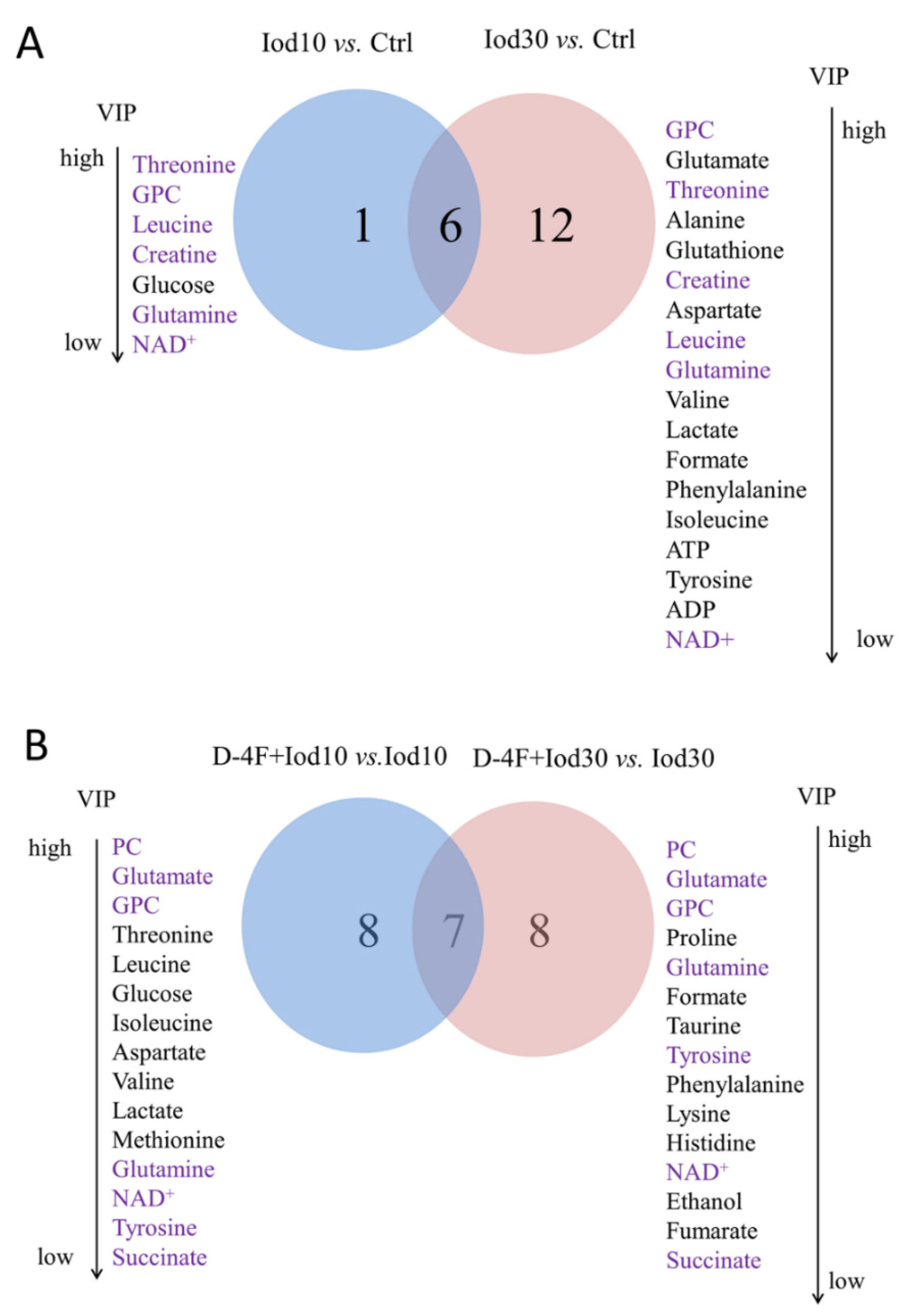
2.4. Significant Metabolites Identified in HUVECs
2.5. Differential Metabolites Identified in HUVECs
2.6. Characteristic Metabolites Identified in HUVECs
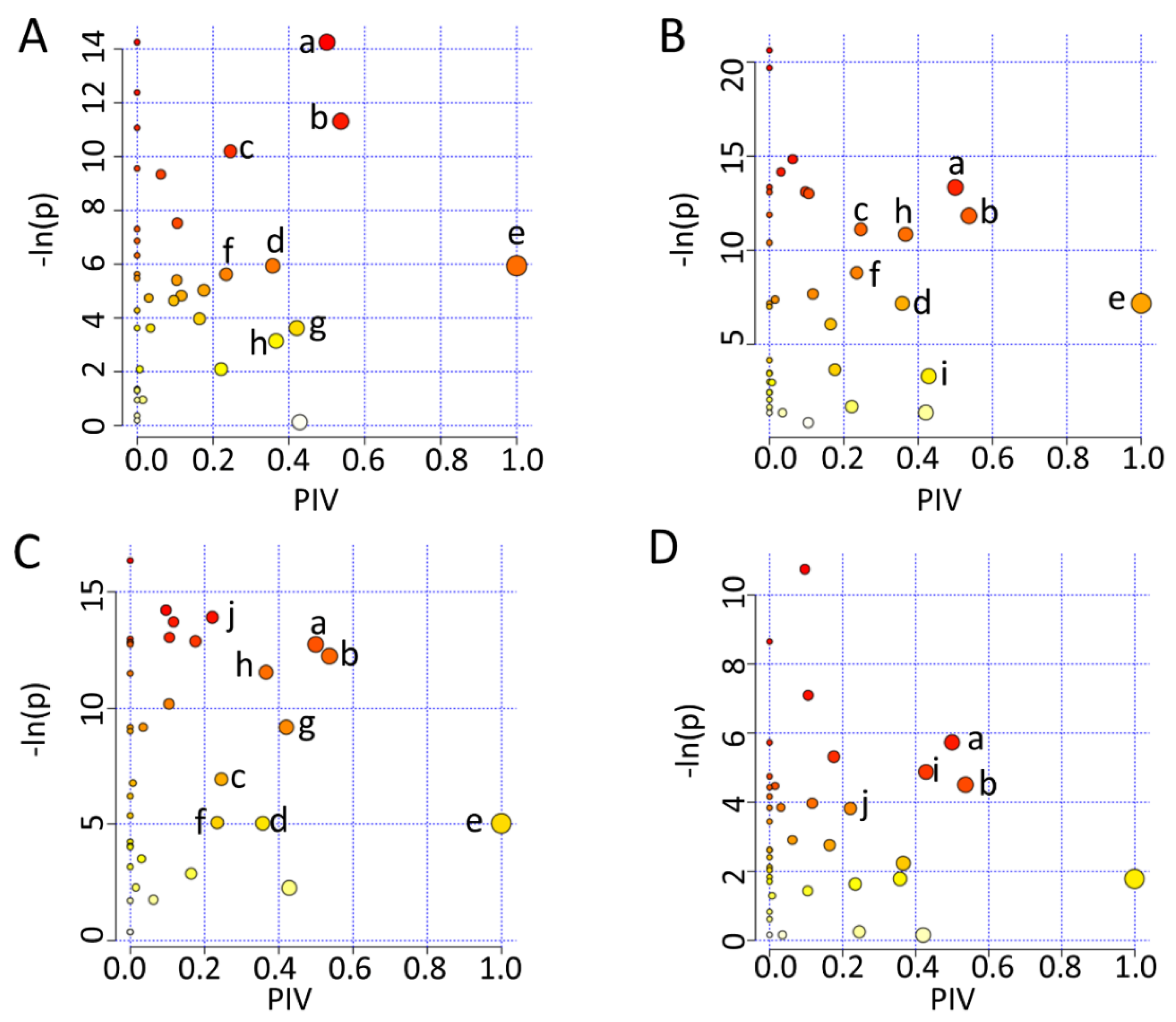
2.7. Significant Altered Metabolic Pathways Identified in HUVECs
3. Discussion
3.1. D-4F Alleviates Iodixanol-Induced Intracellular Oxidative Stress
3.2. D-4F Restores the Energy Production Impaired by Iodixanol
3.3. D-4F Enhances Iodixanol-Inhibited Choline Metabolism
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Metabolite Extraction
4.3. NMR Sample Preparation and NMR Spectrum Acquisition
4.4. NMR Data Preprocessing
4.5. Multivariate Analysis
4.6. Univariate Analysis
4.7. Metabolic Pathway Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
References
- Fähling, M.; Seeliger, E.; Patzak, A.; Persson, P. Understanding and preventing contrast-induced acute kidney injury. Nat. Rev. Nephrol. 2017, 13, 169–180. [Google Scholar] [CrossRef]
- Mamoulakis, C.; Tsarouhas, K.; Fragkiadoulaki, E.; Heretis, I.; Wilks, M.F.; Spandidos, D.; Tsitsimpikou, C.; Tsatsakis, A. Contrast-induced nephropathy: Basic concepts, pathophysiological implications and prevention strategies. Pharmacol. Ther. 2017, 180, 99–112. [Google Scholar] [CrossRef]
- Hoste, E.A.J.; Kellum, J.A.; Selby, N.M.; Zarbock, A.; Palevsky, P.M.; Bagshaw, S.M.; Goldstein, S.L.; Cerdá, J.; Chawla, L.S. Global epidemiology and outcomes of acute kidney injury. Nat. Rev. Nephrol. 2018, 14, 607–625. [Google Scholar] [CrossRef] [PubMed]
- Ramponi, S.; Grotti, A.; Morisetti, A.; Vultaggio, S.; Lorusso, V. Effects of iodinated contrast media on endothelium: An in vitro study. Toxicol. In Vitro 2007, 21, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Li, W.; Qian, M.; Jiang, T.; Guo, P.; Du, Q.; Lin, N.; Xie, X.; Wu, Z.; Lin, D.; et al. D-4F Ameliorates Contrast Media–Induced Oxidative Injuries in Endothelial Cells via the AMPK/PKC Pathway. Front. Pharmacol. 2021, 11, 2226. [Google Scholar] [CrossRef] [PubMed]
- Sudano, I.; Spieker, L.E.; Hermann, F.; Flammer, A.; Corti, R.; Noll, G.; Lüscher, T.F. Protection of endothelial function: Targets for nutritional and pharmacological interventions. J. Cardiovasc. Pharmacol. 2006, 47 (Suppl. 2), S136–S150. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Hao, H.; Leeper, N.J.; Zhu, L. Thrombotic Regulation From the Endothelial Cell Perspectives. Arter. Thromb. Vasc. Biol. 2018, 38, e90–e95. [Google Scholar] [CrossRef] [Green Version]
- Scoditti, E.; Massaro, M.; Montinari, M.R. Endothelial safety of radiological contrast media: Why being concerned. Vasc. Pharmacol. 2013, 58, 48–53. [Google Scholar] [CrossRef]
- Hutcheson, I.R.; Griffith, T.M.; Pitman, M.R.; Towart, R.; Gregersen, M.; Refsum, H.; Karlsson, J.O.G. Iodinated radiographic contrast media inhibit shear stress- and agonist-evoked release of NO by the endothelium. Br. J. Pharmacol. 1999, 128, 451–457. [Google Scholar] [CrossRef] [Green Version]
- Xiang, L.; Xiang, G.; Zhang, J.; Yue, L.; Zhao, L. Contrast agent suppresses endothelium-dependent arterial dilation after digital subtraction angiography procedure in patients with diabetic foot. Endocrine 2013, 46, 505–511. [Google Scholar] [CrossRef]
- Chang, C.-F.; Liu, X.-M.; Peyton, K.J.; Durante, W. Heme oxygenase-1 counteracts contrast media-induced endothelial cell dysfunction. Biochem. Pharmacol. 2014, 87, 303–311. [Google Scholar] [CrossRef] [Green Version]
- Ronda, N.; Potì, F.; Palmisano, A.; Gatti, R.; Orlandini, G.; Maggiore, U.; Cabassi, A.; Regolisti, G.; Fiaccadori, E. Effects of the radiocontrast agent iodixanol on endothelial cell morphology and function. Vasc. Pharmacol. 2013, 58, 39–47. [Google Scholar] [CrossRef]
- Zhao, J.; Huang, Y.; Song, Y.; Zhao, X.; Jin, J.; Wang, J.; Huang, L. Low osmolar contrast medium induces cellular injury and disruption of calcium homeostasis in rat glomerular endothelial cells in vitro. Toxicol. Lett. 2009, 185, 124–131. [Google Scholar] [CrossRef] [PubMed]
- van Capelleveen, J.C.; Brewer, H.B.; Kastelein, J.J.; Hovingh, G.K. Novel therapies focused on the high-density lipoprotein particle. Circ. Res. 2014, 114, 193–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uehara, Y.; Chiesa, G.; Saku, K. High-Density Lipoprotein-Targeted Therapy and Apolipoprotein A-I Mimetic Peptides. Circ. J. 2015, 79, 2523–2528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baotic, I.; Ge, Z.-D.; Sedlic, F.; Coon, A.; Weihrauch, R.; Warltier, D.C.; Kersten, J.R. Apolipoprotein A-1 mimetic D-4F enhances isoflurane-induced eNOS signaling and cardioprotection during acute hyperglycemia. Am. J. Physiol. Circ. Physiol. 2013, 305, H219–H227. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Ding, Z.; Wu, M.; Xu, W.; Qian, M.; Du, Q.; Zhang, L.; Cui, Y.; Zheng, J.; Chang, H.; et al. The apolipoprotein A-I mimetic peptide, D-4F, alleviates ox-LDL-induced oxidative stress and promotes endothelial repair through the eNOS/HO-1 pathway. J. Mol. Cell. Cardiol. 2017, 105, 77–88. [Google Scholar] [CrossRef]
- McGarrah, R.W.; Crown, S.B.; Zhang, G.-F.; Shah, S.H.; Newgard, C.B. Cardiovascular Metabolomics. Circ. Res. 2018, 122, 1238–1258. [Google Scholar] [CrossRef]
- Prieto-Merino, D.; Tillin, T.; Ghorbani, A.; Artati, A.; Wang, Q.; Tiainen, M.; Kangas, A.J.; Kettunen, J.; Kaikkonen, J.; Mikkilä, V.; et al. Metabolite Profiling and Cardiovascular Event Risk Clinical Perspective. Circulation 2015, 131, 774–785. [Google Scholar]
- Xu, W.; Qian, M.; Huang, C.; Cui, P.; Li, W.; Du, Q.; Yi, S.; Shi, X.; Guo, Y.; Zheng, J.; et al. Comparison of Mechanisms of Endothelial Cell Protections Between High-Density Lipoprotein and Apolipoprotein A-I Mimetic Peptide. Front. Pharmacol. 2019, 10, 817. [Google Scholar] [CrossRef] [Green Version]
- McCullough, P.A.; Choi, J.P.; Feghali, G.A.; Schussler, J.M.; Stoler, R.M.; Vallabahn, R.C.; Mehta, A. Contrast-Induced Acute Kidney Injury. J. Am. Coll. Cardiol. 2016, 68, 1465–1473. [Google Scholar] [CrossRef] [Green Version]
- Hwang, S.D.; Kim, Y.J.; Lee, S.H.; Cho, D.K.; Cho, Y.H.; Moon, S.J.; Yoon, S.Y.; Lee, S.C. Iodinated Contrast Media Can Induce Long-Lasting Oxidative Stress in Hemodialysis Patients. Yonsei Med. J. 2013, 54, 1438–1446. [Google Scholar] [CrossRef]
- Heyman, S.N.; Rosen, S.; Khamaisi, M.; Idée, J.-M.; Rosenberger, C. Reactive Oxygen Species and the Pathogenesis of Radiocontrast-Induced Nephropathy. Investig. Radiol. 2010, 45, 188–195. [Google Scholar] [CrossRef]
- Wu, G.; Fang, Y.-Z.; Yang, S.; Lupton, J.R.; Turner, N.D. Glutathione Metabolism and Its Implications for Health. J. Nutr. 2004, 134, 489–492. [Google Scholar] [CrossRef] [Green Version]
- Dan Dunn, J.; Alvarez, L.A.; Zhang, X.; Soldati, T. Reactive oxygen species and mitochondria: A nexus of cellular homeostasis. Redox Biol. 2015, 6, 472–485. [Google Scholar] [CrossRef]
- Unterluggauer, H.; Mazurek, S.; Lener, B.; Hütter, E.; Eigenbrodt, E.; Zwerschke, W.; Jansen-Dürr, P. Premature senescence of human endothelial cells induced by inhibition of glutaminase. Biogerontology 2008, 9, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.L.; Zhang, R.; Anand, P.; Stomberski, C.T.; Qian, Z.; Hausladen, A.; Wang, L.; Rhee, E.P.; Parikh, S.M.; Karumanchi, S.A.; et al. Metabolic reprogramming by the S-nitroso-CoA reductase system protects against kidney injury. Nature 2019, 565, 96–100, Erratum in 2019, 570, E23. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Wang, K.; Zheng, N.; Qiu, Y.; Xie, G.; Su, M.; Jia, W.; Li, H. Metformin suppressed the proliferation of LoVo cells and induced a time-dependent metabolic and transcriptional alteration. Sci. Rep. 2015, 5, 17423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, W.; Wang, R.-S.; Handy, D.E.; Loscalzo, J. NAD(H) and NADP(H) Redox Couples and Cellular Energy Metabolism. Antioxid. Redox Signal. 2018, 28, 251–272. [Google Scholar] [CrossRef] [PubMed]
- Hizoh, I.; Haller, C. Radiocontrast-induced renal tubular cell apoptosis: Hypertonic versus oxidative stress. Investig. Radiol. 2002, 37, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Paik, J.-Y.; Jung, K.-H.; Lee, J.-H.; Park, J.-W.; Lee, K.-H. Reactive oxygen species-driven HIF1α triggers accelerated glycolysis in endothelial cells exposed to low oxygen tension. Nucl. Med. Biol. 2017, 45, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Chaneton, B.; Hillmann, P.; Zheng, L.; Martin, A.C.L.; Maddocks, O.; Chokkathukalam, A.; Coyle, J.E.; Jankevics, A.; Holding, F.P.; Vousden, K.H.; et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nat. Cell Biol. 2012, 491, 458–462. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Mancuso, A.; Tong, X.; Ward, P.; Fan, J.; Rabinowitz, J.D.; Thompson, C.B. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc. Natl. Acad. Sci. USA 2012, 109, 6904–6909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [Green Version]
- De Bock, K.; Georgiadou, M.; Schoors, S.; Kuchnio, A.; Wong, B.W.; Cantelmo, A.R.; Annelies, Q.; Ghesquière, B.; Cauwenberghs, S.; Eelen, G.; et al. Role of PFKFB3-Driven Glycolysis in Vessel Sprouting. Cell 2013, 154, 651–663. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Holt, C.M.; Malik, N.; Shepherd, L.; Morcos, S.K. Effects of radiographic contrast media on proliferation and apoptosis of human vascular endothelial cells. Br. J. Radiol. 2000, 73, 1034–1041. [Google Scholar] [CrossRef]
- Zhang, Z.; Qun, J.; Cao, C.; Wang, J.; Li, W.; Wu, Y.; Du, L.; Zhao, P.; Gong, K. Apolipoprotein A-I mimetic peptide D-4F promotes human endothelial progenitor cell proliferation, migration, adhesion though eNOS/NO pathway. Mol. Biol. Rep. 2011, 39, 4445–4454. [Google Scholar] [CrossRef]
- Eguchi, Y.; Shimizu, S.; Tsujimoto, Y. Intracellular ATP levels determine cell death fate by apoptosis or necrosis. Cancer Res. 1997, 57, 1835–1840. [Google Scholar]
- Leist, M.; Single, B.; Castoldi, A.F.; Kühnle, S.; Nicotera, P. Intracellular Adenosine Triphosphate (ATP) Concentration: A Switch in the Decision Between Apoptosis and Necrosis. J. Exp. Med. 1997, 185, 1481–1486. [Google Scholar] [CrossRef]
- Belenky, P.; Bogan, K.L.; Brenner, C. NAD+ metabolism in health and disease. Trends Biochem. Sci. 2007, 32, 12–19. [Google Scholar] [CrossRef]
- Wang, S.; Xu, T.; Li, W.; Miao, C. Targeting Nicotinamide Phosphoribosyltransferase as a Potential Therapeutic Strategy to Restore Adult Neurogenesis. CNS Neurosci. Ther. 2016, 22, 431–439. [Google Scholar] [CrossRef] [Green Version]
- Mehr, A.P.; Tran, M.; Ralto, K.M.; Leaf, D.; Washco, V.; Messmer, J.; Lerner, A.; Kher, A.; Kim, S.H.; Khoury, C.C.; et al. De novo NAD+ biosynthetic impairment in acute kidney injury in humans. Nat. Med. 2018, 24, 1351–1359. [Google Scholar] [CrossRef]
- Zhang, H.; Ryu, D.; Wu, Y.; Gariani, K.; Wang, X.; Luan, P.; D’Amico, D.; Ropelle, E.R.; Lutolf, M.P.; Aebersold, R.; et al. NAD+ repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 2016, 352, 1436–1443. [Google Scholar] [CrossRef] [Green Version]
- Mehta, R.L.; Cerdá, J.; Burdmann, E.A.; Tonelli, M.; García-García, G.; Jha, V.; Susantitaphong, P.; Rocco, M.; Vanholder, R.; Sever, M.S.; et al. International Society of Nephrology’s 0by25 initiative for acute kidney injury (zero preventable deaths by 2025): A human rights case for nephrology. Lancet 2015, 385, 2616–2643. [Google Scholar] [CrossRef]
- Braidy, N.; Guillemin, G.J.; Mansour, H.; Chan-Ling, T.; Poljak, A.; Grant, R. Age Related Changes in NAD plus Metabolism Oxidative Stress and Sirt1 Activity in Wistar Rats. PLoS ONE 2011, 6, e19194. [Google Scholar] [CrossRef]
- Franke, R.-P.; Fuhrmann, R.; Mrowietz, C.; Rickert, D.; Hiebl, B.; Jung, F. Reduced diagnostic value of lactate dehydrogenase (LDH) in the presence of radiographic contrast media. Clin. Hemorheol. Microcirc. 2010, 45, 123–130. [Google Scholar] [CrossRef]
- Vance, J.E.; Vance, D.E. Phospholipid biosynthesis in mammalian cells. Biochem. Cell Biol. 2004, 82, 113–128. [Google Scholar] [CrossRef]
- Liu, D.; Ji, L.; Tong, X.; Pan, B.; Han, J.Y.; Huang, Y.; Zheng, L. Human apolipoprotein A-I induces cyclooxygenase-2 expression and prostaglandin I-2 release in endothelial cells through ATP-binding cassette transporter A1. Am. J. Physiol. Cell Physiol. 2011, 301, C739–C748. [Google Scholar] [CrossRef] [Green Version]
- Viant, M.R. Revealing the metabolome of animal tissues using 1H nuclear magnetic resonance spectroscopy. Methods Mol. Biol. 2007, 358, 229–246. [Google Scholar]
- Shao, W.; Gu, J.; Huang, C.; Liu, D.; Huang, H.; Huang, Z.; Lin, Z.; Yang, W.; Liu, K.; Lin, D.; et al. Malignancy-associated metabolic profiling of human glioma cell lines using 1H NMR spectroscopy. Mol. Cancer 2014, 13, 197. [Google Scholar] [CrossRef] [Green Version]
| Metabolite | Mean ± Standard Error | Student’s t Test | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Ctrl | Iod10 | D-4F + Iod10 | Iod30 | D-4F + Iod30 | Iod10 | D-4F + Iod10 | Iod30 | D-4F + Iod30 | |
| vs. Ctrl | vs. Iod10 | vs. Ctrl | vs. Iod30 | ||||||
| Amino acid metabolism | |||||||||
| Pantothenate | 0.13 ± 0.01 | 0.13 ± 0.01 | 0.13 ± 0.01 | 0.11 ± 0.00 | 0.13 ± 0.01 | ns | ns | ns | ns |
| Leucine | 1.35 ± 0.02 | 1.43 ± 0.02 | 1.27 ± 0.03 | 1.26 ± 0.00 | 1.28 ± 0.02 | * | *** | *** | ns |
| Isoleucine | 0.60 ± 0.01 | 0.62 ± 0.01 | 0.55 ± 0.02 | 0.55 ± 0.00 | 0.56 ± 0.01 | * | *** | *** | ns |
| Valine | 0.56 ± 0.01 | 0.57 ± 0.00 | 0.52 ± 0.01 | 0.51 ± 0.00 | 0.51 ± 0.01 | ns | *** | *** | ns |
| Threonine | 4.29 ± 0.07 | 4.76 ± 0.07 | 4.41 ± 0.07 | 4.63 ± 0.11 | 4.63 ± 0.08 | *** | ** | * | ns |
| Aspartate | 0.85 ± 0.01 | 0.86 ± 0.02 | 0.79 ± 0.02 | 0.76 ± 0.02 | 0.77 ± 0.01 | ns | * | *** | ns |
| Alanine | 1.57 ± 0.04 | 1.53 ± 0.07 | 1.43 ± 0.02 | 1.38 ± 0.01 | 1.34 ± 0.04 | ns | ns | *** | ns |
| Lysine | 0.14 ± 0.00 | 0.14 ± 0.01 | 0.15 ± 0.01 | 0.14 ± 0.00 | 0.15 ± 0.00 | ns | ns | ns | * |
| Proline | 0.58 ± 0.01 | 0.58 ± 0.01 | 0.62 ± 0.01 | 0.55 ± 0.01 | 0.60 ± 0.01 | ns | ** | ** | **** |
| Phenylalanine | 0.32 ± 0.00 | 0.33 ± 0.01 | 0.32 ± 0.01 | 0.29 ± 0.00 | 0.30 ± 0.01 | ns | * | *** | * |
| Tryosine | 0.36 ± 0.00 | 0.37 ± 0.00 | 0.36 ± 0.00 | 0.33 ± 0.00 | 0.35 ± 0.01 | ** | * | *** | * |
| Fumarate | 0.04 ± 0.00 | 0.04 ± 0.00 | 0.04 ± 0.00 | 0.03 ± 0.00 | 0.03 ± 0.00 | ns | ns | ** | * |
| Succinate | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.02 ± 0.00 | 0.02 ± 0.00 | *** | ** | *** | * |
| Formate | 0.20 ± 0.04 | 0.15 ± 0.02 | 0.11 ± 0.01 | 0.12 ± 0.01 | 0.09 ± 0.01 | ns | ns | ns | * |
| Histidine | 0.07 ± 0.00 | 0.06 ± 0.00 | 0.07 ± 0.00 | 0.07 ± 0.00 | 0.06 ± 0.00 | ns | ns | ns | * |
| Taurine | 0.36 ± 0.01 | 0.36 ± 0.01 | 0.33 ± 0.01 | 0.37 ± 0.01 | 0.33 ± 0.01 | ns | ns | ns | ** |
| Creatine | 0.96 ± 0.01 | 0.89 ± 0.02 | 0.94 ± 0.02 | 0.84 ± 0.01 | 0.86 ± 0.01 | ** | ns | *** | ns |
| Glutathione metabolism | |||||||||
| Glutathione | 1.81 ± 0.02 | 1.75 ± 0.06 | 1.78 ± 0.03 | 1.63 ± 0.03 | 1.64 ± 0.04 | ns | ns | *** | ns |
| Glutamate | 8.22 ± 0.04 | 7.89 ± 0.10 | 8.69 ± 0.08 | 7.66 ± 0.08 | 8.08 ± 0.04 | ** | **** | *** | *** |
| Glutamine | 0.38 ± 0.01 | 0.43 ± 0.01 | 0.47 ± 0.01 | 0.45 ± 0.01 | 0.49 ± 0.01 | *** | ** | *** | ** |
| Methionine | 0.39 ± 0.01 | 0.43 ± 0.01 | 0.38 ± 0.00 | 0.36 ± 0.01 | 0.36 ± 0.01 | ** | **** | * | ns |
| Glycine | 0.10 ± 0.00 | 0.11 ± 0.00 | 0.11 ± 0.00 | 0.12 ± 0.00 | 0.12 ± 0.00 | ns | ns | ** | ns |
| Glucose metabolism | |||||||||
| Glucose | 0.31 ± 0.03 | 0.39 ± 0.02 | 0.23 ± 0.02 | 0.34 ± 0.03 | 0.35 ± 0.02 | * | **** | ns | ns |
| Lactate | 0.36 ± 0.01 | 0.38 ± 0.01 | 0.32 ± 0.02 | 0.42 ± 0.02 | 0.40 ± 0.01 | ns | * | * | ns |
| Glycerophospholipid metabolism | |||||||||
| Choline | 0.13 ± 0.01 | 0.14 ± 0.01 | 0.14 ± 0.01 | 0.14 ± 0.01 | 0.14 ± 0.01 | ns | ns | ns | ns |
| PC | 5.19 ± 0.14 | 5.06 ± 0.05 | 6.06 ± 0.16 | 4.97 ± 0.11 | 5.50 ± 0.08 | ns | **** | ns | ** |
| GPC | 1.45 ± 0.06 | 1.19 ± 0.04 | 0.87 ± 0.03 | 0.92 ± 0.02 | 0.76 ± 0.02 | ** | **** | *** | **** |
| EA | 0.08 ± 0.00 | 0.08 ± 0.00 | 0.08 ± 0.00 | 0.08 ± 0.00 | 0.09 ± 0.00 | ns | ns | ns | * |
| Others | |||||||||
| ATP | 0.30 ± 0.01 | 0.30 ± 0.01 | 0.28 ± 0.02 | 0.25 ± 0.01 | 0.27 ± 0.01 | ns | ns | * | ns |
| ADP | 0.17 ± 0.01 | 0.16 ± 0.00 | 0.16 ± 0.01 | 0.15 ± 0.01 | 0.16 ± 0.00 | ns | ns | ** | ns |
| NAD+ | 0.09 ± 0.00 | 0.09 ± 0.00 | 0.10 ± 0.00 | 0.08 ± 0.00 | 0.09 ± 0.00 | *** | *** | *** | ** |
| NADP+ | 0.07 ± 0.00 | 0.07 ± 0.00 | 0.07 ± 0.00 | 0.06 ± 0.00 | 0.06 ± 0.00 | ns | ns | *** | ns |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, T.; Du, Q.; Huang, C.; Xu, W.; Guo, P.; Li, W.; Xie, X.; Guo, Y.; Liu, D.; Lin, D. NMR-Based Metabolomic Analysis on the Protective Effects of Apolipoprotein A-I Mimetic Peptide against Contrast Media-Induced Endothelial Dysfunction. Molecules 2021, 26, 5123. https://doi.org/10.3390/molecules26175123
Jiang T, Du Q, Huang C, Xu W, Guo P, Li W, Xie X, Guo Y, Liu D, Lin D. NMR-Based Metabolomic Analysis on the Protective Effects of Apolipoprotein A-I Mimetic Peptide against Contrast Media-Induced Endothelial Dysfunction. Molecules. 2021; 26(17):5123. https://doi.org/10.3390/molecules26175123
Chicago/Turabian StyleJiang, Ting, Qian Du, Caihua Huang, Wenqi Xu, Ping Guo, Wei Li, Xianwei Xie, Yansong Guo, Donghui Liu, and Donghai Lin. 2021. "NMR-Based Metabolomic Analysis on the Protective Effects of Apolipoprotein A-I Mimetic Peptide against Contrast Media-Induced Endothelial Dysfunction" Molecules 26, no. 17: 5123. https://doi.org/10.3390/molecules26175123







