Positive Charges in the Brace Region Facilitate the Membrane Disruption of MLKL-NTR in Necroptosis
Abstract
:1. Introduction
2. Results
2.1. MLKL2–154 Reconstituted into Bicelles
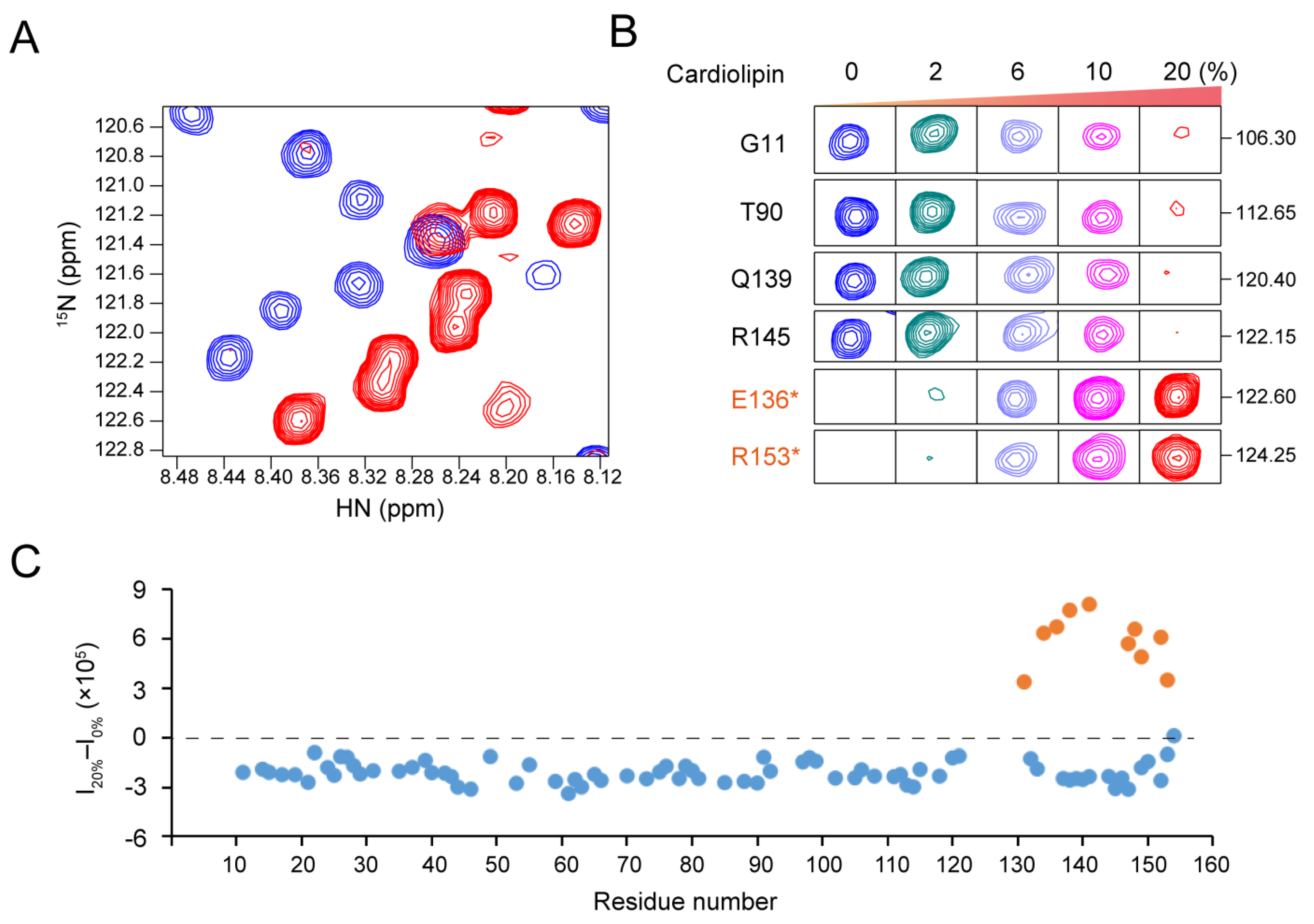
2.2. Negatively Charged Phospholipids Induced the Release of the Brace Region
2.3. The Intrinsic Disordered Properties of H6 Helix
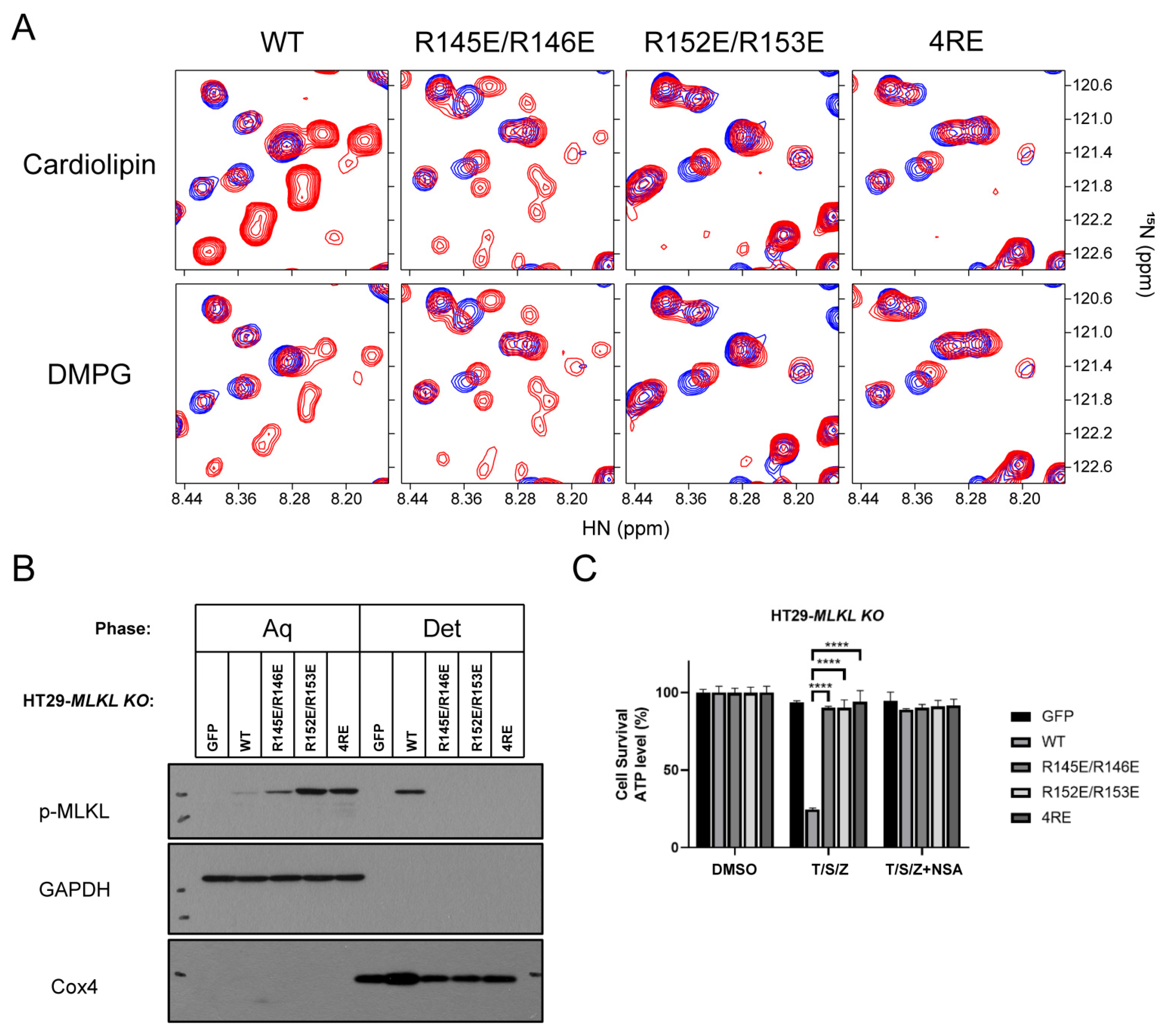
2.4. Polybasic Residues in the C-Terminus of the Brace Region Is Essential for the Release
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Reconstitution of MLKL2–154 into Bicelles
4.3. Titration of Negatively Charged Phospholipids
4.4. NMR Resonance Assignment and Secondary Structure Prediction
4.5. Solvent PRE Analysis
4.6. Lipophilic PRE Analysis
4.7. Molecular Dynamics Simulation
4.8. Triton X-114 Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
References
- Dionísio, P.A.; Amaral, J.D.; Rodrigues, C.M.P. Molecular mechanisms of necroptosis and relevance for neurodegenerative diseases. Int. Rev. Cell. Mol. Biol. 2020, 353, 31–82. [Google Scholar] [CrossRef]
- Sun, L.; Wang, X. A new kind of cell suicide: Mechanisms and functions of programmed necrosis. Trends Biochem. Sci. 2014, 39, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, H.; Wang, Z.; He, S.; Chen, S.; Liao, D.; Wang, L.; Yan, J.; Liu, W.; Lei, X.; et al. Mixed Lineage Kinase Domain-like Protein Mediates Necrosis Signaling Downstream of RIP3 Kinase. Cell 2012, 148, 213–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, J.M.; Czabotar, P.E.; Hildebrand, J.M.; Lucet, I.S.; Zhang, J.G.; Alvarez-Diaz, S.; Lewis, R.; Lalaoui, N.; Metcalf, D.; Webb, A.I.; et al. The pseudokinase mlkl mediates necroptosis via a molecular switch mechanism. Immunity 2013, 39, 443–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Sun, L.; Su, L.; Rizo, J.; Liu, L.; Wang, L.F.; Wang, F.-S.; Wang, X. Mixed Lineage Kinase Domain-like Protein MLKL Causes Necrotic Membrane Disruption upon Phosphorylation by RIP. Mol. Cell 2014, 54, 133–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samson, A.L.; Zhang, Y.; Geoghegan, N.D.; Gavin, X.J.; Davies, K.A.; Mlodzianoski, M.J.; Whitehead, L.W.; Frank, D.; Garnish, S.E.; FitzGibbon, C.; et al. MLKL trafficking and accumulation at the plasma membrane control the kinetics and threshold for necroptosis. Nat. Commun. 2020, 11, 3151. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Tong, A.; Zhang, Q.; Wei, Y.; Wei, X. The molecular mechanisms of MLKL-dependent and MLKL-independent necrosis. J. Mol. Cell Biol. 2020, 13, 3–14. [Google Scholar] [CrossRef]
- Cai, Z.; Liu, Z.-G. Detection of MLKL Oligomerization During Programmed Necrosis. Methods Mol. Biol. 2018, 1857, 85–92. [Google Scholar] [CrossRef]
- Cai, Z.; Jitkaew, S.; Zhao, J.; Chiang, H.-C.; Choksi, S.; Liu, J.; Ward, Y.; Wu, L.-G.; Liu, Z.-G. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat. Cell Biol. 2014, 16, 55–65. [Google Scholar] [CrossRef]
- Petrie, E.J.; Sandow, J.J.; Jacobsen, A.V.; Smith, B.J.; Griffin, M.D.W.; Lucet, I.S.; Dai, W.; Young, S.N.; Tanzer, M.C.; Wardak, A.; et al. Conformational switching of the pseudokinase domain promotes human mlkl tetramerization and cell death by necroptosis. Nat. Commun. 2018, 9, 2422. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.; Zheng, X.; Wang, Z.-A.; Chen, X.; He, W.-T.; Zhang, Y.; Xu, J.-G.; Zhao, H.; Shi, W.; Wang, X.; et al. The MLKL Channel in Necroptosis Is an Octamer Formed by Tetramers in a Dyadic Process. Mol. Cell. Biol. 2017, 37, 37. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Liu, H.; Johnston, A.; Hanna-Addams, S.; Reynoso, E.; Xiang, Y.; Wang, Z. MLKL forms disulfide bond-dependent amyloid-like polymers to induce necroptosis. Proc. Natl. Acad. Sci. USA 2017, 114, E7450–E7459. [Google Scholar] [CrossRef] [Green Version]
- Mahdi, L.K.; Huang, M.; Zhang, X.; Nakano, R.T.; Kopp, L.B.; Saur, I.M.; Jacob, F.; Kovacova, V.; Lapin, D.; Parker, J.E.; et al. Discovery of a Family of Mixed Lineage Kinase Domain-like Proteins in Plants and Their Role in Innate Immune Signaling. Cell Host Microbe 2020, 28, 813–824.e8. [Google Scholar] [CrossRef]
- Dondelinger, Y.; Declercq, W.; Montessuit, S.; Roelandt, R.; Goncalves, A.; Bruggeman, I.; Hulpiau, P.; Weber, K.; Sehon, C.A.; Marquis, R.W.; et al. Mlkl compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Rep. 2014, 7, 971–981. [Google Scholar] [CrossRef] [Green Version]
- McNamara, D.E.; Dovey, C.M.; Hale, A.T.; Quarato, G.; Grace, C.R.; Guibao, C.D.; Diep, J.; Nourse, A.; Cai, C.R.; Wu, H.; et al. Direct Activation of Human MLKL by a Select Repertoire of Inositol Phosphate Metabolites. Cell Chem. Biol. 2019, 26, 863–877. [Google Scholar] [CrossRef] [PubMed]
- Dovey, C.M.; Diep, J.; Clarke, B.P.; Hale, A.T.; McNamara, D.E.; Guo, H.; Brown, N.W., Jr.; Cao, J.Y.; Grace, C.R.; Gough, P.J.; et al. MLKL Requires the Inositol Phosphate Code to Execute Necroptosis. Mol. Cell 2018, 70, 936–948.e7. [Google Scholar] [CrossRef] [Green Version]
- Hildebrand, J.M.; Tanzer, M.C.; Lucet, I.S.; Young, S.N.; Spall, S.K.; Sharma, P.; Pierotti, C.; Garnier, J.-M.; Dobson, R.C.; Webb, A.I.; et al. Activation of the pseudokinase MLKL unleashes the four-helix bundle domain to induce membrane localization and necroptotic cell death. Proc. Natl. Acad. Sci. USA 2014, 111, 15072–15077. [Google Scholar] [CrossRef] [Green Version]
- Tanzer, M.C.; Matti, I.; Hildebrand, J.M.; Young, S.N.; Wardak, A.; Tripaydonis, A.; Petrie, E.J.; Mildenhall, A.L.; Vaux, D.L.; Vince, J.E.; et al. Evolutionary divergence of the necroptosis effector MLKL. Cell Death Differ. 2016, 23, 1185–1197. [Google Scholar] [CrossRef] [Green Version]
- Petrie, E.J.; Birkinshaw, R.W.; Koide, A.; Denbaum, E.; Hildebrand, J.M.; Garnish, S.E.; Davies, K.A.; Sandow, J.J.; Samson, A.L.; Gavin, X.; et al. Identification of mlkl membrane translocation as a checkpoint in necroptotic cell death using monobodies. Proc. Natl. Acad. Sci. USA 2020, 117, 8468–8475. [Google Scholar] [CrossRef]
- Su, L.; Quade, B.; Wang, H.; Sun, L.; Wang, X.; Rizo, J. A Plug Release Mechanism for Membrane Permeation by MLKL. Structure 2014, 22, 1489–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnez, K.H.; Kindlova, M.; Bokil, N.J.; Murphy, J.M.; Sweet, M.J.; Guncar, G. Analysis of the n-terminal region of human mlkl, as well as two distinct mlkl isoforms, reveals new insights into necroptotic cell death. Biosci. Rep. 2015, 36, e00291. [Google Scholar] [CrossRef] [Green Version]
- Murphy, J.M.; Lucet, I.S.; Hildebrand, J.M.; Tanzer, M.C.; Young, S.N.; Sharma, P.; Lessene, G.; Alexander, W.S.; Babon, J.J.; Silke, J.; et al. Insights into the evolution of divergent nucleotide-binding mechanisms among pseudokinases revealed by crystal structures of human and mouse MLKL. Biochem. J. 2014, 457, 369–377. [Google Scholar] [CrossRef]
- Davies, K.A.; Tanzer, M.C.; Griffin, M.D.W.; Mok, Y.F.; Young, S.N.; Qin, R.; Petrie, E.J.; Czabotar, P.E.; Silke, J.; Murphy, J.M. The brace helices of mlkl mediate interdomain communication and oligomerisation to regulate cell death by necroptosis. Cell Death Differ. 2018, 25, 1567–1580. [Google Scholar] [CrossRef]
- Rübbelke, M.; Fiegen, D.; Bauer, M.; Binder, F.; Hamilton, J.; King, J.; Thamm, S.; Nar, H.; Zeeb, M. Locking mixed-lineage kinase domain-like protein in its auto-inhibited state prevents necroptosis. Proc. Natl. Acad. Sci. USA 2020, 117, 33272–33281. [Google Scholar] [CrossRef]
- Quarato, G.; Guy, C.S.; Grace, C.R.; Llambi, F.; Nourse, A.; Rodriguez, D.A.; Wakefield, R.; Frase, S.; Moldoveanu, T.; Green, D.R. Sequential engagement of distinct mlkl phosphatidylinositol-binding sites executes necroptosis. Mol. Cell 2016, 61, 589–601. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Zhou, Z.; Li, L.; Zhong, C.Q.; Zheng, X.; Wu, X.; Zhang, Y.; Ma, H.; Huang, D.; Li, W.; et al. Diverse sequence determinants control human and mouse receptor interacting protein 3 (rip3) and mixed lineage kinase domain-like (mlkl) interaction in necroptotic signaling. J. Biol. Chem. 2013, 288, 16247–16261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Li, W.; Ren, J.; Huang, D.; He, W.T.; Song, Y.; Yang, C.; Li, W.; Zheng, X.; Chen, P.; et al. Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res. 2014, 24, 105–121. [Google Scholar] [CrossRef]
- Bansal, N.; Sciabola, S.; Bhisetti, G. Understanding allosteric interactions in hMLKL protein that modulate necroptosis and its inhibition. Sci. Rep. 2019, 9, 16853. [Google Scholar] [CrossRef]
- Ma, Y.; Poole, K.; Goyette, J.; Gaus, K. Introducing membrane charge and membrane potential to t cell signaling. Front. Immunol. 2017, 8, 1513. [Google Scholar] [CrossRef] [PubMed]
- Guibao, C.D.; Petrinjak, K.; Moldoveanu, T. Uncovering human mixed lineage kinase domain-like activation in necroptosis. Future Med. Chem. 2019, 11, 2831–2844. [Google Scholar] [CrossRef] [PubMed]
- McNamara, D.E.; Quarato, G.; Guy, C.S.; Green, D.R.; Moldoveanu, T. Characterization of MLKL-mediated Plasma Membrane Rupture in Necroptosis. J. Vis. Exp. 2018, e58088. [Google Scholar] [CrossRef] [Green Version]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985, 31, 1695–1697. [Google Scholar] [CrossRef] [Green Version]
- Deng, Z.; Martyna, G.J.; Klein, M.L. Structure and dynamics of bipolarons in liquid ammonia. Phys. Rev. Lett. 1992, 68, 2496–2499. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Xie, E.; Du, L.; Yang, Y.; Wu, B.; Sun, L.; Wang, S.; OuYang, B. Positive Charges in the Brace Region Facilitate the Membrane Disruption of MLKL-NTR in Necroptosis. Molecules 2021, 26, 5194. https://doi.org/10.3390/molecules26175194
Yang Y, Xie E, Du L, Yang Y, Wu B, Sun L, Wang S, OuYang B. Positive Charges in the Brace Region Facilitate the Membrane Disruption of MLKL-NTR in Necroptosis. Molecules. 2021; 26(17):5194. https://doi.org/10.3390/molecules26175194
Chicago/Turabian StyleYang, Yaqing, Encheng Xie, Lingyu Du, Yu Yang, Bin Wu, Liming Sun, Shuqing Wang, and Bo OuYang. 2021. "Positive Charges in the Brace Region Facilitate the Membrane Disruption of MLKL-NTR in Necroptosis" Molecules 26, no. 17: 5194. https://doi.org/10.3390/molecules26175194
APA StyleYang, Y., Xie, E., Du, L., Yang, Y., Wu, B., Sun, L., Wang, S., & OuYang, B. (2021). Positive Charges in the Brace Region Facilitate the Membrane Disruption of MLKL-NTR in Necroptosis. Molecules, 26(17), 5194. https://doi.org/10.3390/molecules26175194





