Structural Insights in Mammalian Sialyltransferases and Fucosyltransferases: We Have Come a Long Way, but It Is Still a Long Way Down
Abstract
:1. Introduction
2. ST and FUT Families
2.1. Twenty STs Are Arranged into Four Families
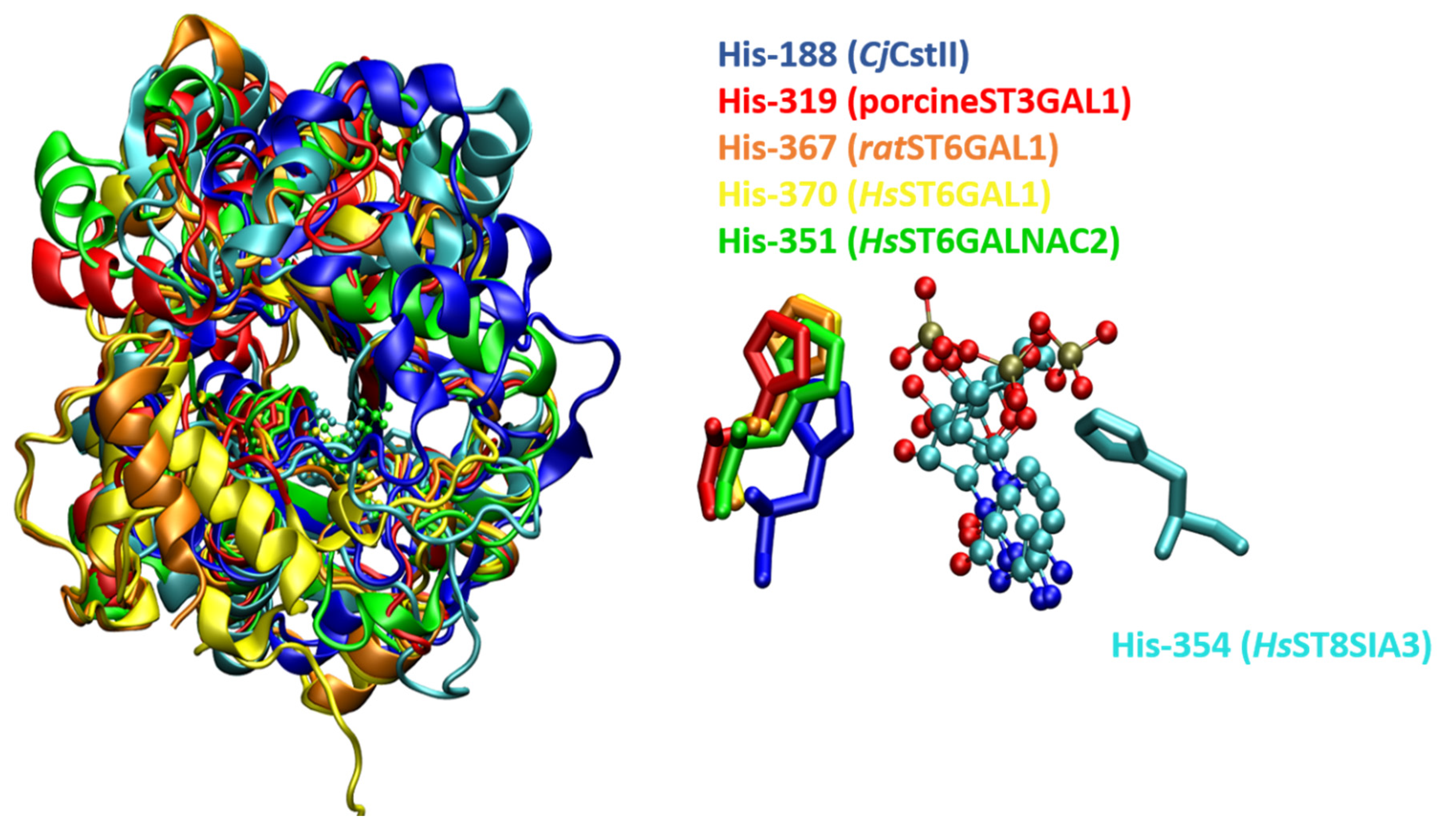
2.2. Sequence Analysis and Conserved Patterns in STs
2.3. Thirteen FUTs Are Organized into Four Families
2.4. Sequence Analysis and Conserved Patterns in FUTs
3. Cellular Localization of STs and FUTs
4. Global Fold Architecture
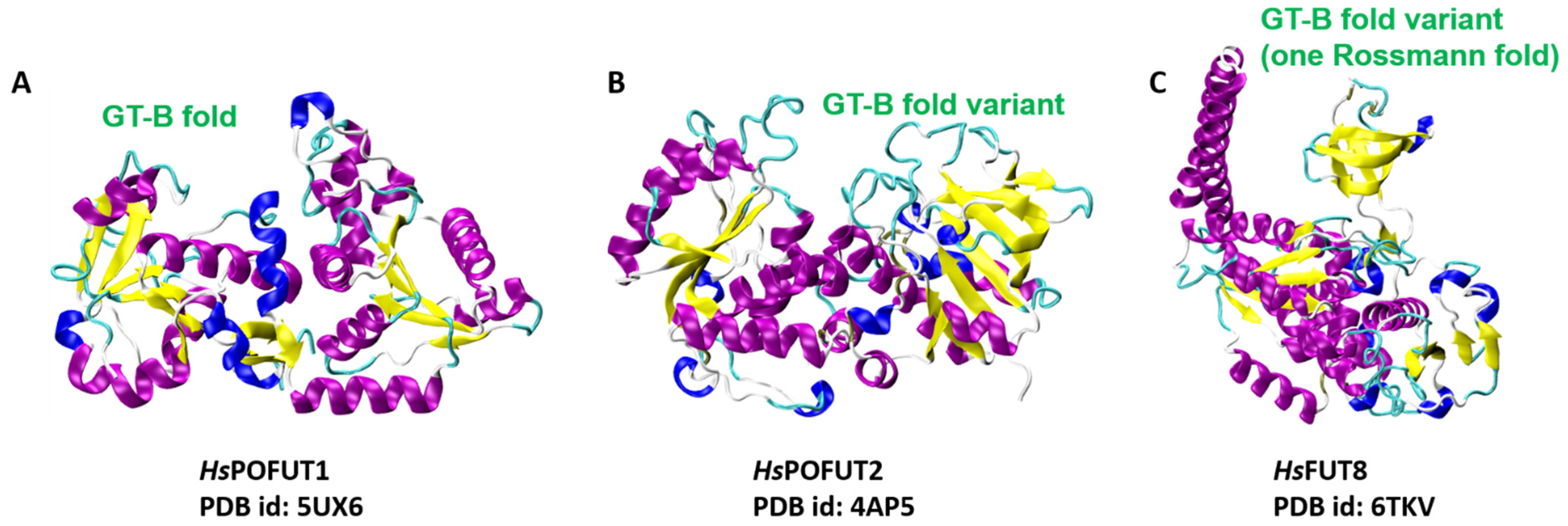
4.1. STs Display Variants of the GT-A Fold
4.2. FUTs Display the GT-B Fold and Variations of It
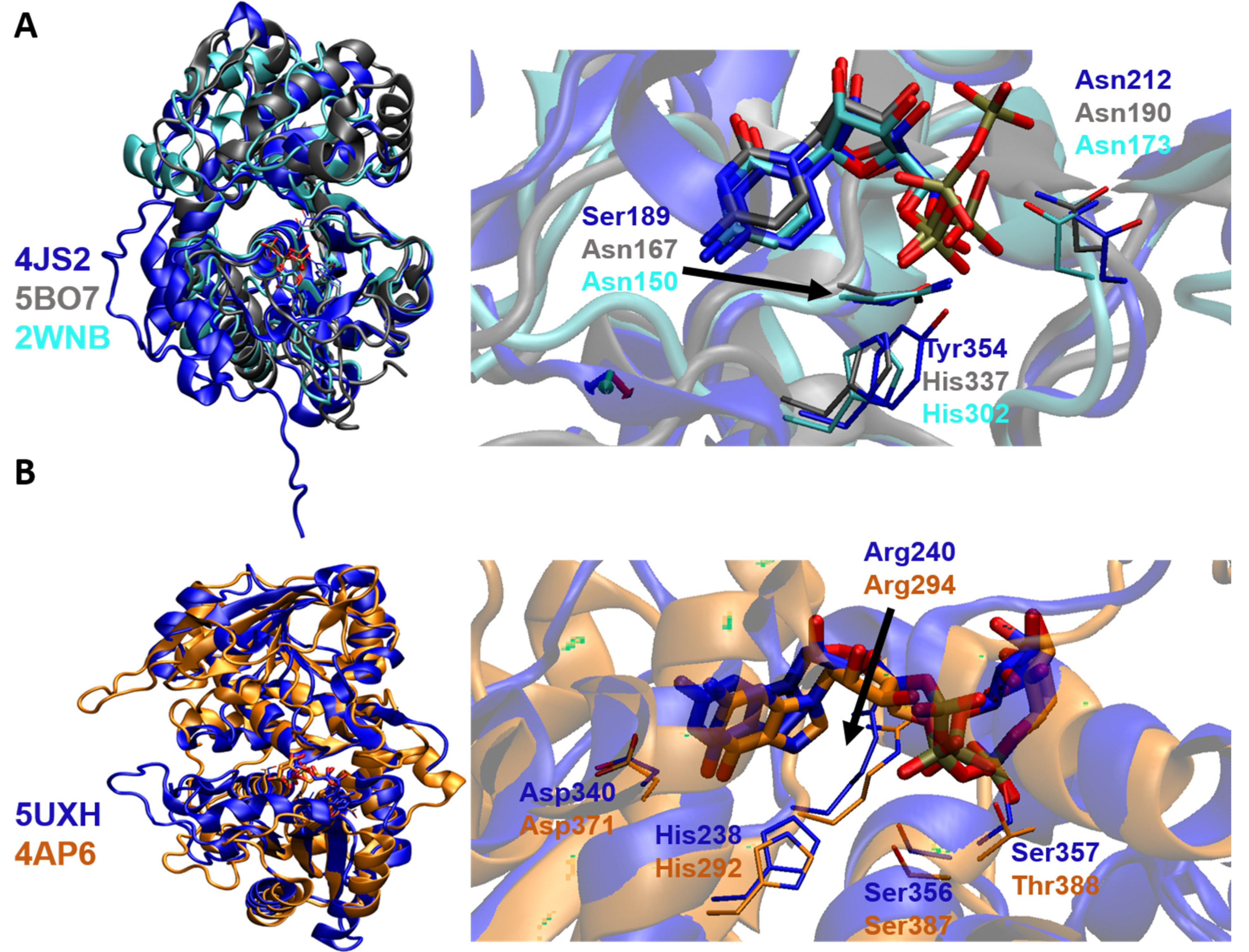
5. Binding Interactions of STs with Natural Donor Substrate CMP-Neu5Ac
6. Binding Interactions between FUT and Its Natural Donor Substrate GDP-Fucose
7. Binding Interactions of STs and FUTs with Their Acceptor Substrates
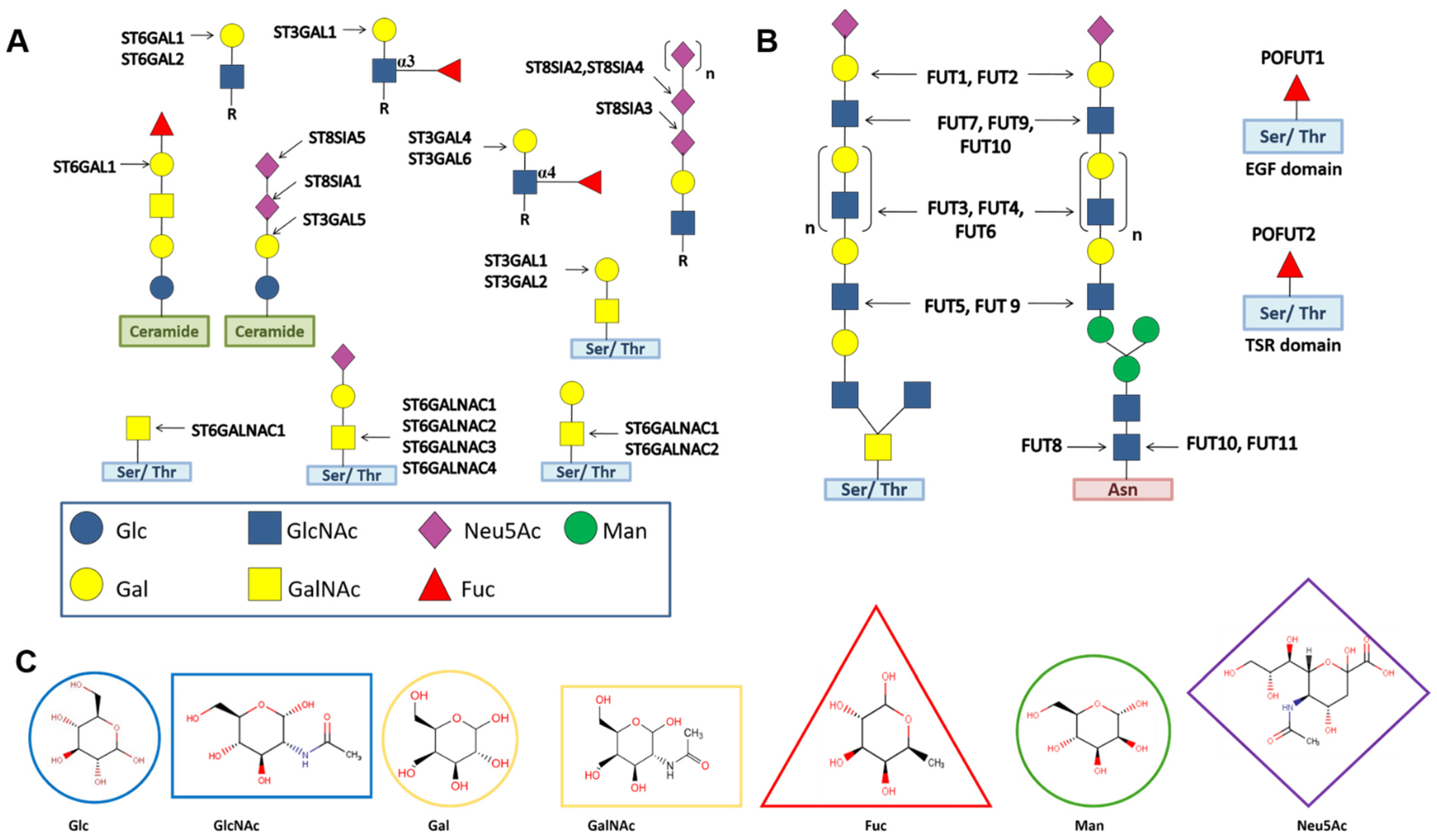
7.1. Glycan-Modifying STs and FUTs
7.2. Protein-Modifying FUTs
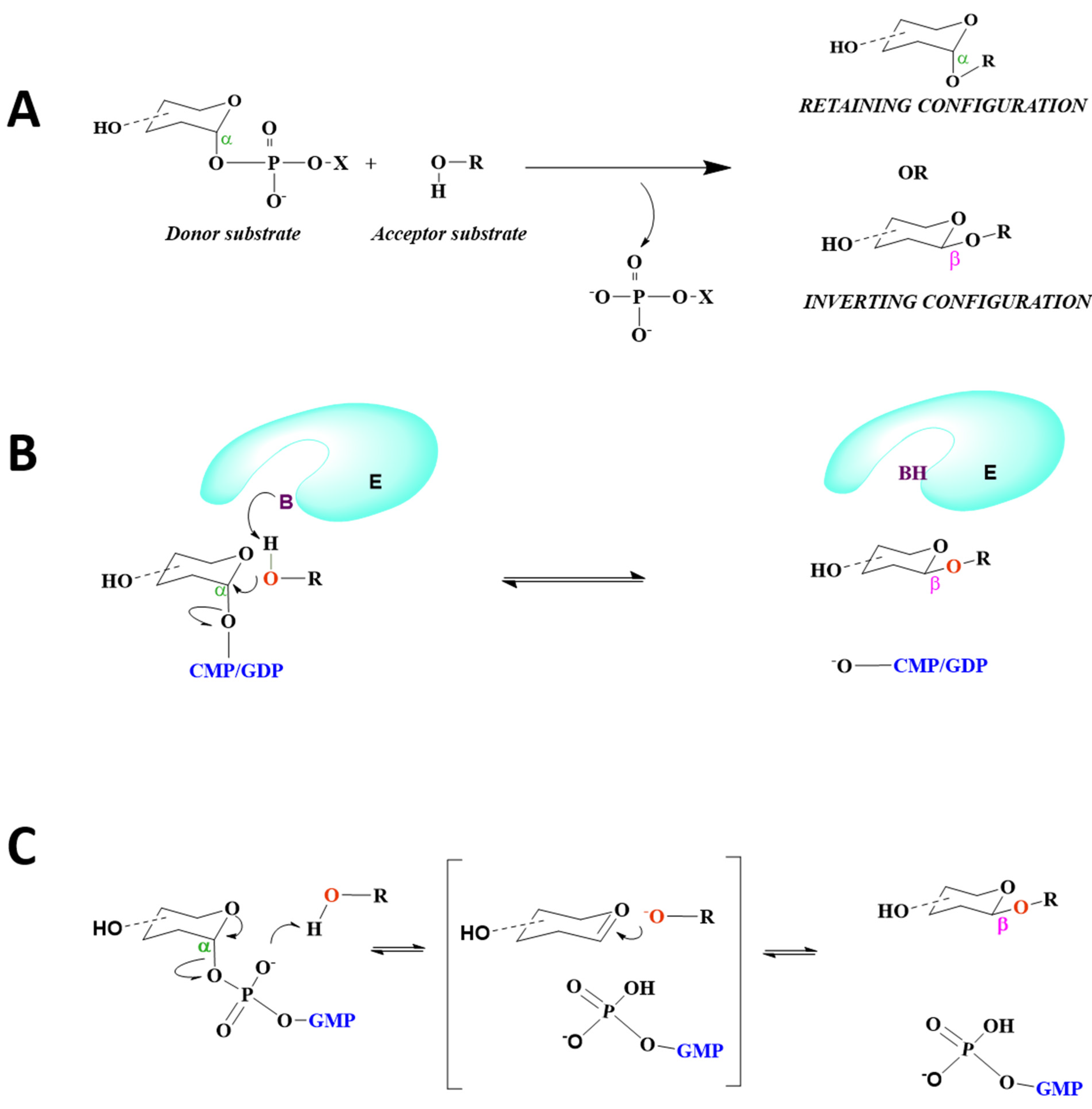
8. Mechanism of Catalysis
8.1. STs Display SN2 Catalysis
8.2. FUTs Display SN1 or SN2 Catalysis
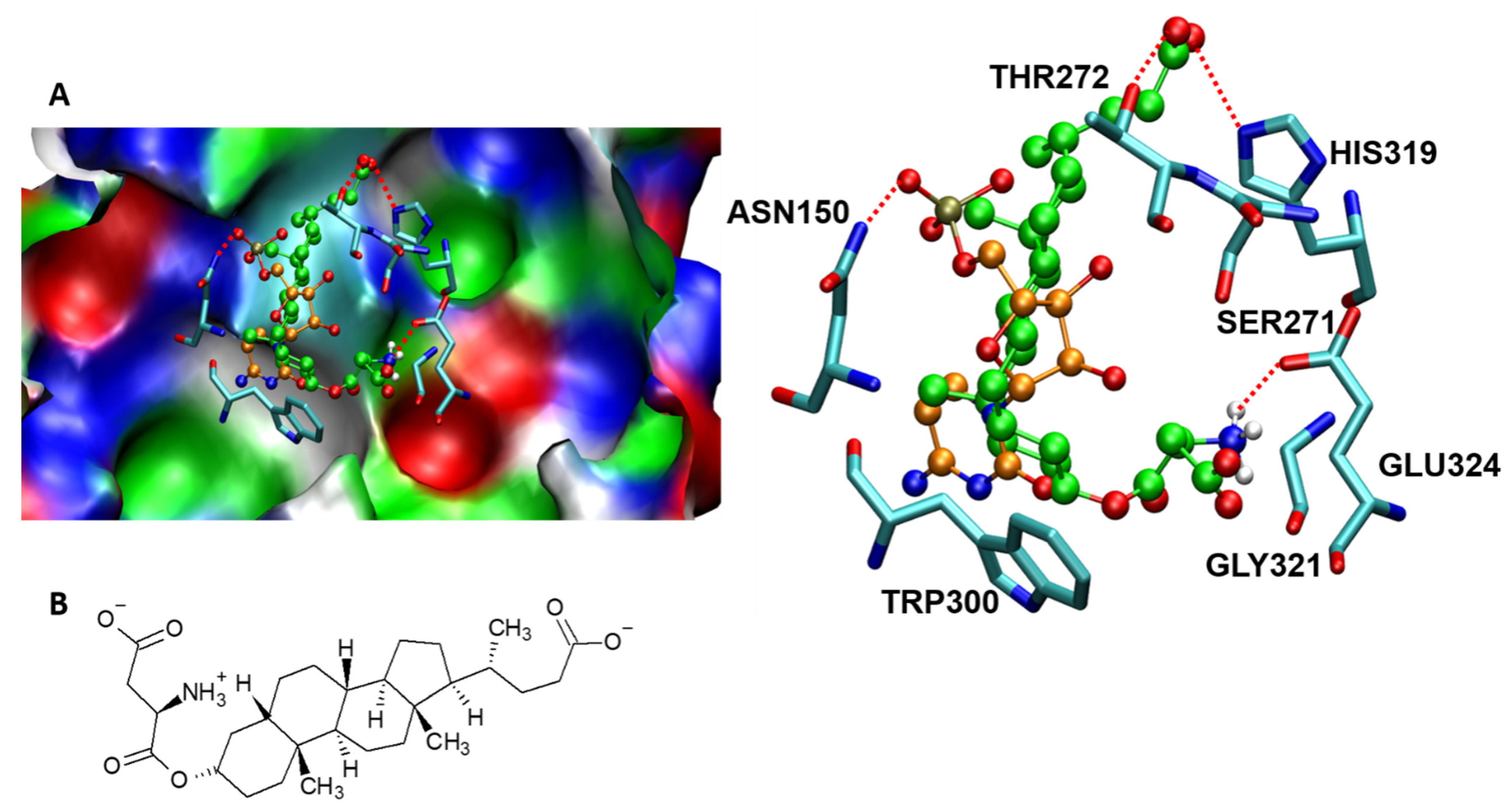
9. Molecular Interactions of Porcine ST3GAL1 with a Potent Inhibitor That Mitigates Tumor Cell Metastasis
10. Structural Modeling of STs and FUTs
11. Conclusions
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Grewal, R.K.; Mahmood, A. A shift in microvillus membrane fucosylation to sialylation by ethanol ingestion in rat intestine. Mol. Cell Biochem. 2009, 331, 19–25. [Google Scholar] [CrossRef]
- Grewal, R.K.; Mahmood, A. Ethanol induced changes in glycosylation of mucins in rat intestine. Ann. Gastroentrol. 2009, 22, 178–183. [Google Scholar]
- Grewal, R.K.; Mahmood, A. The effects of ethanol administration on brush border membrane glycolipids in rat intestine. Alcohol 2010, 44, 515–522. [Google Scholar] [CrossRef]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef]
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell. Biol. 2020, 21, 729–749. [Google Scholar] [CrossRef]
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef] [Green Version]
- Albesa-Jove, D.; Guerin, M.E. The conformational plasticity of glycosyltransferases. Curr. Opin. Struct. Biol. 2016, 40, 23–32. [Google Scholar] [CrossRef]
- Breton, C.; Fournel-Gigleux, S.; Palcic, M.M. Recent structures, evolution and mechanisms of glycosyltransferases. Curr. Opin. Struct. Biol. 2012, 22, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Naumoff, D.G. Hierarchical classification of glycoside hydrolases. Biochemistry 2011, 76, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Pak, J.E.; Satkunarajah, M.; Seetharaman, J.; Rini, J.M. Structural and mechanistic characterization of leukocyte-type core 2 beta1,6-N-acetylglucosaminyltransferase: A metal-ion-independent GT-A glycosyltransferase. J. Mol. Biol. 2011, 414, 798–811. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, 490–495. [Google Scholar] [CrossRef] [Green Version]
- Breton, C.; Snajdrova, L.; Jeanneau, C.; Koca, J.; Imberty, A. Structures and mechanisms of glycosyltransferases. Glycobiology 2006, 16, 29–37. [Google Scholar] [CrossRef]
- Gloster, T.M. Advances in understanding glycosyltransferases from a structural perspective. Curr. Opin. Struct. Biol. 2014, 28, 131–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moremen, K.W.; Ramiah, A.; Stuart, M.; Steel, J.; Meng, L.; Forouhar, F.; Moniz, H.A.; Gahlay, G.; Gao, Z.; Chapla, D.; et al. Expression system for structural and functional studies of human glycosylation enzymes. Nat. Chem Biol 2018, 14, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Videira, P.A.; Marcelo, F.; Grewal, R.K. Glycosyltransferase inhibitors: A promising strategy to pave a path from laboratory to therapy. R. Soc. Chem. 2018, 43, 135–158. [Google Scholar]
- Moremen, K.W.; Haltiwanger, R.S. Emerging structural insights into glycosyltransferase-mediated synthesis of glycans. Nat. Chem. Biol. 2019, 15, 853–864. [Google Scholar] [CrossRef]
- Bull, C.; Stoel, M.A.; den Brok, M.H.; Adema, G.J. Sialic acids sweeten a tumor’s life. Cancer Res. 2014, 74, 3199–3204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dall’Olio, F.; Malagolini, N.; Trinchera, M.; Chiricolo, M. Sialosignaling: Sialyltransferases as engines of self-fueling loops in cancer progression. Biochim. Biophys. Acta 2014, 1840, 2752–2764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobie, C.; Skropeta, D. Insights into the role of sialylation in cancer progression and metastasis. Br. J. Cancer 2021, 124, 76–90. [Google Scholar] [CrossRef]
- Harduin-Lepers, A.; Krzewinski-Recchi, M.A.; Colomb, F.; Foulquier, F.; Groux-Degroote, S.; Delannoy, P. Sialyltransferases functions in cancers. Front. Biosci. 2012, 4, 499–515. [Google Scholar] [CrossRef]
- Munkley, J.; Scott, E. Targeting Aberrant Sialylation to Treat Cancer. Medicines 2019, 6, 102. [Google Scholar] [CrossRef] [Green Version]
- Pearce, O.M.; Laubli, H. Sialic acids in cancer biology and immunity. Glycobiology 2016, 26, 111–128. [Google Scholar] [CrossRef] [Green Version]
- Pietrobono, S.; Stecca, B. Aberrant Sialylation in Cancer: Biomarker and Potential Target for Therapeutic Intervention? Cancers 2021, 13, 2014. [Google Scholar] [CrossRef] [PubMed]
- Blanas, A.; Sahasrabudhe, N.M.; Rodriguez, E.; van Kooyk, Y.; van Vliet, S.J. Fucosylated Antigens in Cancer: An Alliance toward Tumor Progression, Metastasis, and Resistance to Chemotherapy. Front. Oncol. 2018, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Keeley, T.S.; Yang, S.; Lau, E. The Diverse Contributions of Fucose Linkages in Cancer. Cancers 2019, 11, 1241. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Liu, Y.; Wu, L.; Sun, X.L. Sialyltransferase inhibition and recent advances. Biochim. Biophys. Acta 2016, 1, 143–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Audry, M.; Jeanneau, C.; Imberty, A.; Harduin-Lepers, A.; Delannoy, P.; Breton, C. Current trends in the structure-activity relationships of sialyltransferases. Glycobiology 2011, 21, 716–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeanneau, C.; Chazalet, V.; Auge, C.; Soumpasis, D.M.; Harduin-Lepers, A.; Delannoy, P.; Imberty, A.; Breton, C. Structure-function analysis of the human sialyltransferase ST3Gal I: Role of n-glycosylation and a novel conserved sialylmotif. J. Biol Chem. 2004, 279, 13461–13468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, R.Y.; Balaji, P.V. Identification of linkage-specific sequence motifs in sialyltransferases. Glycobiology 2006, 16, 108–116. [Google Scholar] [CrossRef] [Green Version]
- Grewal, R.K.; Pathania, A.S.; Chawla, M. Human Sialyltransferases and its interaction with phytochemicals. Int. J. Pharm. Bio. Sci. 2014, 5, 691–698. [Google Scholar]
- Harrus, D.; Harduin-Lepers, A.; Glumoff, T. Unliganded and CMP-Neu5Ac bound structures of human alpha-2,6-sialyltransferase ST6Gal I at high resolution. J. Struct. Biol. 2020, 212, 107628. [Google Scholar] [CrossRef] [PubMed]
- Datta, A.K.; Chammas, R.; Paulson, J.C. Conserved cysteines in the sialyltransferase sialylmotifs form an essential disulfide bond. J. Biol. Chem. 2001, 276, 15200–15207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, F.V.; Rich, J.R.; Rakic, B.; Buddai, S.; Schwartz, M.F.; Johnson, K.; Bowe, C.; Wakarchuk, W.W.; Defrees, S.; Withers, S.G.; et al. Structural insight into mammalian sialyltransferases. Nat. Struct. Mol. Biol. 2009, 16, 1186–1188. [Google Scholar] [CrossRef] [PubMed]
- Kitazume-Kawaguchi, S.; Kabata, S.; Arita, M. Differential biosynthesis of polysialic or disialic acid Structure by ST8Sia II and ST8Sia IV. J. Biol. Chem. 2001, 276, 15696–15703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harduin-Lepers, A. Comprehensive Analysis of Sialyltransferases in Vertebrate Genomes. Glycobiology Insights 2010, 2. [Google Scholar] [CrossRef]
- Sarnesto, A.; Kohlin, T.; Hindsgaul, O.; Thurin, J.; Blaszczyk-Thurin, M. Purification of the secretor-type beta-galactoside alpha 1----2-fucosyltransferase from human serum. J. Biol. Chem. 1992, 267, 2737–2744. [Google Scholar] [CrossRef]
- Sarnesto, A.; Kohlin, T.; Thurin, J.; Blaszczyk-Thurin, M. Purification of H gene-encoded beta-galactoside alpha 1----2 fucosyltransferase from human serum. J. Biol. Chem. 1990, 265, 15067–15075. [Google Scholar] [CrossRef]
- Dupuy, F.; Germot, A.; Julien, R.; Maftah, A. Structure/function study of Lewis alpha3- and alpha3/4-fucosyltransferases: The alpha1,4 fucosylation requires an aromatic residue in the acceptor-binding domain. Glycobiology 2004, 14, 347–356. [Google Scholar] [CrossRef] [Green Version]
- Dupuy, F.; Petit, J.M.; Mollicone, R.; Oriol, R.; Julien, R.; Maftah, A. A single amino acid in the hypervariable stem domain of vertebrate alpha1,3/1,4-fucosyltransferases determines the type 1/type 2 transfer. Characterization of acceptor substrate specificity of the lewis enzyme by site-directed mutagenesis. J. Biol. Chem. 1999, 274, 12257–12262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, B.; Simala-Grant, J.L.; Taylor, D.E. Fucosylation in prokaryotes and eukaryotes. Glycobiology 2006, 16, 158–184. [Google Scholar] [CrossRef] [Green Version]
- de Vries, T.; Knegtel, R.M.; Holmes, E.H.; Macher, B.A. Fucosyltransferases: Structure/function studies. Glycobiology 2001, 11, 119–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, Z.; Lin, Y.N.; Lin, C.H. Development of fucosyltransferase and fucosidase inhibitors. Chem Soc. Rev. 2013, 42, 4459–4475. [Google Scholar] [CrossRef]
- Mollicone, R.; Moore, S.E.; Bovin, N.; Garcia-Rosasco, M.; Candelier, J.J.; Martinez-Duncker, I.; Oriol, R. Activity, splice variants, conserved peptide motifs, and phylogeny of two new alpha1,3-fucosyltransferase families (FUT10 and FUT11). J. Biol. Chem. 2009, 284, 4723–4738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishihara, S.; Hiraga, T.; Ikehara, Y.; Kudo, T.; Iwasaki, H.; Morozumi, K.; Akamatsu, S.; Tachikawa, T.; Narimatsu, H. Molecular mechanisms of expression of Lewis b antigen and other type I Lewis antigens in human colorectal cancer. Glycobiology 1999, 9, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, I.G.; Carrascal, M.; Mineiro, A.G.; Bugalho, A.; Borralho, P.; Silva, Z.; Dall’olio, F.; Videira, P.A. Carcinoembryonic antigen is a sialyl Lewis x/a carrier and an Eselectin ligand in nonsmall cell lung cancer. Int. J. Oncol. 2019, 55, 1033–1048. [Google Scholar]
- Carrascal, M.A.; Silva, M.; Ferreira, J.A.; Azevedo, R.; Ferreira, D.; Silva, A.M.N.; Ligeiro, D.; Santos, L.L.; Sackstein, R.; Videira, P.A. A functional glycoproteomics approach identifies CD13 as a novel E-selectin ligand in breast cancer. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2069–2080. [Google Scholar] [CrossRef]
- Yanagidani, S.; Uozumi, N.; Ihara, Y.; Miyoshi, E.; Yamaguchi, N.; Taniguchi, N. Purification and cDNA cloning of GDP-L-Fuc:N-acetyl-beta-D-glucosaminide:alpha1-6 fucosyltransferase (alpha1-6 FucT) from human gastric cancer MKN45 cells. J. Biochem. 1997, 121, 626–632. [Google Scholar] [CrossRef]
- Holdener, B.C.; Haltiwanger, R.S. Protein O-fucosylation: Structure and function. Curr. Opin. Struct. Biol. 2019, 56, 78–86. [Google Scholar] [CrossRef]
- Shao, L.; Haltiwanger, R.S. O-fucose modifications of epidermal growth factor-like repeats and thrombospondin type 1 repeats: Unusual modifications in unusual places. Cell Mol. Life. Sci. 2003, 60, 241–250. [Google Scholar] [CrossRef]
- Bastian, K.; Scott, E.; Elliott, D.J.; Munkley, J. FUT8 Alpha-(1,6)-Fucosyltransferase in Cancer. Int. J. Mol. Sci. 2021, 22, 455. [Google Scholar] [CrossRef]
- Jarva, M.A.; Dramicanin, M.; Lingford, J.P.; Mao, R.; John, A.; Jarman, K.E.; Grinter, R.; Goddard-Borger, E.D. Structural basis of substrate recognition and catalysis by fucosyltransferase 8. J. Biol. Chem. 2020, 295, 6677–6688. [Google Scholar] [CrossRef] [Green Version]
- Oriol, R.; Mollicone, R.; Cailleau, A.; Balanzino, L.; Breton, C. Divergent evolution of fucosyltransferase genes from vertebrates, invertebrates, and bacteria. Glycobiology 1999, 9, 323–334. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Duncker, I.; Mollicone, R.; Candelier, J.J.; Breton, C.; Oriol, R. A new superfamily of protein-O-fucosyltransferases, alpha2-fucosyltransferases, and alpha6-fucosyltransferases: Phylogeny and identification of conserved peptide motifs. Glycobiology 2003, 13, 1–5. [Google Scholar] [CrossRef]
- Woodcock, S.; Mornon, J.P.; Henrissat, B. Detection of secondary structure elements in proteins by hydrophobic cluster analysis. Protein Eng. 1992, 5, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Chazalet, V.; Uehara, K.; Geremia, R.A.; Breton, C. Identification of essential amino acids in the AzorhizobiumcaulinodansfucosyltransferaseNodZ. J. Bacteriol. 2001, 183, 7067–7075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brzezinski, K.; Dauter, Z.; Jaskolski, M. Structures of NodZ alpha1,6-fucosyltransferase in complex with GDP and GDP-fucose. Acta Crystallogr D Biol. Crystallogr. 2012, 68, 160–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocha, J.; Ciceron, F.; de Sanctis, D.; Lelimousin, M.; Chazalet, V.; Lerouxel, O.; Breton, C. Structure of Arabidopsis thaliana FUT1 Reveals a Variant of the GT-B Class Fold and Provides Insight into Xyloglucan Fucosylation. Plant. Cell 2016, 28, 2352–2364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urbanowicz, B.R.; Bharadwaj, V.S.; Alahuhta, M.; Pena, M.J.; Lunin, V.V.; Bomble, Y.J.; Wang, S.; Yang, J.Y.; Tuomivaara, S.T.; Himmel, M.E.; et al. Structural, mutagenic and in silico studies of xyloglucan fucosylation in Arabidopsis thaliana suggest a water-mediated mechanism. Plant. J. 2017, 91, 931–949. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.I.; Keusch, J.J.; Klein, D.; Hess, D.; Hofsteenge, J.; Gut, H. Structure of human POFUT2: Insights into thrombospondin type 1 repeat fold and O-fucosylation. EMBO J. 2012, 31, 3183–3197. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Han, K.; Pak, J.E.; Satkunarajah, M.; Zhou, D.; Rini, J.M. Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1. Nat. Chem. Biol. 2017, 13, 757–763. [Google Scholar] [CrossRef]
- Valero-Gonzalez, J.; Leonhard-Melief, C.; Lira-Navarrete, E.; Jimenez-Oses, G.; Hernandez-Ruiz, C.; Pallares, M.C.; Yruela, I.; Vasudevan, D.; Lostao, A.; Corzana, F.; et al. A proactive role of water molecules in acceptor recognition by protein O-fucosyltransferase 2. Nat. Chem Biol. 2016, 12, 240–246. [Google Scholar] [CrossRef] [Green Version]
- Ihara, H.; Ikeda, Y.; Toma, S.; Wang, X.; Suzuki, T.; Gu, J.; Miyoshi, E.; Tsukihara, T.; Honke, K.; Matsumoto, A.; et al. Crystal structure of mammalian alpha1,6-fucosyltransferase, FUT8. Glycobiology 2007, 17, 455–466. [Google Scholar] [CrossRef]
- McMillan, B.J.; Zimmerman, B.; Egan, E.D.; Lofgren, M.; Xu, X.; Hesser, A.; Blacklow, S.C. Structure of human POFUT1, its requirement in ligand-independent oncogenic Notch signaling, and functional effects of Dowling-Degos mutations. Glycobiology 2017, 27, 777–786. [Google Scholar] [CrossRef] [Green Version]
- Harduin-Lepers, A.; Vallejo-Ruiz, V.; Krzewinski-Recchi, M.A.; Samyn-Petit, B.; Julien, S.; Delannoy, P. The human sialyltransferase family. Biochimie 2001, 83, 727–737. [Google Scholar] [CrossRef]
- Stanley, P. Golgi glycosylation. Cold Spring Harb. Perspect. Biol. 2011, 3, a005199. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Haltiwanger, R.S. O-fucosylation of notch occurs in the endoplasmic reticulum. J. Biol. Chem. 2005, 280, 11289–11294. [Google Scholar] [CrossRef] [Green Version]
- Lizak, C.; Gerber, S.; Numao, S.; Aebi, M.; Locher, K.P. X-ray structure of a bacterial oligosaccharyltransferase. Nature 2011, 474, 350–355. [Google Scholar] [CrossRef]
- Matsumoto, S.; Shimada, A.; Nyirenda, J.; Igura, M.; Kawano, Y.; Kohda, D. Crystal structures of an archaeal oligosaccharyltransferase provide insights into the catalytic cycle of N-linked protein glycosylation. Proc. Natl. Acad. Sci. USA 2013, 110, 17868–17873. [Google Scholar] [CrossRef] [Green Version]
- Petrou, V.I.; Herrera, C.M.; Schultz, K.M.; Clarke, O.B.; Vendome, J.; Tomasek, D.; Banerjee, S.; Rajashankar, K.R.; Belcher Dufrisne, M.; Kloss, B.; et al. Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation. Science 2016, 351, 608–612. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Soto, M.E.; Reising, S.; Schlosser, A.; Seibel, J. Structural and functional role of disulphide bonds and substrate binding residues of the human beta-galactoside alpha-2,3-sialyltransferase 1 (hST3Gal1). Sci. Rep. 2019, 9, 17993. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, B.; Benz, J.; Greif, M.; Engel, A.M.; Sobek, H.; Rudolph, M.G. The structure of human alpha-2,6-sialyltransferase reveals the binding mode of complex glycans. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1826–1838. [Google Scholar] [CrossRef]
- Volkers, G.; Worrall, L.J.; Kwan, D.H.; Yu, C.C.; Baumann, L.; Lameignere, E.; Wasney, G.A.; Scott, N.E.; Wakarchuk, W.; Foster, L.J.; et al. Structure of human ST8SiaIII sialyltransferase provides insight into cell-surface polysialylation. Nat. Struct Mol. Biol. 2015, 22, 627–635. [Google Scholar] [CrossRef]
- Meng, L.; Forouhar, F.; Thieker, D.; Gao, Z.; Ramiah, A.; Moniz, H.; Xiang, Y.; Seetharaman, J.; Milaninia, S.; Su, M.; et al. Enzymatic basis for N-glycan sialylation: Structure of rat alpha2,6-sialyltransferase (ST6GAL1) reveals conserved and unique features for glycan sialylation. J. Biol. Chem. 2013, 288, 34680–34698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lira-Navarrete, E.; Hurtado-Guerrero, R. A perspective on structural and mechanistic aspects of protein O-fucosylation. Acta Crystallogr. F Struct Biol. Commun. 2018, 74, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Garcia, A.; Ceballos-Laita, L.; Serna, S.; Artschwager, R.; Reichardt, N.C.; Corzana, F.; Hurtado-Guerrero, R. Structural basis for substrate specificity and catalysis of alpha1,6-fucosyltransferase. Nat. Commun. 2020, 11, 973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, C.P.; Watts, A.G.; Lairson, L.L.; Gilbert, M.; Lim, D.; Wakarchuk, W.W.; Withers, S.G.; Strynadka, N.C. Structural analysis of the sialyltransferaseCstII from Campylobacter jejuni in complex with a substrate analog. Nat. Struct. Mol. Biol. 2004, 11, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Kotzler, M.P.; Blank, S.; Bantleon, F.I.; Spillner, E.; Meyer, B. Donor substrate binding and enzymatic mechanism of human core alpha1,6-fucosyltransferase (FUT8). Biochim Biophys. Acta 2012, 1820, 1915–1925. [Google Scholar] [CrossRef] [PubMed]
- Boruah, B.M.; Kadirvelraj, R.; Liu, L.; Ramiah, A.; Li, C.; Zong, G.; Bosman, G.P.; Yang, J.Y.; Wang, L.X.; Boons, G.J.; et al. Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases. J. Biol. Chem. 2020, 295, 17027–17045. [Google Scholar] [CrossRef] [PubMed]
- Kotzler, M.P.; Blank, S.; Bantleon, F.I.; Wienke, M.; Spillner, E.; Meyer, B. Donor assists acceptor binding and catalysis of human alpha1,6-fucosyltransferase. ACS Chem. Biol. 2013, 8, 1830–1840. [Google Scholar] [CrossRef]
- Schneider, M.; Al-Shareffi, E.; Haltiwanger, R.S. Biological functions of fucose in mammals. Glycobiology 2017, 27, 601–618. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.Y.; Tang, Y.A.; Huang, S.M.; Juan, H.F.; Wu, L.W.; Sun, Y.C.; Wang, S.C.; Wu, K.W.; Balraj, G.; Chang, T.T.; et al. A novel sialyltransferase inhibitor suppresses FAK/paxillin signaling and cancer angiogenesis and metastasis pathways. Cancer Res. 2011, 71, 473–483. [Google Scholar] [CrossRef] [Green Version]
- Breton, C.; Oriol, R.; Imberty, A. Sequence alignment and fold recognition of fucosyltransferases. Glycobiology 1996, 6, 7–12. [Google Scholar]
- Strecker, C.; Baerenfaenger, M.; Miehe, M.; Spillner, E.; Meyer, B. In Silico Evaluation of the Binding Site of Fucosyltransferase 8 and First Attempts to Synthesize an Inhibitor with Drug-Like Properties. Chembiochem 2020, 21, 1923–1931. [Google Scholar] [CrossRef]





| Enzyme Family | Enzyme | CAZy Family | Global Fold | Donor Substrate | Preferred Acceptor Substrate | Upregulated Expression in Cell Lines or Tissues |
|---|---|---|---|---|---|---|
| ST3GAL | ST3GAL1 | GT29 | GT-A | CMP-Neu5Ac | Galβ1, 3GalNAc | Melanoma, breast, ovarian, and liver cancers |
| ST3GAL2 | Galβ1, 3GalNAc | - | ||||
| ST3GAL3 | Galβ1, 3(4)GlcNAc | Melanoma, glioma, breast, and pancreatic cancers | ||||
| ST3GAL4 | Galβ1, 4(3)GlcNAc | Gastric cancer | ||||
| ST3GAL5 | Galβ1, 4Glc-ceramide | - | ||||
| ST3GAL6 | Galβ1, 4GlcNAc | Multiple myeloma and gastric cancer | ||||
| ST6GAL | ST6GAL1 | Galβ1, 4GlcNAc | Liver, gastric, colon, bone, pancreatic, lung, prostate, breast, and ovarian cancers | |||
| ST6GAL2 | Galβ1, 4GlcNAc | Breast cancer | ||||
| ST6GALNAC | STGALNAC1 | GalβNAc-Ser/Thr and Galβ1,3GalNAc- Ser/Thr | Liver and gastric cancers | |||
| STGALNAC2 | Galβ1, 3GalNAc-Ser/Thr | Thyroid and colorectal cancers | ||||
| STGALNAC3 | Siaα2, 3Galβ1, 3GalNAc-ceramide Siaα2, 3Galβ1, 3GalNAc-Ser/Thr | - | ||||
| STGALNAC4 | Siaα2, 3Galβ1, 3GalNAc-Ser/Thr | - | ||||
| STGALNAC5 | GM1b | - | ||||
| STGALNAC6 | All α-series gangliosides | - | ||||
| ST8SIA | ST8SIA1 | Siaα2,3Galβ1,4Glc-ceramide | Breast cancer | |||
| ST8SIA2 | (Siaα2,8)nSiaα2,3Gal | - | ||||
| ST8SIA3 | Siaα2,3Galβ1,4GlcNAc | - | ||||
| ST8SIA4 | (Siaα2,8)nSiaα2, 3Gal | Breast cancer, acute myeloid leukemia | ||||
| ST8SIA5 | GM1b, GT1b, GD1a, GD3 | - | ||||
| ST8SIA6 | Siaα2, 3Gal | Breast, lung, and liver cancers | ||||
| α1,2 FUT | FUT1 | GT11 | GT-B | GDP-Fuc | Galβ1, 4GlcNAc | Bladder, breast, epidermoid, ovarian, and prostate cancers |
| FUT2 | Galβ1, 3GlcNAc | - | ||||
| α1,3/4 FUT | FUT3 | GT10 | (Both sialyl- and non-sialyl-) Galβ1,3GlcNAc and Galβ1,4GlcNAc | Pancreatic, gastric, colorectal, oral, and head and neck cancers | ||
| FUT4 | Galβ1, 4GlcNAc | Melanoma, breast, and lung cancers | ||||
| FUT5 | (Both sialyl- and non-sialyl-) Galβ1,3GlcNAc and Galβ1,4GlcNAc | Colorectal and gastric cancers | ||||
| FUT6 | (Both sialyl- and non-sialyl-) Galβ1, 4GlcNAc | Liver, colorectal, prostate, oral, andhead and neck cancers; prostate cancer metastasis to bone | ||||
| FUT7 | (sialyl-) Galβ1, 4GlcNAc | Liver, prostate, and lung cancers | ||||
| FUT9 | Galβ1, 4GlcNAc | - | ||||
| FUT10 | GlcNAc β1, 4GlcNAc-Asn | - | ||||
| FUT11 | GlcNAc β1, 4GlcNAc-Asn | - | ||||
| α1,6 FUT | FUT8 | GT23 | GlcNAcβ1, 2Manα1, 6[GlcNAcβ1,2Manα1,3]Manβ1,4GlcNAcβ1,4GlcNAc-Asn | Melanoma, lung, liver, breast, prostate, ovarian, cervical, and colorectal cancers | ||
| O-FUT | POFUT1 | GT65 | EGF-like repeats | Liver cancer | ||
| O-FUT | POFUT2 | GT68 | Thrombospondin type 1 repeats |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grewal, R.K.; Shaikh, A.R.; Gorle, S.; Kaur, M.; Videira, P.A.; Cavallo, L.; Chawla, M. Structural Insights in Mammalian Sialyltransferases and Fucosyltransferases: We Have Come a Long Way, but It Is Still a Long Way Down. Molecules 2021, 26, 5203. https://doi.org/10.3390/molecules26175203
Grewal RK, Shaikh AR, Gorle S, Kaur M, Videira PA, Cavallo L, Chawla M. Structural Insights in Mammalian Sialyltransferases and Fucosyltransferases: We Have Come a Long Way, but It Is Still a Long Way Down. Molecules. 2021; 26(17):5203. https://doi.org/10.3390/molecules26175203
Chicago/Turabian StyleGrewal, Ravneet Kaur, Abdul Rajjak Shaikh, Suresh Gorle, Manjeet Kaur, Paula Alexendra Videira, Luigi Cavallo, and Mohit Chawla. 2021. "Structural Insights in Mammalian Sialyltransferases and Fucosyltransferases: We Have Come a Long Way, but It Is Still a Long Way Down" Molecules 26, no. 17: 5203. https://doi.org/10.3390/molecules26175203






