Isotope-Labeled RNA Building Blocks for NMR Structure and Dynamics Studies
Abstract
1. Introduction
2. Stable Isotope Labeling of RNA Building Blocks
2.1. Commercial Isotopes Sources
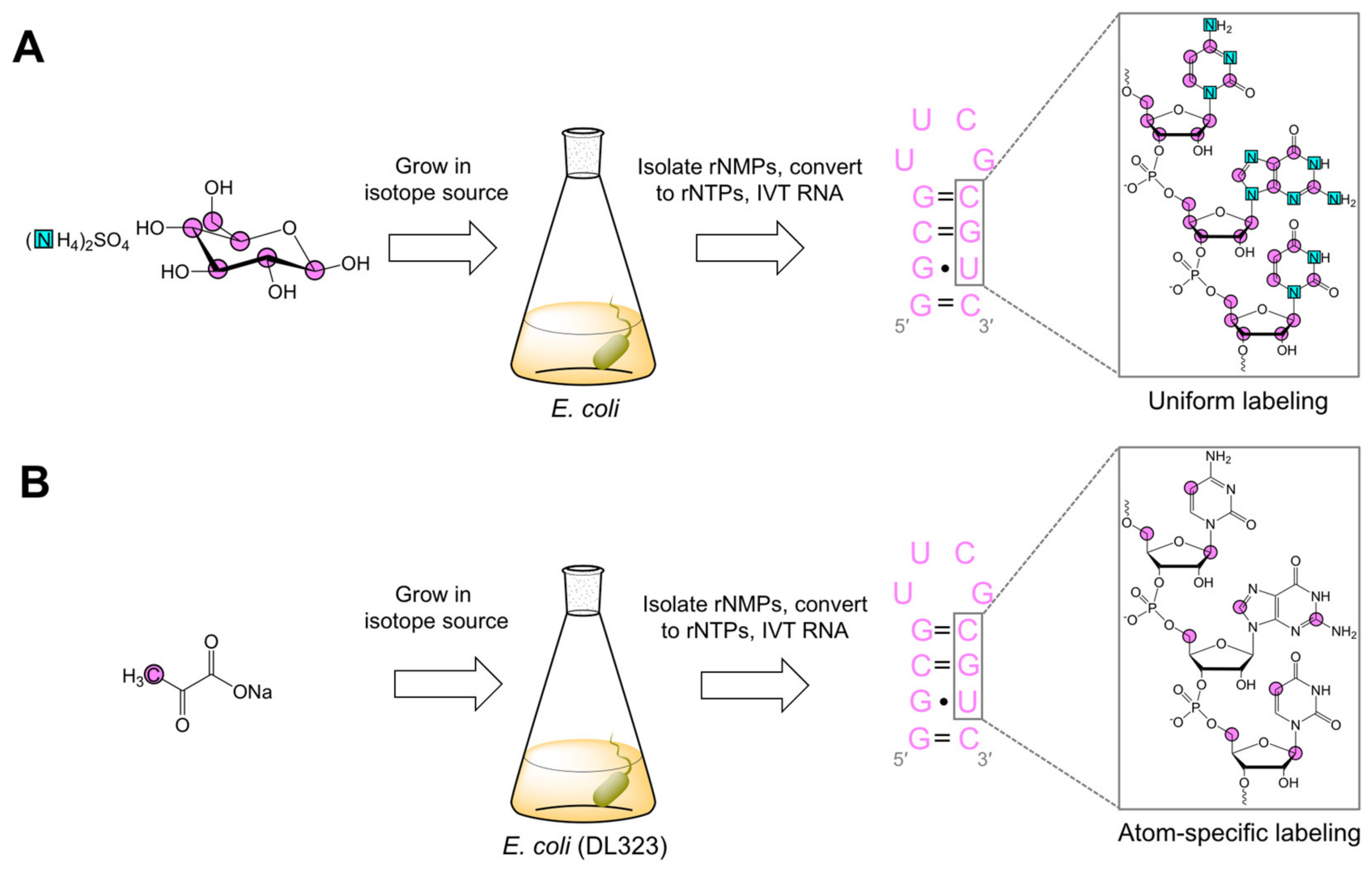
2.2. Biomass Labeling
2.2.1. Biomass Uniform Labeling
2.2.2. Biomass Atom-Specific Labeling
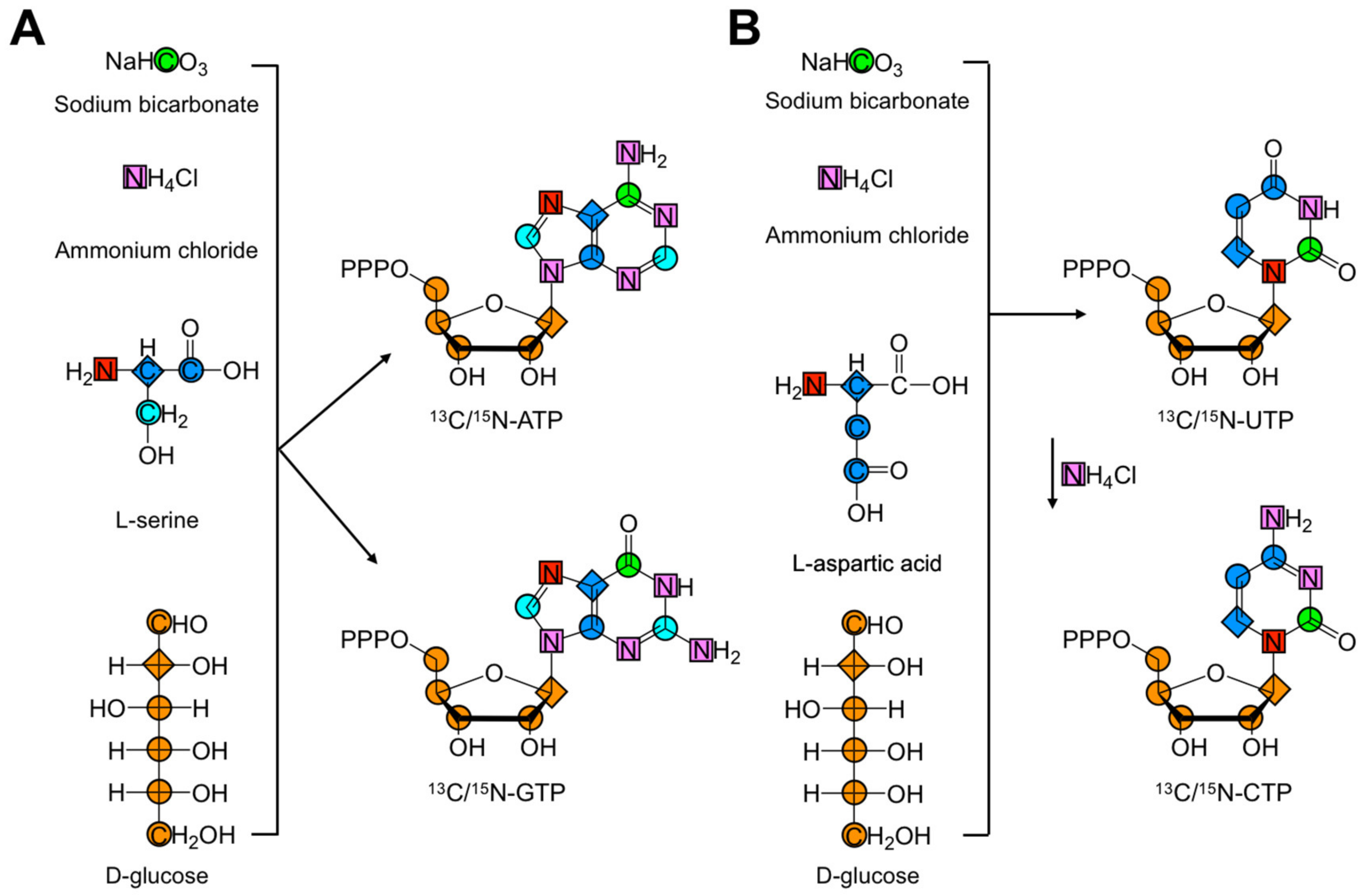
2.3. Ribonucleotide De Novo Biosynthesis
2.3.1. Purine De Novo Biosynthesis
2.3.2. Pyrimidine De Novo Biosynthesis
2.4. Chemo-Enzymatic Labeling
2.5. RNA Phosphoramidite Labeling
3. RNA Preparation Methods
3.1. Chemical Solid-Phase RNA Synthesis
3.2. T7 RNA Polymerase-Based In Vitro Transcription
| 5′-End Sequences a | 5′-End Heterogeneity (<1%) |
|---|---|
| GGG | No |
| GAG | No |
| GCG | No |
| GUG | Yes |
| GGA | Yes |
| GAA | Yes |
| GCA b | Yes |
| GUA | Yes |
| GGC | No |
| GAC | Yes |
| GCC | No |
| GUC | Yes |
| GGU | No |
| GAU | Yes |
| GCU | Yes |
| GUU | Yes |
| AGG | No |
| AAG | Yes |
| ACG | Yes |
| AUG b | Yes |
| AGA | No |
| AAA | No |
| ACA | No |
| AUA | No |
| AGC | Yes |
| AAC | No |
| ACC | No |
| AUC | No |
| AGU b | Yes |
| AAU | No |
| ACU | Yes |
| AUU | Yes |
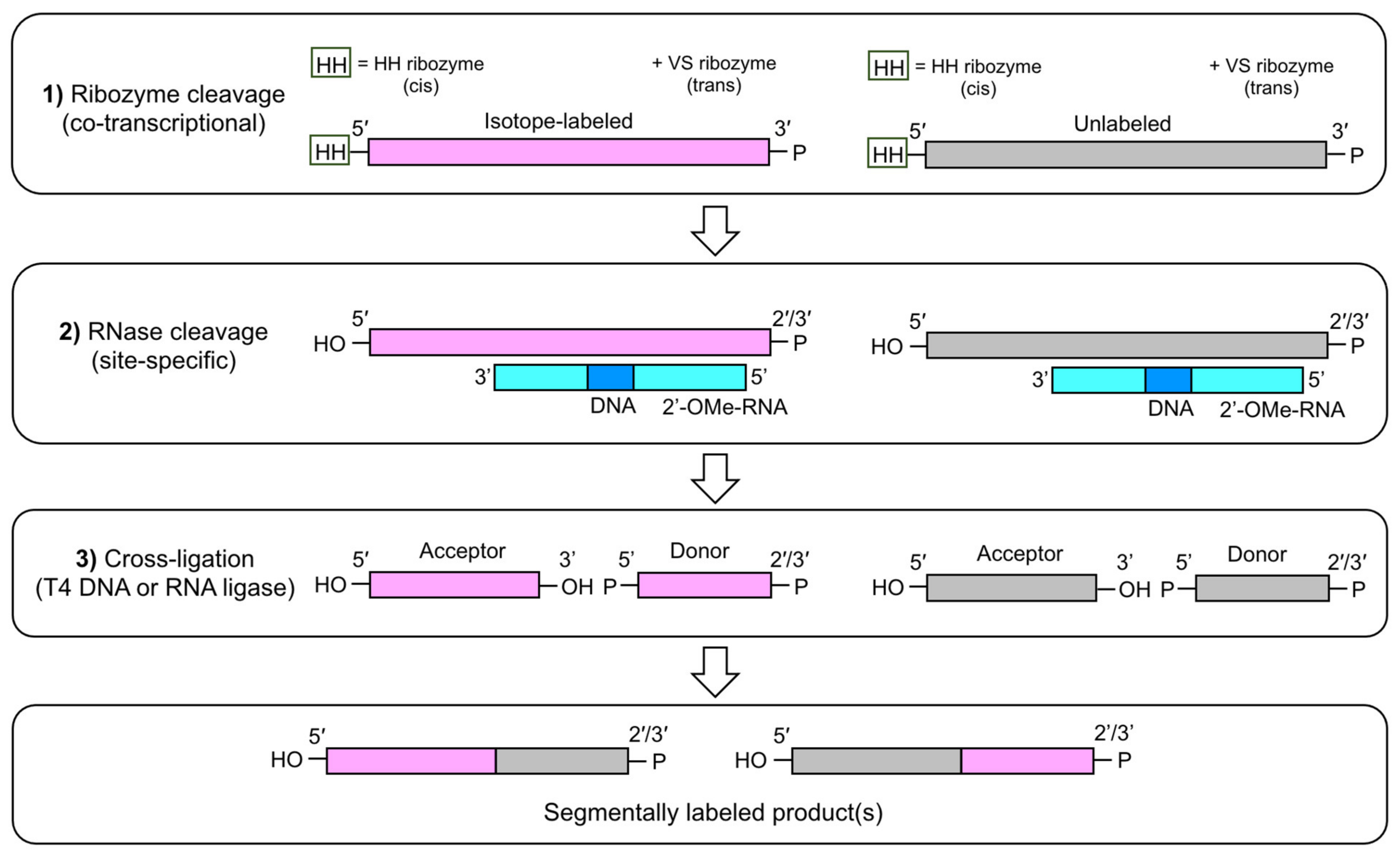
3.3. Enzymatic Ligation
3.3.1. T4 DNA and RNA Ligation
3.3.2. Segmental RNA Labeling
3.4. Enzymatic Position-Specific RNA Labeling
3.4.1. Position-Selective Labeling of RNA (PLOR)
3.4.2. Chemo-Enzymatic Position-Specific Labeling
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambrost, V. The C. elegans Heterochronic Gene lin-4 Encodes Small RNAs with Antisense Complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Mironov, A.S.; Gusarov, I.; Rafikov, R.; Lopez, L.E.; Shatalin, K.; Kreneva, R.A.; Perumov, D.A.; Nudler, E. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell 2002, 111, 747–756. [Google Scholar] [CrossRef]
- Winkler, W.; Nahvi, A.; Breaker, R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 2002, 419, 952–956. [Google Scholar] [CrossRef]
- Grundy, F.J.; Henkin, T.M. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell 1993, 74, 475–482. [Google Scholar] [CrossRef]
- Steitz, T.A.; Steitz, J.A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. USA 1993, 90, 6498–6502. [Google Scholar] [CrossRef] [PubMed]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-Splicing RNA: Autoexcision and Autocyclization of the Ribosomal RNA Intervening Sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef]
- Zappulla, D.C.; Cech, T.R. RNA as a flexible scaffold for proteins: Yeast telomerase and beyond. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 217–224. [Google Scholar] [CrossRef]
- Yik, J.H.N.; Chen, R.; Nishimura, R.; Jennings, J.L.; Link, A.J.; Zhou, Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 2003, 12, 971–982. [Google Scholar] [CrossRef]
- D’Souza, V.; Summers, M.F. How retroviruses select their genomes. Nat. Rev. Microbiol. 2005, 3, 643–655. [Google Scholar] [CrossRef]
- Tycowski, K.T.; Guo, Y.E.; Lee, N.; Moss, W.N.; Vallery, T.K.; Xie, M.; Steitz, J.A. Viral noncoding RNAs: More surprises. Genes Dev. 2015, 29, 567–584. [Google Scholar] [CrossRef]
- Ganser, L.R.; Kelly, M.L.; Herschlag, D.; Al-Hashimi, H.M. The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell Biol. 2019, 20, 25–27. [Google Scholar] [CrossRef]
- Marušič, M.; Schlagnitweit, J.; Petzold, K. RNA Dynamics by NMR Spectroscopy. ChemBioChem 2019, 20, 1–27. [Google Scholar] [CrossRef]
- Zhao, B.; Zhang, Q. Characterizing excited conformational states of RNA by NMR spectroscopy. Curr. Opin. Struct. Biol. 2015, 30, 134–146. [Google Scholar] [CrossRef] [PubMed][Green Version]
- LeBlanc, R.M.; Longhini, A.P.; Tugarinov, V.; Dayie, T.K. NMR probing of invisible excited states using selectively labeled RNAs. J. Biomol. NMR 2018, 71, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Strebitzer, E.; Nußbaumer, F.; Kremser, J.; Tollinger, M.; Kreutz, C. Studying sparsely populated conformational states in RNA combining chemical synthesis and solution NMR spectroscopy. Methods 2018, 148, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Wijmenga, S.S.; Buuren, B.N.M. Van The use of NMR methods for conformational studies of nucleic acids. Prog. Nucl. Magn. Reson. Spectrosc. 1998, 32, 287–387. [Google Scholar] [CrossRef]
- Dayie, K.T. Key labeling technologies to tackle sizeable problems in RNA structural biology. Int. J. Mol. Sci. 2008, 9, 1214–1240. [Google Scholar] [CrossRef]
- Tolbert, T.J.; Williamson, J.R. Preparation of specifically deuterated and 13C-labeled RNA for NMR studies using enzymatic synthesis. J. Am. Chem. Soc. 1997, 119, 12100–12108. [Google Scholar] [CrossRef]
- Lu, K.; Miyazaki, Y.; Summers, M.F. Isotope labeling strategies for NMR studies of RNA. J. Biomol. NMR 2010, 46, 113–125. [Google Scholar] [CrossRef]
- Nikonowicz, E.P.; Sirr, A.; Legault, P.; Jucker, F.M.; Baer, L.M.; Pardi, A. Preparation of 13C and 15N labelled RNAs for heteronuclear multi-dimensional NMR studies. Nucleic Acids Res. 1992, 20, 4507–4513. [Google Scholar] [CrossRef]
- Batey, R.T.; Inada, M.; Kujawinski, E.; Puglisi, J.D.; Williamson, J.R. Preparation of isotopically labeled ribonucleotides for multidimensional NMR spectroscopy of RNA. Nucleic Acids Res. 1992, 20, 4515–4523. [Google Scholar] [CrossRef]
- Barnwal, R.P.; Yang, F.; Varani, G. Applications of NMR to structure determination of RNAs large and small. Arch. Biochem. Biophys. 2017, 628, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Milligan, J.; Groebe, D.; Whherell, G.; Uhlenbeck, O. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987, 15, 8783–8798. [Google Scholar] [CrossRef] [PubMed]
- Milligan, J.F.; Uhlenbeck, O.C. Synthesis of Small RNAs Using T7 RNA Polymerase. Methods Enzymol. 1989, 180, 51–62. [Google Scholar] [PubMed]
- Schultheisz, H.L.; Szymczyna, B.R.; Scott, L.G.; Williamson, J.R. Pathway engineered enzymatic de novo purine nucleotide synthesis. ACS Chem. Biol. 2008, 3, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Schultheisz, H.L.; Szymczyna, B.R.; Scott, L.G.; Williamson, J.R. Enzymatic de novo pyrimidine nucleotide synthesis. J. Am. Chem. Soc. 2011, 133, 297–304. [Google Scholar] [CrossRef] [PubMed]
- SantaLucia, J.; Shen, L.X.; Cai, Z.; Lewis, H.; Tinoco, I. Synthesis and NMR of RNA with selective isotopic enrichment in the bases. Nucleic Acids Res. 1995, 23, 4913–4921. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, L.J.; LeBlanc, R.M.; Longhini, A.P.; Keane, S.C.; Jain, N.; Yildiz, Z.F.; Tolbert, B.S.; D’Souza, V.M.; Summers, M.F.; Kreutz, C.; et al. Regio-Selective Chemical-Enzymatic Synthesis of Pyrimidine Nucleotides Facilitates RNA Structure and Dynamics Studies. ChemBioChem 2014, 15, 1573–1577. [Google Scholar] [CrossRef]
- Longhini, A.P.; Leblanc, R.M.; Becette, O.; Salguero, C.; Wunderlich, C.H.; Johnson, B.A.; D’souza, V.M.; Kreutz, C.; Dayie, T.K. Chemo-enzymatic synthesis of site-specific isotopically labeled nucleotides for use in NMR resonance assignment, dynamics and structural characterizations. Nucleic Acids Res. 2015, 44, 52. [Google Scholar] [CrossRef]
- Ogilvie, K.K.; Theriault, N.; Sadana, K.L. Synthesis of Oligoribonucleotides. J. Am. Chem. Soc. 1977, 99, 7741–7743. [Google Scholar] [CrossRef] [PubMed]
- Ogilvie, K.K.; Sadana, K.L.; Thompson, E.A.; Quilliam, M.A.; Westmore, J.B. The use of silyl groups in protecting the hydroxyl functions of ribonucleosides. Tetrahedron Lett. 1974, 15, 2861–2863. [Google Scholar] [CrossRef]
- Reese, C.B. The Chemical Synthesis of Oligo- and Poly-ribonucleotides. In Nucleic Acids and Molecular Biology; Springer: Berlin/Heidelberg, Germany, 1989; pp. 164–181. [Google Scholar] [CrossRef]
- Beaucage, S.L.; Reese, C.B. Recent advances in the chemical synthesis of RNA. In Current Protocols in Nucleic Acid Chemistry; John Wiley & Sons: Hoboken, NJ, USA, 2009; Volume 38, pp. 2.16.1–2.16.31. [Google Scholar] [CrossRef]
- Becette, O.; Olenginski, L.T.; Dayie, T.K. Solid-phase chemical synthesis of stable isotope-labeled RNA to aid structure and dynamics studies by NMR spectroscopy. Molecules 2019, 24, 3476. [Google Scholar] [CrossRef]
- Abbreviations and Symbols for the Description of Conformations of Polynucleotide Chains: Recommendations 1982. Eur. J. Biochem. 1983, 131, 9–15. [CrossRef]
- Markley, J.L.; Bax, A.; Arata, Y.; Hilbers, C.W.; Kaptein, R.; Sykes, B.D.; Wright, P.E.; Wüthrich, K. Recommendations for the presentation of NMR structures of proteins and nucleic acids. J. Mol. Biol. 1998, 280, 933–952. [Google Scholar] [CrossRef]
- Weickhmann, A.K.; Keller, H.; Duchardt-Ferner, E.; Strebitzer, E.; Juen, M.A.; Kremser, J.; Wurm, J.P.; Kreutz, C.; Wöhnert, J. NMR resonance assignments for the SAM/SAH-binding riboswitch RNA bound to S-adenosylhomocysteine. Biomol. NMR Assign. 2018, 12, 329–334. [Google Scholar] [CrossRef]
- Weickhmann, A.K.; Keller, H.; Wurm, J.P.; Strebitzer, E.; Juen, M.A.; Kremser, J.; Weinberg, Z.; Kreutz, C.; Duchardt-Ferner, E.; Wöhnert, J. The structure of the SAM/SAH-binding riboswitch. Nucleic Acids Res. 2019, 47, 2654–2665. [Google Scholar] [CrossRef]
- Hoard, D.E.; Ott, D.G. Conversion of Mono-and Oligodeoxyribonucleotides to 5′-Triphosphates1. J. Am. Chem. Soc. 1965, 87, 1785–1788. [Google Scholar] [CrossRef] [PubMed]
- Simon, E.S.; Grabowski, S.; Whitesides, G.M. Convenient Syntheses of Cytidine 5′-Triphosphate, Guanosine 5′-Triphosphate, and Uridine 5′-Triphosphate and Their Use in the Preparation of UDP-glucose, UDP-glucuronic Acid, and GDP-mannose. J. Org. Chem. 1990, 55, 1834–1841. [Google Scholar] [CrossRef]
- Michnicka, M.J.; King, G.C.; Harper, J.W. Selective Isotopic Enrichment of Synthetic RNA: Application to the HIV-1 TAR Element. Biochemistry 1993, 32, 395–400. [Google Scholar] [CrossRef]
- Thakur, C.S.; Dayie, T.K. Asymmetry of 13C labeled 3-pyruvate affords improved site specific labeling of RNA for NMR spectroscopy. J. Biomol. NMR 2012, 52, 65–77. [Google Scholar] [CrossRef] [PubMed][Green Version]
- LeMaster, D.M.; Kushlan, D.M. Dynamical mapping of E. coli thioredoxin via 13C NMR relaxation analysis. J. Am. Chem. Soc. 1996, 118, 9255–9264. [Google Scholar] [CrossRef]
- Hines, J.V.; Landry, S.M.; Varani, G.; Tinoco, I. Carbon-Proton Scalar Couplings in RNA: 3D Heteronuclear and 2D Isotope-Edited NMR of a 13C-Labeled Extra-Stable Hairpin. J. Am. Chem. Soc. 1994, 116, 5823–5831. [Google Scholar] [CrossRef]
- Hoffman, D.W.; Holland, J.A. Preparation of carbon-13 labeled ribonucleotides using acetate as an isotope source. Nucleic Acids Res. 1995, 23, 3361–3362. [Google Scholar] [CrossRef]
- Fraenkel, D.G. Selection of Escherichia coli mutants lacking glucose-6-phosphate dehydrogenase or gluconate-6-phosphate dehydrogenase. J. Bacteriol. 1968, 95, 1267–1271. [Google Scholar] [CrossRef]
- Johnson, J.E.; Julien, K.R.; Hoogstraten, C.G. Alternate-site isotopic labeling of ribonucleotides for NMR studies of ribose conformational dynamics in RNA. J. Biomol. NMR 2006, 35, 261–274. [Google Scholar] [CrossRef]
- Vorbrüggen, H.; Krolikiewicz, K.; Bennua, B. Nucleoside syntheses, XXII1) Nucleoside synthesis with trimethylsilyl triflate and perchlorate as catalysts. Chem. Ber. 1981, 114, 1234–1255. [Google Scholar] [CrossRef]
- Földesi, A.; Nilson, F.P.R.; Glemarec, C.; Gioeli, C.; Chattopadhyaya, J. Synthesis of 1′,2′,3′,4′,5′,5″-2H6-β-D-ribonucleosides and 1′, 2′,2″,3′,4′,5′,5″-2H7-β-D-2′-deoxyribonucleosides for selective suppression of proton resonances in partially-deuterated oligo-DNA, oligo-RNA and in 2,5A core (1H-NMR window). Tetrahedron 1992, 48, 9033–9072. [Google Scholar] [CrossRef]
- Toyama, A.; Takino, Y.; Takeuchi, H.; Harada, I. Ultraviolet Resonance Raman Spectra of Ribosyl C(1′)-Deuterated Purine Nucleosides: Evidence of Vibrational Coupling between Purine and Ribose Rings. J. Am. Chem. Soc. 1993, 115, 11092–11098. [Google Scholar] [CrossRef]
- Cook, G.P.; Greenberg, M.M. A General Synthesis of C2′-Deuteriated Ribonucleosides. J. Org. Chem. 1994, 59, 4704–4706. [Google Scholar] [CrossRef]
- Kline, P.C.; Serianni, A.S. 13C-Enriched Ribonucleosides: Synthesis and Application of 13C-1H and 13C-13C Spin-Coupling Constants ToAssess Furanose and A-Glycoside Bond Conformations. J. Am. Chem. Soc. 1990, 112, 7373–7381. [Google Scholar] [CrossRef]
- Lunn, F.A.; MacDonnell, J.E.; Bearne, S.L. Structural requirements for the activation of Escherichia coli CTP synthase by the allosteric effector GTP are stringent, but requirements for inhibition are lax. J. Biol. Chem. 2008, 283, 2010–2020. [Google Scholar] [CrossRef]
- Arthur, P.K.; Alvarado, L.J.; Dayie, T.K. Expression, purification and analysis of the activity of enzymes from the pentose phosphate pathway. Protein Expr. Purif. 2011, 76, 229–237. [Google Scholar] [CrossRef]
- LeBlanc, R.M.; Longhini, A.P.; Le Grice, S.F.J.; Johnson, B.A.; Dayie, T.K. Combining asymmetric 13C-labeling and isotopic filter/edit NOESY: A novel strategy for rapid and logical RNA resonance assignment. Nucleic Acids Res. 2017, 45, e146. [Google Scholar] [CrossRef] [PubMed]
- Olenginski, L.T.; Dayie, T.K. Chemo-enzymatic synthesis of [2-13C, 7-15N]-ATP for facile NMR analysis of RNA. Mon. für Chem. 2020, 151, 1467–1473. [Google Scholar] [CrossRef]
- Taiwo, K.M.; Becette, O.B.; Zong, G.; Chen, B.; Zavalij, P.Y.; Dayie, T.K. Chemo-enzymatic synthesis of 13C- and 19F-labeled uridine-5′-triphosphate for RNA NMR probing. Mon. für Chem.-Chem. Mon. 2021, 1, 3. [Google Scholar] [CrossRef]
- Zhang, W.; Turney, T.; Surjancev, I.; Serianni, A.S. Enzymatic synthesis of ribo- and 2′-deoxyribonucleosides from glycofuranosyl phosphates: An approach to facilitate isotopic labeling. Carbohydr. Res. 2017, 449, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Longhini, A.; Nußbaumer, F.; Kreutz, C.; Dinman, J.; Dayie, T.K. CCR5 RNA Pseudoknots: Residue and Site-Specific Labeling correlate Internal Motions with microRNA Binding. Chem.-A Eur. J. 2018, 24, 5462–5468. [Google Scholar] [CrossRef]
- Wenter, P.; Pitsch, S. Synthesis of Selectively15N-Labeled 2′-O-{[(Triisopropylsilyl)oxy]methyl}(=tom)-Protected Ribonucleoside Phosphoramidites and Their Incorporation into a Bistable 32Mer RNA Sequence. Helv. Chim. Acta 2003, 86, 3955–3974. [Google Scholar] [CrossRef]
- Zhang, X.; Gaffney, B.L.; Jones, R.A. 15N NMR of a Specifically Labeled RNA Fragment Containing Intrahelical GU Wobble Pairs. J. Am. Chem. Soc. 1997, 119, 6432–6433. [Google Scholar] [CrossRef]
- Zhang, X.; Gaffney, B.L.; Jones, R.A. 15N NMR of RNA fragments containing specifically labeled GU and GC pairs. J. Am. Chem. Soc. 1998, 120, 615–618. [Google Scholar] [CrossRef]
- Shallop, A.J.; Gaffney, B.L.; Jones, R.A. Use of Both Direct and Indirect 13C Tags for Probing Nitrogen Interactions in Hairpin Ribozyme Models by 15N NMRI. Nucleosides Nucleotides Nucleic Acids 2004, 23, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Gaffney, B.L.; Jones, R.A. Regioselective 2′-Silylation of Purine Ribonucleosides for Phosphoramidite RNA Synthesis. Curr. Protoc. Nucleic Acid Chem. 2001, 6, 2.8.1–2.8.13. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Neuner, S.; Kreutz, C.; Micura, R. The synthesis of 15N(7)-Hoogsteen face-labeled adenosine phosphoramidite for solid-phase RNA synthesis. Mon. für Chem. 2017, 148, 149–155. [Google Scholar] [CrossRef]
- Neuner, S.; Santner, T.; Kreutz, C.; Micura, R. The “speedy” Synthesis of Atom-Specific 15N Imino/Amido-Labeled RNA. Chem.-A Eur. J. 2015, 21, 11634–11643. [Google Scholar] [CrossRef] [PubMed]
- Kremser, J.; Strebitzer, E.; Plangger, R.; Juen, M.A.; Nußbaumer, F.; Glasner, H.; Breuker, K.; Kreutz, C. Chemical synthesis and NMR spectroscopy of long stable isotope labelled RNA. Chem. Commun. 2017, 53, 12938–12941. [Google Scholar] [CrossRef] [PubMed]
- Juen, M.A.; Wunderlich, C.H.; Nußbaumer, F.; Tollinger, M.; Kontaxis, G.; Konrat, R.; Hansen, D.F.; Kreutz, C. Excited States of Nucleic Acids Probed by Proton Relaxation Dispersion NMR Spectroscopy. Angew. Chem. Int. Ed. 2016, 55, 12008–12012. [Google Scholar] [CrossRef] [PubMed]
- Wunderlich, C.H.; Spitzer, R.; Santner, T.; Fauster, K.; Tollinger, M.; Kreutz, C. Synthesis of (6-13C)pyrimidine nucleotides as spin-labels for RNA dynamics. J. Am. Chem. Soc. 2012, 134, 7558–7569. [Google Scholar] [CrossRef]
- D’Souza, V.; Dey, A.; Habib, D.; Summers, M.F. NMR structure of the 101-nucleotide core encapsidation signal of the moloney murine leukemia virus. J. Mol. Biol. 2004, 337, 427–442. [Google Scholar] [CrossRef]
- Zhang, K.; Keane, S.C.; Su, Z.; Irobalieva, R.N.; Chen, M.; Van, V.; Sciandra, C.A.; Marchant, J.; Heng, X.; Schmid, M.F.; et al. Structure of the 30 kDa HIV-1 RNA Dimerization Signal by a Hybrid Cryo-EM, NMR, and Molecular Dynamics Approach. Structure 2018, 26, 490–498.e3. [Google Scholar] [CrossRef] [PubMed]
- Keane, S.C.; Heng, X.; Lu, K.; Kharytonchyk, S.; Ramakrishnan, V.; Carter, G.; Barton, S.; Hosic, A.; Florwick, A.; Santos, J.; et al. Structure of the HIV-1 RNA packaging signal. Science 2015, 348, 917–921. [Google Scholar] [CrossRef]
- Marchant, J.; Bax, A.; Summers, M.F. Accurate Measurement of Residual Dipolar Couplings in Large RNAs by Variable Flip Angle NMR. J. Am. Chem. Soc. 2018, 140, 6978–6983. [Google Scholar] [CrossRef] [PubMed]
- Beaucage, S.L.; Caruthers, M.H. Deoxynucleoside phosphoramidites-A new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981, 22, 1859–1862. [Google Scholar] [CrossRef]
- Scaringe, S.A.; Kitchen, D.; Kaiser, R.J.; Marshall, W.S. Preparation of 5′-silyl-2′-orthoester ribonucleosides for use in oligoribonucleotide synthesis. Curr. Protoc. Nucleic Acid Chem. 2004, 16, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, M.E.; Breaker, R.R.; Asteriadis, G.T.; deBear, J.S.; Gough, G.R. Rapid synthesis of oligoribonucleotides using 2′-O-(o-nitrobenzyloxymethyl)-protected monomers. Bioorg. Med. Chem. Lett. 1992, 2, 1019–1024. [Google Scholar] [CrossRef]
- Pitsch, S.; Weiss, P.; Jenny, L.; Stutz, A.; Wu, X. Reliable Chemical Synthesis of Oligoribonucleotides (RNA) with 2′-O-[(Triisopropylsilyl)oxy]methyl(2′-O-tom)-Protected Phosphoramidites. Helv. Chim. Acta 2001, 84, 3773–3795. [Google Scholar] [CrossRef]
- Shiba, Y.; Masuda, H.; Watanabe, N.; Ego, T.; Takagaki, K.; Ishiyama, K.; Ohgi, T.; Yano, J. Chemical synthesis of a very long oligoribonucleotide with 2-cyanoethoxymethyl (CEM) as the 2′-O-protecting group: Structural identification and biological activity of a synthetic 110mer precursor-microRNA candidate. Nucleic Acids Res. 2007, 35, 3287–3296. [Google Scholar] [CrossRef]
- Krieg, P.A.; Melton, D.A. In Vitro RNA Synthesis with SP6 RNA Polymerase. Methods Enzymol. 1987, 155, 397–415. [Google Scholar] [CrossRef]
- Pokrovskaya, I.D.; Gurevich, V.V. In Vitro transcription: Preparative RNA yields in analytical scale reactions. Anal. Biochem. 1994, 220, 420–423. [Google Scholar] [CrossRef] [PubMed]
- William Studier, F.; Rosenberg, A.H.; Dunn, J.J.; Dubendorff, J.W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990, 185, 60–89. [Google Scholar] [CrossRef]
- Coleman, T.M.; Wang, G.; Huang, F. Superior 5′ homogeneity of RNA from ATP-initiated transcription under the T7 phi 2.5 promoter. Nucleic Acids Res. 2004, 32, e14. [Google Scholar] [CrossRef] [PubMed]
- Pleiss, J.A.; Derrick, M.L.; Uhlenbeck, O.C. T7 RNA polymerase produces 5′ end heterogeneity during in vitro transcription from certain templates. RNA 1998, 4, 1313–1317. [Google Scholar] [CrossRef] [PubMed]
- Helm, M.; Brule, H.; Giege, R.; Florentz, C. More mistakes by T7 RNA polymerase at the 5′ ends of in vitro- transcribed RNAs. RNA 1999, 5, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Krupp, G. RNA synthesis: Strategies for the use of bacteriophage RNA polymerases. Gene 1988, 72, 75–89. [Google Scholar] [CrossRef]
- Ferré-D’Amaré, A.R.; Doudna, J.A. Use of cis- and trans-ribozymes to remove 5′ and 3′ heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 1996, 24, 977–978. [Google Scholar] [CrossRef] [PubMed]
- Grosshans, C.A.; Cech, T.R. A hammerhead ribozyme allows synthesis of a new form of the Tetrahymena ribozyme homogeneous in length with a 3′ end blocked for transesterification. Nucleic Acids Res. 1991, 19, 3875–3880. [Google Scholar] [CrossRef]
- Kao, C.; Zheng, M.; Rüdisser, S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3′ terminus of RNAs transcribed by T7 RNA polymerase. RNA 1999, 5, 1268–1272. [Google Scholar] [CrossRef]
- Brieba, L.G.; Sousa, R. Roles of Histidine 784 and Tyrosine 639 in Ribose Discrimination by T7 RNA Polymerase. Biochemistry 2000, 39, 919–923. [Google Scholar] [CrossRef]
- Padilla, R.; Sousa, R. A Y639F/H784A T7 RNA polymerase double mutant displays superior properties for synthesizing RNAs with non-canonical NTPs. Nucleic Acids Res. 2002, 30, e138. [Google Scholar] [CrossRef]
- Sousa, R.; Padilla, R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995, 14, 4609–4621. [Google Scholar] [CrossRef] [PubMed]
- Kostyuk, D.A.; Dragan, S.M.; Lyakhov, D.L.; Rechinsky, V.O.; Tunitskaya, V.L.; Chernov, B.K.; Kochetkov, S.N. Mutants of T7 RNA polymerase that are able to synthesize both RNA and DNA. FEBS Lett. 1995, 369, 165–168. [Google Scholar] [CrossRef]
- Wu, M.Z.; Asahara, H.; Tzertzinis, G.; Roy, B. Synthesis of low immunogenicity RNA with high-temperature in vitro transcription. RNA 2020, 26, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yu, P.; Leproust, E.; Gao, X. An efficient and economic site-specific deuteration strategy for NMR studies of homologous oligonucleotide repeat sequences. Nucleic Acids Res. 1997, 25, 4758–4763. [Google Scholar] [CrossRef] [PubMed]
- Guilleres, J.; Lopez, P.J.; Proux, F.; Launay, H.; Dreyfus, M. A mutation in T7 RNA polymerase that facilitates promoter clearance. Proc. Natl. Acad. Sci. USA 2005, 102, 5958–5963. [Google Scholar] [CrossRef]
- Salvail-Lacoste, A.; Di Tomasso, G.; Piette, B.L.; Legault, P. Affinity purification of T7 RNA transcripts with homogeneous ends using ARiBo and CRISPR tags. RNA 2013, 19, 1003–1014. [Google Scholar] [CrossRef]
- Moore, M.J.; Query, C.C. Joining of RNAs by splinted ligation. Methods Enzymol. 2000, 317, 109–123. [Google Scholar] [CrossRef]
- Porecha, R.; Herschlag, D. RNA radiolabeling. In Methods in Enzymology; Academic Press Inc.: Cambridge, MA, USA, 2013; Volume 530, pp. 255–279. ISBN 9780124200371. [Google Scholar]
- Romaniuk, P.J.; Uhlenbeck, O.C. Joining of RNA molecules with RNA ligase. Methods Enzymol. 1983, 100, 52–59. [Google Scholar] [CrossRef]
- Bain, J.D.; Switzer, C. Regioselective ligation of oligoribonucleotides using DNA splints. Nucleic Acids Res. 1992, 20, 4372. [Google Scholar] [CrossRef]
- Stark, M.R.; Pleiss, J.A.; Deras, M.; Scaringe, S.A.; Rader, S.D. An RNA ligase-mediated method for the efficient creation of large, synthetic RNAs. RNA 2006, 12, 2014–2019. [Google Scholar] [CrossRef]
- Kim, I.; Lukavsky, P.J.; Puglisi, J.D. NMR study of 100 kDa HCV IRES RNA, using segmental isotope labeling. J. Am. Chem. Soc. 2002, 124, 9338–9339. [Google Scholar] [CrossRef] [PubMed]
- Tzakos, A.G.; Easton, L.E.; Lukavsky, P.J. Complementary segmental labeling of large RNAs: Economic preparation and simplified NMR spectra for measurement of more RDCs. J. Am. Chem. Soc. 2006, 128, 13344–13345. [Google Scholar] [CrossRef]
- Nelissen, F.H.T.; van Gammeren, A.J.; Tessari, M.; Girard, F.C.; Heus, H.A.; Wijmenga, S.S. Multiple segmental and selective isotope labeling of large RNA for NMR structural studies. Nucleic Acids Res. 2008, 36, e89. [Google Scholar] [CrossRef] [PubMed]
- Duss, O.; Maris, C.; Von Schroetter, C.; Dé, F.; Allain, H.-T. A fast, efficient and sequence-independent method for flexible multiple segmental isotope labeling of RNA using ribozyme and RNase H cleavage. Nucleic Acids Res. 2010, 38, e188. [Google Scholar] [CrossRef]
- Liu, Y.; Holmstrom, E.; Zhang, J.; Yu, P.; Wang, J.; Dyba, M.A.; Chen, D.; Ying, J.; Lockett, S.; Nesbitt, D.J.; et al. Synthesis and applications of RNAs with position-selective labelling and mosaic composition. Nature 2015, 522, 368–372. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, P.; Dyba, M.; Sousa, R.; Stagno, J.R.; Wang, Y.X. Applications of PLOR in labeling large RNAs at specific sites. Methods 2016, 103, 4–10. [Google Scholar] [CrossRef]
- Stagno, J.R.; Yu, P.; Dyba, M.A.; Wang, Y.X.; Liu, Y. Heavy-atom labeling of RNA by PLOR for de novo crystallographic phasing. PLoS ONE 2019, 14, e0215555. [Google Scholar] [CrossRef]
- Liu, Y.; Holmstrom, E.; Yu, P.; Tan, K.; Zuo, X.; Nesbitt, D.J.; Sousa, R.; Stagno, J.R.; Wang, Y.X. Incorporation of isotopic, fluorescent, and heavy-atom-modified nucleotides into RNAs by position-selective labeling of RNA. Nat. Protoc. 2018, 13, 987–1005. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, M.; Liu, Y. Optimization and characterization of position-selective labelling of RNA (PLOR) for diverse RNA and DNA sequences. RNA Biol. 2020, 17, 1009–1017. [Google Scholar] [CrossRef]
- Martin, C.T.; Muller, D.K.; Coleman, J.E. Processivity in Early Stages of Transcription by T7 RNA Polymerase. Biochemistry 1988, 27, 3966–3974. [Google Scholar] [CrossRef] [PubMed]
- Imburgio, D.; Rong, M.; Ma, K.; McAllister, W.T. Studies of promoter recognition and start site selection by T7 RNA polymerase using a comprehensive collection of promoter variants. Biochemistry 2000, 39, 10419–10430. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, P.R.; Weitzmann, C.J.; Ofengand, J. SP6 RNA polymerase stutters when initiating from an AAA… sequence. Nucleic Acids Res. 1991, 19, 4669–4673. [Google Scholar] [CrossRef] [PubMed]
- Keyhani, S.; Goldau, T.; Blümler, A.; Heckel, A.; Schwalbe, H. Chemo-Enzymatic Synthesis of Position-Specifically Modified RNA for Biophysical Studies including Light Control and NMR Spectroscopy. Angew. Chem. Int. Ed. 2018, 57, 12017–12021. [Google Scholar] [CrossRef] [PubMed]
- Kappel, K.; Zhang, K.; Su, Z.; Watkins, A.M.; Kladwang, W.; Li, S.; Pintilie, G.; Topkar, V.V.; Rangan, R.; Zheludev, I.N.; et al. Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures. Nat. Methods 2020, 17, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Zhang, K.; Kappel, K.; Li, S.; Palo, M.Z.; Pintilie, G.D.; Rangan, R.; Luo, B.; Wei, Y.; Das, R.; et al. Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 Å resolution. Nature 2021, 596, 603. [Google Scholar] [CrossRef] [PubMed]
- Seffernick, J.T.; Lindert, S. Hybrid methods for combined experimental and computational determination of protein structure. J. Chem. Phys. 2020, 153, 240901. [Google Scholar] [CrossRef]
- Shimada, I.; Ueda, T.; Kofuku, Y.; Eddy, M.T.; Wüthrich, K. GPCR drug discovery: Integrating solution NMR data with crystal and cryo-EM structures. Nat. Rev. Drug Discov. 2018, 18, 59–82. [Google Scholar] [CrossRef]
- Gauto, D.F.; Estrozi, L.F.; Schwieters, C.D.; Effantin, G.; Macek, P.; Sounier, R.; Sivertsen, A.C.; Schmidt, E.; Kerfah, R.; Mas, G.; et al. Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex. Nat. Commun. 2019, 10, 2697. [Google Scholar] [CrossRef]
- Pratap Barnwal, R.; Loh, E.; Godin, K.S.; Yip, J.; Lavender, H.; Tang, C.M.; Varani, G. Structure and mechanism of a molecular rheostat, an RNA thermometer that modulates immune evasion by Neisseria meningitidis. Nucleic Acids Res. 2016, 44, 9426–9437. [Google Scholar] [CrossRef]




| Building Block | Price a ($) | Supplier b |
|---|---|---|
| Uniformly 2H-labeled rNTPs | ||
| rNTP (N = A, C, G, or U) | 1300 | Silantes |
| Selectively 2H-labeled rNTPs | ||
| [3′,4′,5′,5″-2H4]-X (X = ATP or GTP) | 800 | CIL |
| [2-2H]-ATP | 1200 | CIL |
| [5,1′,2′,3′,4′,5′,5″-2H7]-CTP | 1800 | CIL |
| [5,3′,4′,5′,5″-2H5]-Y- (CTP or UTP) | 800 | CIL |
| [5,6-2H2]-CTP | 1800 | CIL |
| [5,1′,2′,3′,4′,5′,5″-2H7]-UTP | 1400 | CIL |
| Uniformly 13C-labeled rNTPs and Amidites | ||
| rNTP (N = A, C, G, or U) | 1400 | Silantes |
| N (N = A, C, G, or U Amidites) | 6600 | Silantes |
| Selectively 13C-labeled rNTPs and Amidites | ||
| [8-13C]- X (X = ATP or GTP) | 1400 | Silantes |
| [8-13C]-A | 900 | Silantes |
| [2,8-13C2]-A | 2700 | Silantes |
| [8-13C]-G | 1000 | INNotope |
| Selectively 2H/13C-labeled rNTPs and Amidites | ||
| [6-13C-5-2H]- Y (Y = CTP or UTP) | 1600 | Silantes |
| [6-13C-5-2H]-C or U | 1000 | INNotope |
| Uniformly 15N-labeled rNTPs and Amidites | ||
| rNTP (N = A, C, G, or U) | 900 | CIL |
| A, or G, or C, or U | 1400 | Silantes |
| Selectively 15N-labeled Amidites | ||
| [1-15N]-A | 1000 | INNotope |
| [1-15N]-G | 1100 | INNotope |
| [3-15N]-C- or U | 1000 | Silantes |
| [1,3-15N2]-C | 1200 | Silantes |
| [1,3,4-15N3]-C | 1000 | INNotope |
| [1,3-15N2]-U | 1000 | INNotope |
| Uniformly 2H/15N-labeled rNTPs | ||
| rNTP (N = A, C, G, or U) | 5600 | Silantes |
| Uniformly 13C/15N-labeled rNTPs and Amidites | ||
| rNTP (N = A, C, G, or U) | 1100 | CIL |
| N (N = A, C, G, or U Amidites) | 5300 | Silantes |
| Enzyme a | Gene | EC Number | PDB ID | Organism b |
|---|---|---|---|---|
| Hexokinase | hxk1/2 | 2.7.1.1 | 1HKG | Saccharomyces cerevisiae |
| Glucokinase | glk | 2.7.1.2 | 1Q18 | Escherichia coli |
| Glucose-6-phosphate dehydrogenase | zwf1 | 1.1.1.49 | 2BHL | Homo sapiens |
| Phosphogluconate dehydrogenase | gndA | 1.1.1.44 | 2ZYA | Escherichia coli K-12 |
| Ribose-5-phosphate isomerase | rpiA | 5.3.1.6 | 1O8B | Escherichia coli |
| Ribose-phosphate diphosphate kinase | prsA | 2.7.6.1 | 3Q89 | Staphylococcus aureus |
| Amido phosphoribosyl-transferase | purF | 2.4.2.14 | IECF | Escherichia coli |
| Phosphoribosylamine-glycine ligase | purD | 6.3.4.13 | 5VEV | Neisseria gonnorhea |
| Phosphoribosylglycinamide formyltransferase | purN | 2.1.2.2 | 3P9X | Bacillus halodurans |
| Phosphoribosylformylglycinamidine synthase | purL | 6.3.5.3 | 1VK3 | Thermotoga maritima |
| Phosphoribosylformylglycinamidine cyclo-ligase | purM | 6.3.3.1 | 5VK4 | Neisseria gonorrhoea |
| Phosphoribosylamino-imidazole carboxylase (catalytic subunit) | purE | 4.1.1.21 | 4GRD | Burkholderia cenocepacia |
| Phosphoribosylamino-imidazole carboxylase (ATPase subunit) | purK | 4.1.1.21 | 2Z04 | Aquifex aeolicus |
| Phosphoribosylamino-imidazole-succinocarboxamide synthase | purC | 6.3.2.6 | 3NUA | Clostridium perfringens |
| Adenylosuccinate lyase | purB | 4.3.2.2 | 3GZH | Escherichia coli |
| Phosphoribosylamino-imidazole-carboxamide formyltransferase | purH | 2.1.2.3 | 1ZCZ | Thermotoga maritima |
| Inosine-monophosphate cyclohydrolase | purH | 3.5.4.10 | 2IU0 | Gallus gallus |
| Adenylosuccinate synthase | purA | 6.3.4.4 | 2J91 | Homo sapiens |
| Inosine-monophosphate dehydrogenase | guaB | 1.1.1.205 | 1B30 | Homo sapiens |
| Guanosine-monophosphate synthase | guaA | 6.3.5.2 | 2YWB | Thermus thermophilus |
| Adenylate kinase | plsA | 2.7.4.3 | 1E4Y | Escherichia coli |
| Creatine phosphokinase | ckmT | 2.7.3.2 | 2CRK | Oryctolagus cuniculus |
| Guanylate kinase | spoR | 2.7.4.8 | 2ANC | Escherichia coli |
| Glycine hydroxymethyltransferase | glyA | 2.1.2.1 | 5VMB | Acinetobacter baumannii |
| Methylene-tetrahydrofolate dehydrogenase | folD | 1.5.1.5 | 1B0A | Escherichia coli K-12 |
| Methenyl-tetrahydrofolate cyclohydrolase | folD | 3.5.4.9 | 5TCA | Homo sapiens |
| Aspartate ammonia-lyase | aspA | 4.3.1.1 | 1JSW | Escherichia coli |
| Glutamate dehydrogenase (NAD(P)+) | glud1/ghA | 1.4.1.3 | 4fcc | Escherichia coli |
| Glutamine syntethase | glnA | 6.3.1.2 | 4IS4 | Medicago truncatula |
| Inorganic diphosphatase | ppa | 3.6.1.1 | 1IPW | Escherichia coli |
| Enzyme a | Gene | EC Number | PDB ID | Availability | Organism a |
|---|---|---|---|---|---|
| T7 RNA polymerase | 1 | 2.7.7.6 | 1ARO | Thermo Fisher Scientific | Escherichia coli |
| T4 DNA ligase | 1.3 | 6.5.1.1 | 5WFY | Sigma-Aldrich | Escherichia coli |
| T4 Polynucleotide kinase | pseT | 2.7.1.78 | 1LY1 | Thermo Fisher Scientific | Escherichia coli |
| T4 RNA ligase | 63 | 6.5.1.3 | 5TT6 | Thermo Fisher Scientific | Escherichia coli |
| Alkaline phosphatase | ALPL | 3.1.3.1 | 1K7H | New England Biolabs | Pandalus borealis |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olenginski, L.T.; Taiwo, K.M.; LeBlanc, R.M.; Dayie, T.K. Isotope-Labeled RNA Building Blocks for NMR Structure and Dynamics Studies. Molecules 2021, 26, 5581. https://doi.org/10.3390/molecules26185581
Olenginski LT, Taiwo KM, LeBlanc RM, Dayie TK. Isotope-Labeled RNA Building Blocks for NMR Structure and Dynamics Studies. Molecules. 2021; 26(18):5581. https://doi.org/10.3390/molecules26185581
Chicago/Turabian StyleOlenginski, Lukasz T., Kehinde M. Taiwo, Regan M. LeBlanc, and Theodore K. Dayie. 2021. "Isotope-Labeled RNA Building Blocks for NMR Structure and Dynamics Studies" Molecules 26, no. 18: 5581. https://doi.org/10.3390/molecules26185581
APA StyleOlenginski, L. T., Taiwo, K. M., LeBlanc, R. M., & Dayie, T. K. (2021). Isotope-Labeled RNA Building Blocks for NMR Structure and Dynamics Studies. Molecules, 26(18), 5581. https://doi.org/10.3390/molecules26185581






