Sample Preparation and Diagnostic Methods for a Variety of Settings: A Comprehensive Review
Abstract
1. Introduction
2. Medical Diagnosis
2.1. Diagnostic Methods and Their Importance
2.2. Obstacles and Considerations for Diagnostic Methods
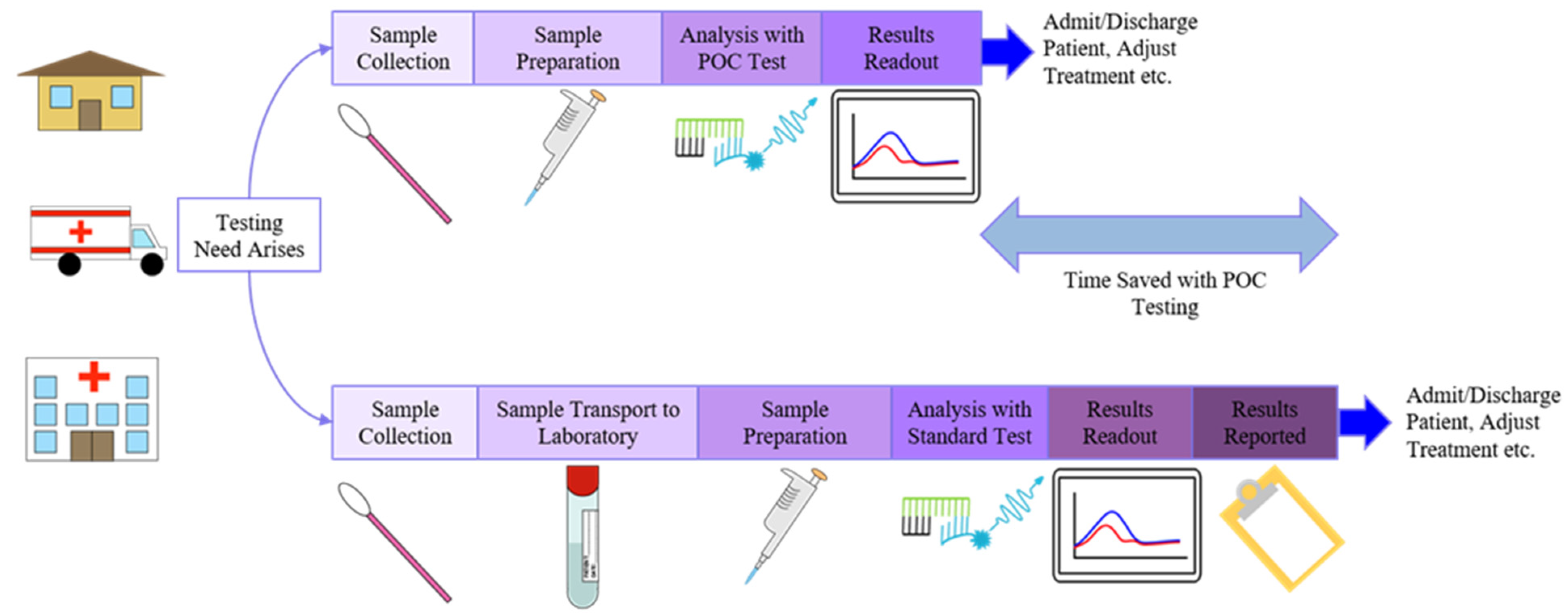
2.3. Point-of-Care Diagnostics
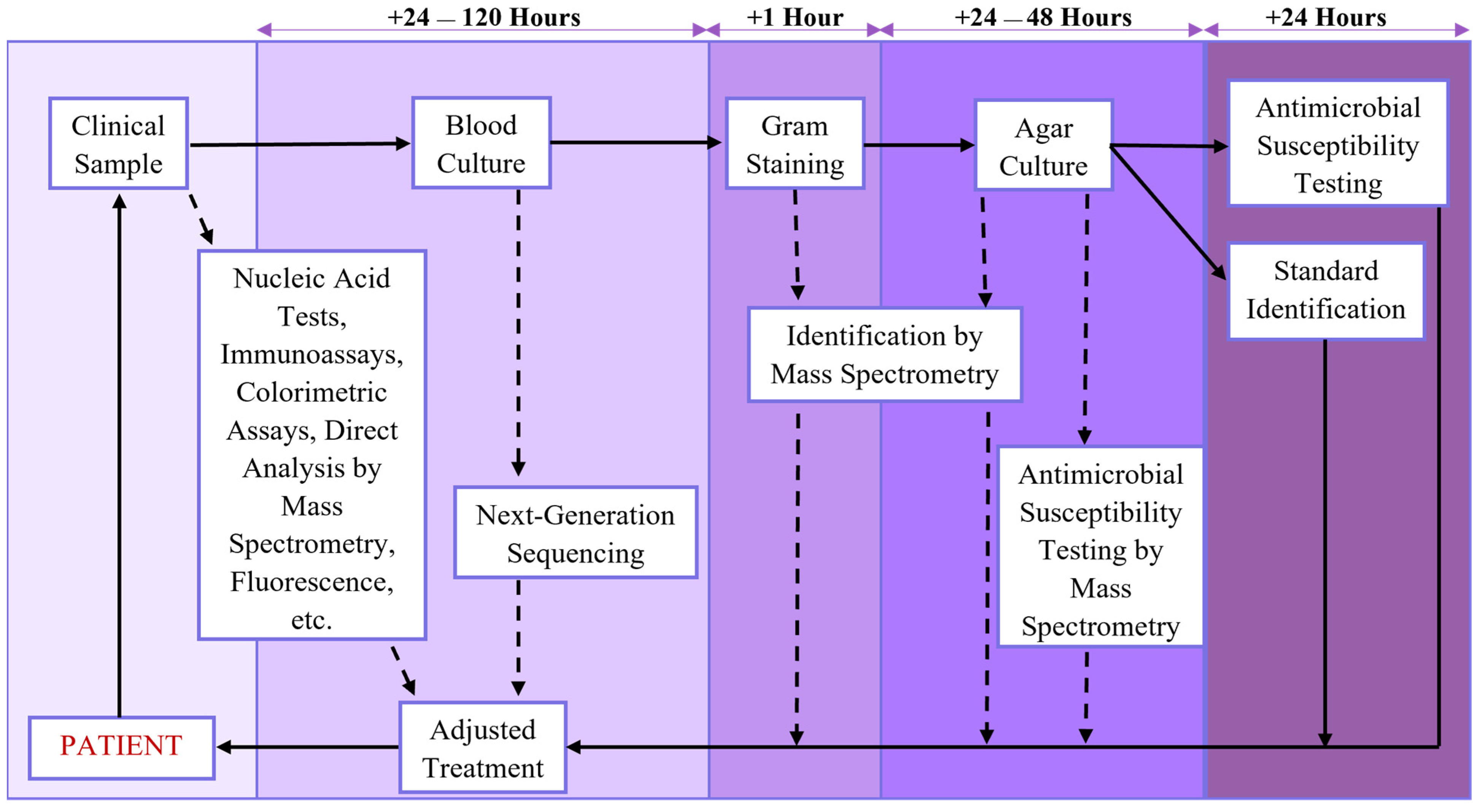
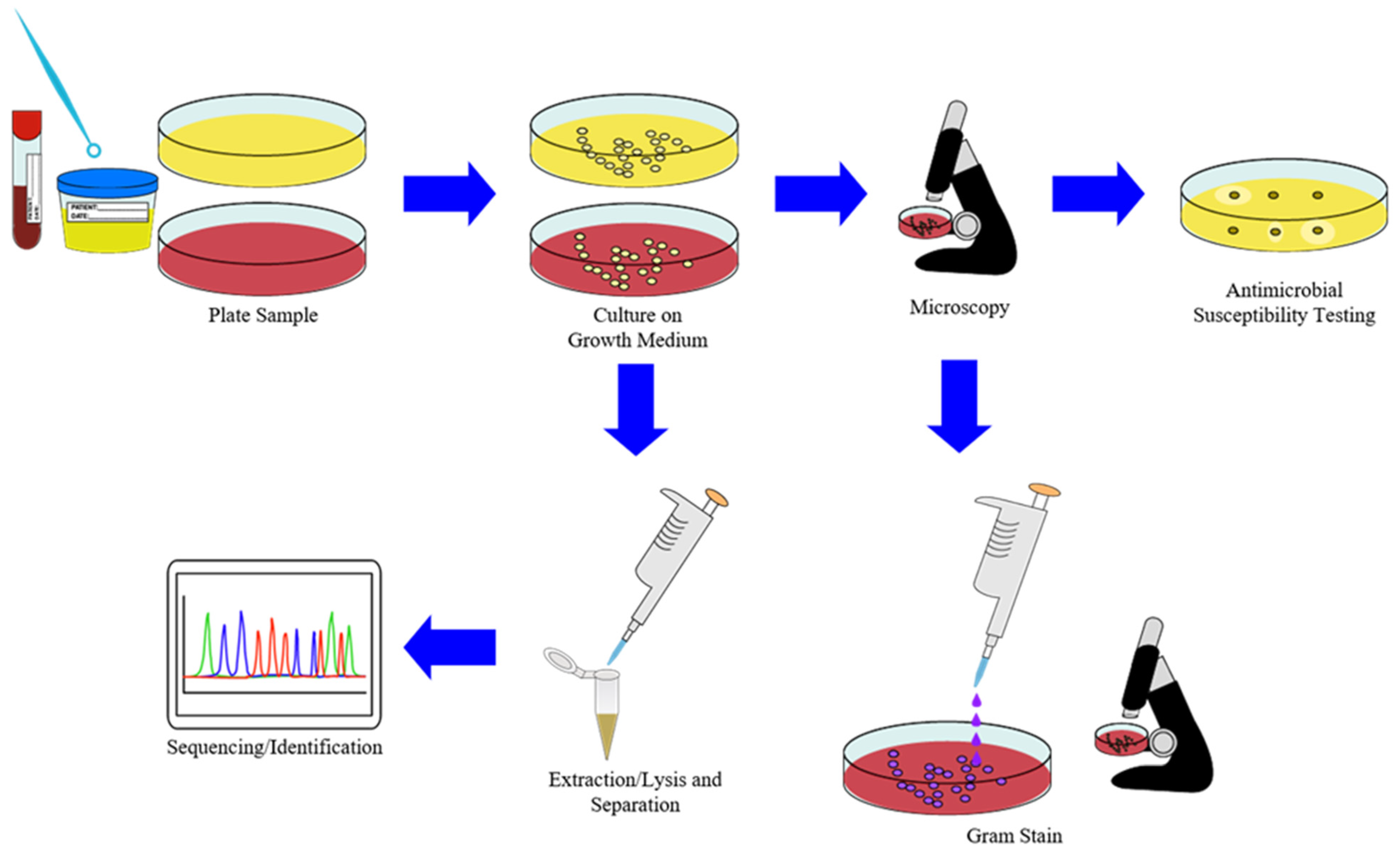
2.4. Sample Preparation in Diagnostics
3. Biosensors
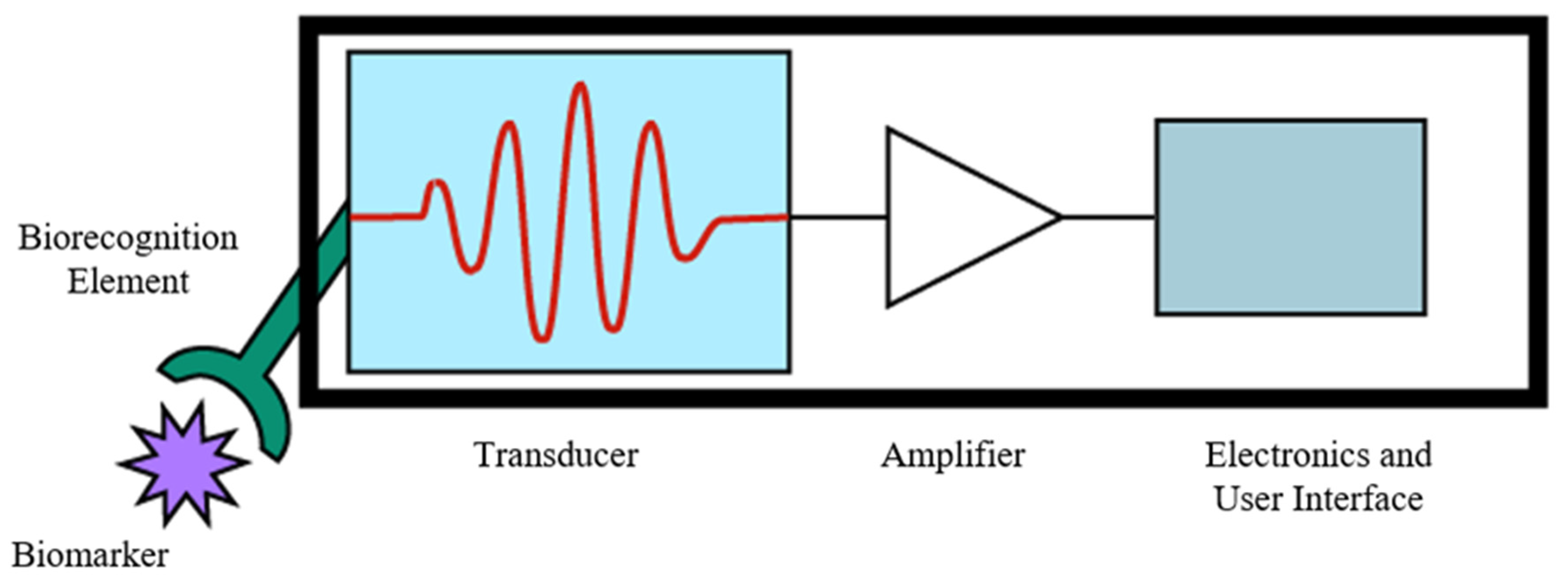
3.1. Biorecognition Elements
3.2. Signal Transduction Methods
3.3. Progress in Biosensors for POCT
4. Integrated and Portable Sample Preparation Devices
4.1. Integrated Sample Preparation Systems
4.2. Standalone Sample Preparation Systems
5. High-Throughput Diagnostic Methods in Laboratory Settings
Modified Microplates and Laboratory Automation for Diagnostic Sample Preparation
6. Closing Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Y.; Guo, Z.; Wang, X.; Qiu, C. Sample preparation. J. Chromatogr. A 2008, 1184, 191–219. [Google Scholar] [CrossRef]
- Danzer, K. Analytical Chemistry: Theoretical and Metrological Fundamentals; Springer: Berlin/Heidelberg, Germany, 2007; pp. 23–26. [Google Scholar]
- Hyötyläinen, T. Critical evaluation of sample pretreatment techniques. Anal. Bioanal. Chem. 2009, 394, 743–758. [Google Scholar] [CrossRef] [PubMed]
- Ramos, L. Critical overview of selected contemporary sample preparation techniques. J. Chromatogr. A 2012, 1221, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, S.; Dieh, D. Sampling and sample preparation. In Modern Instrumental Analysis; Ahuja, S., Jespersen, N., Eds.; Comprehensive analytical chemistry; Elsevier: Amsterdam, The Netherlands, 2006; Volume 47, pp. 15–40. [Google Scholar]
- Pawliszyn, J. Sampling and Sample Preparation for Field and Laboratory; Pawliszyn, J., Ed.; Comprehensive analytical chemistry; Elsevier: Amsterdam, The Netherlands, 2002; Volume 37. [Google Scholar]
- Wuethrich, A.; Haddad, P.R.; Quirino, J.P. The electric field—An emerging driver in sample preparation. TrAC Trends Anal. Chem. 2016, 80, 604–611. [Google Scholar] [CrossRef]
- Xu, L.; Qi, X.; Li, X.; Bai, Y.; Liu, H. Recent advances in applications of nanomaterials for sample preparation. Talanta 2016, 146, 714–726. [Google Scholar] [CrossRef] [PubMed]
- Moreda-Piñeiro, J.; Moreda-Piñeiro, A. Recent advances in combining microextraction techniques for sample pre-treatment. TrAC Trends Anal. Chem. 2015, 71, 265–274. [Google Scholar] [CrossRef]
- Buszewski, B.; Szultka, M. Past, present, and future of solid phase extraction: A review. Crit. Rev. Anal. Chem. 2012, 42, 198–213. [Google Scholar] [CrossRef]
- Teo, C.C.; Chong, W.P.K.; Ho, Y.S. Development and application of microwave-assisted extraction technique in biological sample preparation for small molecule analysis. Metabolomics 2013, 9, 1109–1128. [Google Scholar] [CrossRef]
- Xue, G.; Lung, K.R. Automation and sample preparation. In Sample Preparation of Pharmaceutical Dosage Forms; Nickerson, B., Ed.; Springer: Boston, MA, USA, 2011; pp. 285–307. [Google Scholar]
- Plebani, M.; Sciacovelli, L.; Aita, A.; Padoan, A.; Chiozza, M.L. Quality indicators to detect pre-analytical errors in laboratory testing. Clin. Chim. Acta 2014, 432, 44–48. [Google Scholar] [CrossRef]
- Płotka-Wasylka, J.; Szczepańska, N.; de la Guardia, M.; Namieśnik, J. Miniaturized solid-phase extraction techniques. TrAC Trends Anal. Chem. 2015, 73, 19–38. [Google Scholar] [CrossRef]
- Destandau, E.; Michel, T.; Elfakir, C. CHAPTER 4. Microwave-assisted extraction. In Green Chemistry Series; Rostagno, M.A., Prado, J.M., Eds.; Royal Society of Chemistry: Cambridge, UK, 2013; pp. 113–156. [Google Scholar]
- Adeli, K. Laboratory medicine—A hidden treasure in healthcare. Clin. Biochem. 2017, 50, 645–647. [Google Scholar] [CrossRef] [PubMed]
- Bossuyt, P.M.; Reitsma, J.B.; Linnet, K.; Moons, K.G. Beyond diagnostic accuracy: The clinical utility of diagnostic tests. Clin. Chem. 2012, 58, 1636–1643. [Google Scholar] [CrossRef]
- Ferraro, S.; Panteghini, M. The role of laboratory in ensuring appropriate test requests. Clin. Biochem. 2017, 50, 555–561. [Google Scholar] [CrossRef]
- Joseph, L.; Cankovic, M.; Caughron, S.; Chandra, P.; Emmadi, R.; Hagenkord, J.; Hallam, S.; Jewell, K.E.; Klein, R.D.; Pratt, V.M.; et al. The spectrum of clinical utilities in molecular pathology testing procedures for inherited conditions and cancer. J. Mol. Diagn. 2016, 18, 605–619. [Google Scholar] [CrossRef]
- Fauci, A.S. The perpetual challenge of infectious diseases. N. Engl. J. Med. 2012, 366, 454–461. [Google Scholar] [CrossRef]
- Rentschler, S.; Kaiser, L.; Deigner, H.-P. Emerging options for the diagnosis of bacterial infections and the characterization of antimicrobial resistance. Int. J. Mol. Sci. 2021, 22, 456. [Google Scholar] [CrossRef]
- Schlossberg, D. Clinical Infectious Disease, 2nd ed.; Cambridge University Press: Cambridge, UK, 2015. [Google Scholar]
- Kaya, A.; Ergul, N.; Kaya, S.Y.; Kilic, F.; Yilmaz, M.H.; Besirli, K.; Ozaras, R. The management and the diagnosis of fever of unknown origin. Expert Rev. Anti-Infect. Ther. 2013, 11, 805–815. [Google Scholar] [CrossRef]
- Tenover, F.C. The role for rapid molecular diagnostic tests for infectious diseases in precision medicine. Expert Rev. Precis. Med. Drug Dev. 2018, 3, 69–77. [Google Scholar] [CrossRef]
- Srivastava, S.; Singh, P.K.; Vatsalya, V.; Karch, R.C. Developments in the diagnostic techniques of infectious diseases: Rural and urban prospective. Adv. Infect. Dis. 2018, 8, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.B.; Petrich, A.; Smieja, M. Molecular diagnosis of respiratory virus infections. Crit. Rev. Clin. Lab. Sci. 2011, 48, 217–249. [Google Scholar] [CrossRef] [PubMed]
- Caliendo, A.M.; Gilbert, D.N.; Ginocchio, C.C.; Hanson, K.E.; May, L.; Quinn, T.C.; Tenover, F.C.; Alland, D.; Blaschke, A.J.; Bonomo, R.A.; et al. Better tests, better care: Improved diagnostics for infectious diseases. Clin. Infect. Dis. 2013, 57 (Suppl. S3), S139–S170. [Google Scholar] [CrossRef] [PubMed]
- Goff, D.A.; Jankowski, C.; Tenover, F.C. Using rapid diagnostic tests to optimize antimicrobial selection in antimicrobial stewardship programs. Pharmacotherapy 2012, 32, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Greatorex, J.; Ellington, M.J.; Koser, C.U.; Rolfe, K.J.; Curran, M.D. New methods for identifying infectious diseases. Br. Med. Bull. 2014, 112, 27–35. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cui, F.; Rhee, M.; Singh, A.; Tripathi, A. Microfluidic sample preparation for medical diagnostics. Annu. Rev. Biomed. Eng. 2015, 17, 267–286. [Google Scholar] [CrossRef] [PubMed]
- Lim, W.Y.; Goh, B.T.; Khor, S.M. Microfluidic Paper-based analytical devices for potential use in quantitative and direct detection of disease biomarkers in clinical analysis. J. Chromatogr. B 2017, 1060, 424–442. [Google Scholar] [CrossRef]
- Nilghaz, A.; Guan, L.; Tan, W.; Shen, W. Advances of paper-based microfluidics for diagnostics—The original motivation and current status. ACS Sens. 2016, 1, 1382–1393. [Google Scholar] [CrossRef]
- Gong, M.M.; Sinton, D. Turning the page: Advancing paper-based microfluidics for broad diagnostic application. Chem. Rev. 2017, 117, 8447–8480. [Google Scholar] [CrossRef]
- Ming, T.; Luo, J.; Liu, J.; Sun, S.; Xing, Y.; Wang, H.; Xiao, G.; Deng, Y.; Cheng, Y.; Yang, Z.; et al. Paper-based microfluidic aptasensors. Biosens. Bioelectron. 2020, 170, 112649. [Google Scholar] [CrossRef]
- Boers, S.A.; Jansen, R.; Hays, J.P. Understanding and overcoming the pitfalls and biases of Next-Generation Sequencing (NGS) methods for use in the routine clinical microbiological diagnostic laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1059–1070. [Google Scholar] [CrossRef]
- Grumaz, S.; Stevens, P.; Grumaz, C.; Decker, S.O.; Weigand, M.A.; Hofer, S.; Brenner, T.; von Haeseler, A.; Sohn, K. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 2016, 8, 73. [Google Scholar] [CrossRef]
- Buchan, B.W.; Ledeboer, N.A. Emerging technologies for the clinical microbiology laboratory. Clin. Microbiol. Rev. 2014, 27, 783–822. [Google Scholar] [CrossRef]
- França, R.F.O.; Silva, C.C.; Paula, S.O. Recent advances in molecular medicine techniques for the diagnosis, prevention, and control of infectious diseases. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 723–728. [Google Scholar] [CrossRef]
- Swiner, D.J.; Jackson, S.; Burris, B.J.; Badu-Tawiah, A.K. Applications of mass spectrometry for clinical diagnostics: The influence of turnaround time. Anal. Chem. 2020, 92, 183–202. [Google Scholar] [CrossRef]
- Van Belkum, A.; Dunne, W.M. Next-generation antimicrobial susceptibility testing. J. Clin. Microbiol. 2013, 51, 2018–2024. [Google Scholar] [CrossRef]
- Alexovič, M.; Urban, P.L.; Tabani, H.; Sabo, J. Recent Advances in robotic protein sample preparation for clinical analysis and other biomedical applications. Clin. Chim. Acta 2020, 507, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Choi, S. Powering point-of-care diagnostic devices. Biotechnol. Adv. 2016, 34, 321–330. [Google Scholar] [CrossRef]
- Sow, W.T.; Ye, F.; Zhang, C.; Li, H. Smart materials for point-of-care testing: From sample extraction to analyte sensing and readout signal generator. Biosens. Bioelectron. 2020, 170, 112682. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Roy, I.; Yang, C.; Prasad, P.N. Nanochemistry and nanomedicine for nanoparticle-based diagnostics and therapy. Chem. Rev. 2016, 116, 2826–2885. [Google Scholar] [CrossRef]
- Lee, H.; Shin, T.-H.; Cheon, J.; Weissleder, R. Recent developments in magnetic diagnostic systems. Chem. Rev. 2015, 115, 10690–10724. [Google Scholar] [CrossRef]
- Tokel, O.; Inci, F.; Demirci, U. Advances in plasmonic technologies for point of care applications. Chem. Rev. 2014, 114, 5728–5752. [Google Scholar] [CrossRef] [PubMed]
- Zarei, M. Portable biosensing devices for point-of-care diagnostics: Recent developments and applications. TrAC Trends Anal. Chem. 2017, 91, 26–41. [Google Scholar] [CrossRef]
- Mauriz, E.; Dey, P.; Lechuga, L.M. Advances in nanoplasmonic biosensors for clinical applications. Analyst 2019, 144, 7105–7129. [Google Scholar] [CrossRef] [PubMed]
- Romeo, A.; Leung, T.S.; Sánchez, S. Smart biosensors for multiplexed and fully integrated point-of-care diagnostics. Lab Chip 2016, 16, 1957–1961. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Huang, X.; Guo, J.; Ma, X. Automatic smartphone-based microfluidic biosensor system at the point of care. Biosens. Bioelectron. 2018, 110, 78–88. [Google Scholar] [CrossRef]
- Li, M.; Diamandis, E.P. Technology-driven diagnostics: From smart doctor to smartphone. Crit. Rev. Clin. Lab. Sci. 2016, 53, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Ong, D.S.Y.; Poljak, M. Smartphones as mobile microbiological laboratories. Clin. Microbiol. Infect. 2020, 26, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, V.K.; Bakthavathsalam, P.; Bergquist, P.L.; Sunna, A. Smartphone technology facilitates point-of-care nucleic acid diagnosis: A beginner’s guide. Crit. Rev. Clin. Lab. Sci. 2021, 58, 77–100. [Google Scholar] [CrossRef]
- MacEachern, S.J.; Forkert, N.D. Machine learning for precision medicine. Genome 2021, 64, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 2018, 172, 1122–1131.e9. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.; Wu, G.; Suk, H.-I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef]
- Christaki, E. New technologies in predicting, preventing and controlling emerging infectious diseases. Virulence 2015, 6, 558–565. [Google Scholar] [CrossRef]
- Blaschke, A.J.; Hersh, A.L.; Beekmann, S.E.; Ince, D.; Polgreen, P.M.; Hanson, K.E. Unmet diagnostic needs in infectious disease. Diagn. Microbiol. Infect. Dis. 2015, 81, 57–59. [Google Scholar] [CrossRef][Green Version]
- Ford Carleton, P.; Schachter, S.; Parrish, J.A.; Collins, J.M.; Crocker, J.B.; Dixon, R.F.; Edgman-Levitan, S.; Lewandrowski, K.B.; Stahl, J.E.; Klapperich, C.; et al. National Institute of biomedical imaging and bioengineering point-of-care technology research network: Advancing precision medicine. IEEE J. Transl. Eng. Health Med. 2016, 4, 1–14. [Google Scholar] [CrossRef]
- McLaren, Z.M.; Sharp, A.; Hessburg, J.P.; Sarvestani, A.S.; Parker, E.; Akazili, J.; Johnson, T.R.B.; Sienko, K.H. Cost effectiveness of medical devices to diagnose pre-eclampsia in low-resource settings. Dev. Eng. 2017, 2, 99–106. [Google Scholar] [CrossRef]
- Kost, G.J.; Tran, N.K.; Tuntideelert, M.; Kulrattanamaneeporn, S.; Peungposop, N. Katrina, the tsunami, and point-of-care testing: Optimizing rapid response diagnosis in disasters. Am. J. Clin. Pathol. 2006, 126, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Hattori, T.; Chagan-Yasutan, H.; Shiratori, B.; Egawa, S.; Izumi, T.; Kubo, T.; Nakajima, C.; Suzuki, Y.; Niki, T.; Alisjahbana, B.; et al. Development of point-of-care testing for disaster-related infectious diseases. Tohoku J. Exp. Med. 2016, 238, 287–293. [Google Scholar] [CrossRef]
- Manoto, S.; Lugongolo, M.; Govender, U.; Mthunzi-Kufa, P. Point of care diagnostics for HIV in resource limited settings: An overview. Medicina 2018, 54, 3. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhang, H.; Xu, Y.; Laššáková, S.; Korabečná, M.; Neužil, P. PCR Past, present and future. BioTechniques 2020, 69, 317–325. [Google Scholar] [CrossRef]
- Kiselev, D.; Matsvay, A.; Abramov, I.; Dedkov, V.; Shipulin, G.; Khafizov, K. Current Trends in diagnostics of viral infections of unknown etiology. Viruses 2020, 12, 211. [Google Scholar] [CrossRef] [PubMed]
- Niemz, A.; Ferguson, T.M.; Boyle, D.S. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011, 29, 240–250. [Google Scholar] [CrossRef]
- Cummins, B.M.; Ligler, F.S.; Walker, G.M. Point-of-care diagnostics for niche applications. Biotechnol. Adv. 2016, 34, 161–176. [Google Scholar] [CrossRef]
- Juliano, M.; Wason, C. Comparison of point-of-care versus laboratory troponin testing in an emergency department setting. Mil. Med. 2017, 182, e1938–e1940. [Google Scholar] [CrossRef]
- Luppa, P.B.; Bietenbeck, A.; Beaudoin, C.; Giannetti, A. Clinically relevant analytical techniques, organizational concepts for application and future perspectives of point-of-care testing. Biotechnol. Adv. 2016, 34, 139–160. [Google Scholar] [CrossRef]
- Lindholm, C.; Altimiras, J. Point-of-care devices for physiological measurements in field conditions. A smorgasbord of instruments and validation procedures. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2016, 202, 99–111. [Google Scholar] [CrossRef]
- Hart, R.; Mauk, M.; Liu, C.; Qiu, X.; Thompson, J.; Chen, D.; Malamud, D.; Abrams, W.; Bau, H. Point-of-care oral-based diagnostics: Point-of-care oral-based diagnostics. Oral Dis. 2011, 17, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Guner, H.; Ozgur, E.; Kokturk, G.; Celik, M.; Esen, E.; Topal, A.E.; Ayas, S.; Uludag, Y.; Elbuken, C.; Dana, A. A smartphone based Surface Plasmon Resonance Imaging (SPRi) platform for on-site biodetection. Sens. Actuators B Chem. 2017, 239, 571–577. [Google Scholar] [CrossRef]
- Shehadul Islam, M.; Aryasomayajula, A.; Selvaganapathy, P. A review on macroscale and microscale cell lysis methods. Micromachines 2017, 8, 83. [Google Scholar] [CrossRef]
- Sabatier, M.; Bal, A.; Destras, G.; Regue, H.; Quéromès, G.; Cheynet, V.; Lina, B.; Bardel, C.; Brengel-Pesce, K.; Navratil, V.; et al. Comparison of nucleic acid extraction methods for a viral metagenomics analysis of respiratory viruses. Microorganisms 2020, 8, 1539. [Google Scholar] [CrossRef] [PubMed]
- Vanspauwen, M.J.; Wolffs, P.F.G.; Franssen, F.M.E.; Bruggeman, C.A.; Wouters, E.F.M.; Linssen, C.F.M. Comparison of three different techniques for the isolation of viral RNA in sputum. J. Clin. Virol. 2014, 61, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Emaus, M.N.; Varona, M.; Eitzmann, D.R.; Hsieh, S.-A.; Zeger, V.R.; Anderson, J.L. Nucleic acid extraction: Fundamentals of sample preparation methodologies, current advancements, and future endeavors. TrAC Trends Anal. Chem. 2020, 130, 115985. [Google Scholar] [CrossRef]
- Kidd, S.E.; Chen, S.C.-A.; Meyer, W.; Halliday, C.L. A New age in molecular diagnostics for invasive fungal disease: Are we ready? Front. Microbiol. 2020, 10, 2903. [Google Scholar] [CrossRef]
- Kaya, H.O.; Cetin, A.E.; Azimzadeh, M.; Topkaya, S.N. Pathogen detection with electrochemical biosensors: Advantages, challenges and future perspectives. J. Electroanal. Chem. 2021, 882, 114989. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Mauk, M.; Chen, D.; Qiu, X.; Kim, J.; Gale, B.; Bau, H.H. A PCR Reactor with an integrated alumina membrane for nucleic acid isolation. Analyst 2010, 135, 2408. [Google Scholar] [CrossRef] [PubMed]
- Mulberry, G.; Vuillier, A.; Vaidya, M.; Sugaya, K.; Kim, B.N. Handheld battery-operated sample preparation device for QPCR nucleic acid detections using simple contactless pouring. Anal. Methods 2018, 10, 4671–4679. [Google Scholar] [CrossRef]
- Yetisen, A.K.; Akram, M.S.; Lowe, C.R. Paper-based microfluidic point-of-care diagnostic devices. Lab Chip 2013, 13, 2210. [Google Scholar] [CrossRef]
- Noah, N.M. Biosensors and Nanotechnology: Applications in Health Care Diagnostics; Altintas, Z., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018. [Google Scholar]
- Srinivasan, B.; Tung, S. Development and applications of portable biosensors. J. Lab. Autom. 2015, 20, 365–389. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Xie, Q.; Yang, D.; Xiao, H.; Fu, Y.; Tan, Y.; Yao, S. Recent advances in electrochemical glucose biosensors: A review. RSC Adv. 2013, 3, 4473. [Google Scholar] [CrossRef]
- Zhou, W.; Jimmy Huang, P.-J.; Ding, J.; Liu, J. Aptamer-based biosensors for biomedical diagnostics. Analyst 2014, 139, 2627. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, J.; Boyd, B.J. Peptide-based biosensors. Talanta 2015, 136, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Crapnell, R.D.; Dempsey-Hibbert, N.C.; Peeters, M.; Tridente, A.; Banks, C.E. Molecularly imprinted polymer based electrochemical biosensors: Overcoming the challenges of detecting vital biomarkers and speeding up diagnosis. Talanta Open 2020, 2, 100018. [Google Scholar] [CrossRef]
- Bauch, M.; Toma, K.; Toma, M.; Zhang, Q.; Dostalek, J. Plasmon-enhanced fluorescence biosensors: A review. Plasmonics 2014, 9, 781–799. [Google Scholar] [CrossRef] [PubMed]
- Choi, N.; Dang, H.; Das, A.; Sim, M.S.; Chung, I.Y.; Choo, J. SERS biosensors for ultrasensitive detection of multiple biomarkers expressed in cancer cells. Biosens. Bioelectron. 2020, 164, 112326. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.T. Plasmonic biosensors. WIREs Nanomed. Nanobiotechnol. 2015, 7, 152–168. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Leustean, L.; Inci, F.; Zheng, M.; Demirci, U.; Wang, S. Plasmonic-based platforms for diagnosis of infectious diseases at the point-of-care. Biotechnol. Adv. 2019, 37, 107440. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.J.; Saha, T.; Tey, B.T.; Tan, W.S.; Ooi, C.W. Quartz crystal microbalance-based biosensors as rapid diagnostic devices for infectious diseases. Biosens. Bioelectron. 2020, 168, 112513. [Google Scholar] [CrossRef]
- Masson, J.-F. Surface plasmon resonance clinical biosensors for medical diagnostics. ACS Sens. 2017, 2, 16–30. [Google Scholar] [CrossRef]
- Jiang, Z.; Feng, B.; Xu, J.; Qing, T.; Zhang, P.; Qing, Z. Graphene biosensors for bacterial and viral pathogens. Biosens. Bioelectron. 2020, 166, 112471. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, T.; Pan, Y.; Ciacchi, L.C.; Xu, B.; Wei, G. AFM-based force spectroscopy for bioimaging and biosensing. RSC Adv. 2016, 6, 12893–12912. [Google Scholar] [CrossRef]
- Sarkar, D.; Liu, W.; Xie, X.; Anselmo, A.C.; Mitragotri, S.; Banerjee, K. MoS2 field-effect transistor for next-generation label-free biosensors. ACS Nano 2014, 8, 3992–4003. [Google Scholar] [CrossRef]
- Vu, C.-A.; Chen, W.-Y. Field-effect transistor biosensors for biomedical applications: Recent advances and future prospects. Sensors 2019, 19, 4214. [Google Scholar] [CrossRef]
- Rivet, C.; Lee, H.; Hirsch, A.; Hamilton, S.; Lu, H. Microfluidics for medical diagnostics and biosensors. Chem. Eng. Sci. 2011, 66, 1490–1507. [Google Scholar] [CrossRef]
- Yanik, A.A.; Cetin, A.E.; Huang, M.; Artar, A.; Mousavi, S.H.; Khanikaev, A.; Connor, J.H.; Shvets, G.; Altug, H. Seeing protein monolayers with naked eye through plasmonic fano resonances. Proc. Natl. Acad. Sci. USA 2011, 108, 11784–11789. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, Q. Biosensors and bioelectronics on smartphone for portable biochemical detection. Biosens. Bioelectron. 2016, 75, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Emaminejad, S.; Nyein, H.Y.Y.; Challa, S.; Chen, K.; Peck, A.; Fahad, H.M.; Ota, H.; Shiraki, H.; Kiriya, D.; et al. Fully integrated wearable sensor arrays for multiplexed in situ perspiration analysis. Nature 2016, 529, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Campbell, A.S.; de Ávila, B.E.-F.; Wang, J. Wearable biosensors for healthcare monitoring. Nat. Biotechnol. 2019, 37, 389–406. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Anwar, T.B.; Mulchandani, A. Current status, advances, challenges and perspectives on biosensors for COVID-19 diagnosis in resource-limited settings. Sens. Actuators Rep. 2021, 3, 100025. [Google Scholar] [CrossRef]
- Lee, S.H.; Park, S.; Kim, B.N.; Kwon, O.S.; Rho, W.-Y.; Jun, B.-H. Emerging ultrafast nucleic acid amplification technologies for next-generation molecular diagnostics. Biosens. Bioelectron. 2019, 141, 111448. [Google Scholar] [CrossRef] [PubMed]
- Ünal, B.; Camci-Unal, G.; Mahmud, K. Paper-based microfluidic devices: Low-cost platforms for rapid biochemical detection. Mil. Med. 2021, 186 (Suppl. S1), 716–721. [Google Scholar] [CrossRef]
- Cho, B.; Lee, S.H.; Song, J.; Bhattacharjee, S.; Feng, J.; Hong, S.; Song, M.; Kim, W.; Lee, J.; Bang, D.; et al. Nanophotonic cell lysis and polymerase chain reaction with gravity-driven cell enrichment for rapid detection of pathogens. ACS Nano 2019, 13, 13866–13874. [Google Scholar] [CrossRef]
- Kolluri, N.; Albarran, N.; Fan, A.; Olson, A.; Sagar, M.; Young, A.; Gomez-Marquez, J.; Klapperich, C.M. SNAPflex: A Paper-and-plastic device for instrument-free RNA and DNA extraction from whole blood. Lab Chip 2020, 20, 3386–3398. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.; Balhoff, J.; Landwehr, G.; Rahman, S.; Vaithiyanathan, M.; Melvin, A. Microfluidic and paper-based devices for disease detection and diagnostic research. Int. J. Mol. Sci. 2018, 19, 2731. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.W.; Phillips, S.T.; Whitesides, G.M.; Carrilho, E. Diagnostics for the Developing world: Microfluidic paper-based analytical devices. Anal. Chem. 2010, 82, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.J.; Lee, E.-H.; Yoon, Y.; Chua, B.; Son, A. Portable lysis apparatus for rapid single-step DNA extraction of Bacillus subtilis. J. Appl. Microbiol. 2016, 120, 379–387. [Google Scholar] [CrossRef]
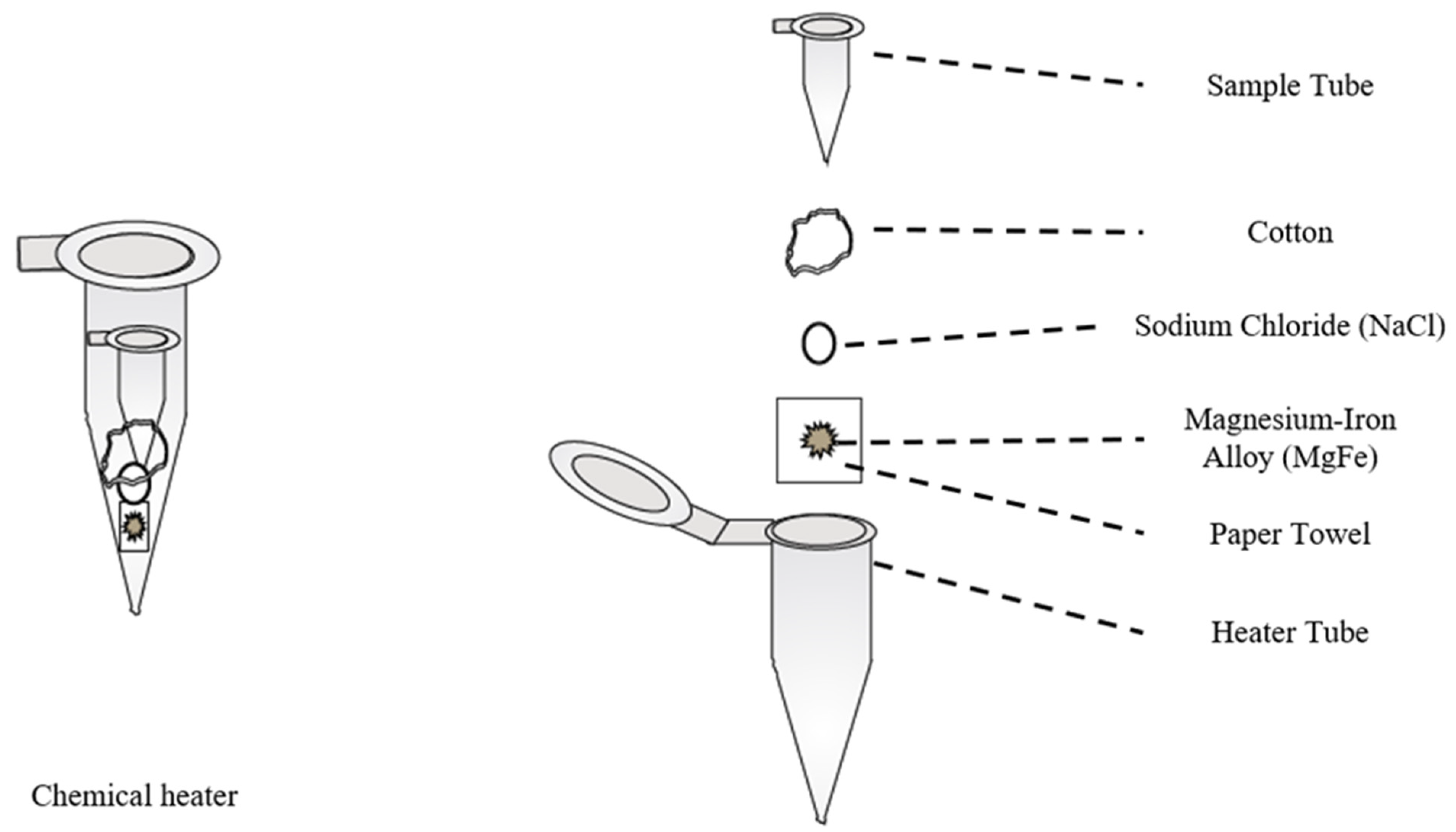
- Buser, J.R.; Zhang, X.; Byrnes, S.A.; Ladd, P.D.; Heiniger, E.K.; Wheeler, M.D.; Bishop, J.D.; Englund, J.A.; Lutz, B.; Weigl, B.H.; et al. A disposable chemical heater and dry enzyme preparation for lysis and extraction of DNA and RNA from microorganisms. Anal. Methods 2016, 8, 2880–2886. [Google Scholar] [CrossRef]
- Lee, E.-H.; Chua, B.; Son, A. Micro corona discharge based cell lysis method suitable for inhibitor resistant bacterial sensing systems. Sens. Actuators B Chem. 2015, 216, 17–23. [Google Scholar] [CrossRef]
- You, M.; Li, Z.; Feng, S.; Gao, B.; Yao, C.; Hu, J.; Xu, F. Ultrafast photonic PCR based on photothermal nanomaterials. Trends Biotechnol. 2020, 38, 637–649. [Google Scholar] [CrossRef]
- Maffert, P.; Reverchon, S.; Nasser, W.; Rozand, C.; Abaibou, H. New nucleic acid testing devices to diagnose infectious diseases in resource-limited settings. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1717–1731. [Google Scholar] [CrossRef]
- Kwon, L.; Long, K.D.; Wan, Y.; Yu, H.; Cunningham, B.T. Medical diagnostics with mobile devices: Comparison of intrinsic and extrinsic sensing. Biotechnol. Adv. 2016, 34, 291–304. [Google Scholar] [CrossRef]
- Snodgrass, R.; Gardner, A.; Semeere, A.; Kopparthy, V.L.; Duru, J.; Maurer, T.; Martin, J.; Cesarman, E.; Erickson, D. A portable device for nucleic acid quantification powered by sunlight, a flame or electricity. Nat. Biomed. Eng. 2018, 2, 657–665. [Google Scholar] [CrossRef]
- Zhang, L.; Ding, B.; Chen, Q.; Feng, Q.; Lin, L.; Sun, J. Point-of-care-testing of nucleic acids by microfluidics. TrAC Trends Anal. Chem. 2017, 94, 106–116. [Google Scholar] [CrossRef]
- Akyazi, T.; Basabe-Desmonts, L.; Benito-Lopez, F. Review on microfluidic paper-based analytical devices towards commercialisation. Anal. Chim. Acta 2018, 1001, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Boobphahom, S.; Nguyet Ly, M.; Soum, V.; Pyun, N.; Kwon, O.-S.; Rodthongkum, N.; Shin, K. Recent advances in microfluidic paper-based analytical devices toward high-throughput screening. Molecules 2020, 25, 2970. [Google Scholar] [CrossRef] [PubMed]
- Creran, B.; Li, X.; Duncan, B.; Kim, C.S.; Moyano, D.F.; Rotello, V.M. Detection of bacteria using inkjet-printed enzymatic test strips. ACS Appl. Mater. Interfaces 2014, 6, 19525–19530. [Google Scholar] [CrossRef]
- Yamada, K.; Suzuki, K.; Citterio, D. Text-displaying colorimetric paper-based analytical device. ACS Sens. 2017, 2, 1247–1254. [Google Scholar] [CrossRef]
- Cruz, D.F.; Fontes, C.M.; Semeniak, D.; Huang, J.; Hucknall, A.; Chilkoti, A.; Mikkelsen, M.H. Ultrabright fluorescence readout of an inkjet-printed immunoassay using plasmonic nanogap cavities. Nano Lett. 2020, 20, 4330–4336. [Google Scholar] [CrossRef]
- Verplaetse, R.; Henion, J. Quantitative Determination of opioids in whole blood using fully automated dried blood spot desorption coupled to on-line SPE-LC-MS/MS: Fully automated DBS-SPE-LC-MS/MS determination of opioid drugs in whole blood. Drug Test. Anal. 2016, 8, 30–38. [Google Scholar] [CrossRef]
- Santaus, T.M.; Melendez, J.H.; Negesse, M.Y.; Harvey, A.; Cyr, M.; Ladd, P.; Geddes, C.D. Lyse-ItTM: A rapid platform for cellular lysing and tunable DNA/protein fragmentation. In Microwave Effects on DNA and Proteins; Geddes, C.D., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 275–296. [Google Scholar] [CrossRef]
- Van Tongeren, S.P.; Degener, J.E.; Harmsen, H.J.M. Comparison of three rapid and easy bacterial DNA extraction methods for use with quantitative real-time PCR. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 1053–1061. [Google Scholar] [CrossRef] [PubMed]
- Shetty, P.; Ghosh, D.; Paul, D. Thermal lysis and isothermal amplification of mycobacterium tuberculosis H37Rv in one tube. J. Microbiol. Methods 2017, 143, 1–5. [Google Scholar] [CrossRef]
- Lee, E.-H.; Lim, H.J.; Son, A.; Chua, B. A Disposable Bacterial Lysis Cartridge (BLC) suitable for an in situ water-borne pathogen detection system. Analyst 2015, 140, 7776–7783. [Google Scholar] [CrossRef] [PubMed]
- Aslan, K.; Previte, M.J.R.; Zhang, Y.; Gallagher, T.; Baillie, L.; Geddes, C.D. Extraction and detection of DNA from Bacillus anthracis spores and the vegetative cells within 1 min. Anal. Chem. 2008, 80, 4125–4132. [Google Scholar] [CrossRef]
- Previte, M.J.R.; Geddes, C.D. Microwave-triggered chemiluminescence with planar geometrical aluminum substrates: Theory, simulation and experiment. J. Fluoresc. 2007, 17, 279–287. [Google Scholar] [CrossRef]
- Previte, M.J.R.; Aslan, K.; Geddes, C.D. Spatial and temporal control of microwave triggered chemiluminescence: A protein detection platform. Anal. Chem. 2007, 79, 7042–7052. [Google Scholar] [CrossRef] [PubMed]
- Melendez, J.H.; Santaus, T.M.; Brinsley, G.; Kiang, D.; Mali, B.; Hardick, J.; Gaydos, C.A.; Geddes, C.D. Microwave-accelerated method for ultra-rapid extraction of neisseria gonorrhoeae DNA for downstream detection. Anal. Biochem. 2016, 510, 33–40. [Google Scholar] [CrossRef]
- Santaus, T.M.; Li, S.; Saha, L.; Chen, W.H.; Bhagat, S.; Stine, O.C.; Geddes, C.D. A Comparison of lyse-it to other cellular sample preparation, bacterial lysing, and DNA fragmentation technologies. PLoS ONE 2019, 14, e0220102. [Google Scholar] [CrossRef]
- Santaus, T.M.; Li, S.; Ladd, P.; Harvey, A.; Cole, S.; Stine, O.C.; Geddes, C.D. Rapid sample preparation with lyse-it® for listeria monocytogenes and vibrio cholerae. PLoS ONE 2018, 13, e0201070. [Google Scholar] [CrossRef]
- Santaus, T.M.; Zhang, F.; Li, S.; Stine, O.C.; Geddes, C.D. Effects of Lyse-it on endonuclease fragmentation, function and activity. PLoS ONE 2019, 14, e0223008. [Google Scholar] [CrossRef]
- Santaus, T.M.; Greenberg, K.; Suri, P.; Geddes, C.D. Elucidation of a non-thermal mechanism for DNA/RNA fragmentation and protein degradation when using lyse-it. PLoS ONE 2019, 14, e0225475. [Google Scholar] [CrossRef] [PubMed]
- Delazar, A.; Nahar, L.; Hamedeyazdan, S.; Sarker, S.D. Microwave-assisted extraction in natural products isolation. In Natural Products Isolation; Sarker, S.D., Nahar, L., Eds.; Methods in molecular biology; Humana Press: Totowa, NJ, USA, 2012; Volume 864, pp. 89–115. [Google Scholar]
- Llompart, M.; Garcia-Jares, C.; Celeiro, M.; Dagnac, T. Microwave-assisted extraction. In Reference Module in Chemistry, Molecular Sciences and Chemical Engineering; Elsevier: Amsterdam, The Netherlands, 2018; p. B9780124095472144000. [Google Scholar]
- Sanchez-Prado, L.; Garcia-Jares, C.; Llompart, M. Microwave-assisted extraction: Application to the determination of emerging pollutants in solid samples. J. Chromatogr. A 2010, 1217, 2390–2414. [Google Scholar] [CrossRef] [PubMed]
- Ahirwar, R.; Tanwar, S.; Bora, U.; Nahar, P. Microwave non-thermal effect reduces elisa timing to less than 5 minutes. RSC Adv. 2016, 6, 20850–20857. [Google Scholar] [CrossRef]
- Byrnes, S.; Fan, A.; Trueb, J.; Jareczek, F.; Mazzochette, M.; Sharon, A.; Sauer-Budge, A.F.; Klapperich, C.M. A portable, pressure driven, room temperature nucleic acid extraction and storage system for point of care molecular diagnostics. Anal. Methods 2013, 5, 3177. [Google Scholar] [CrossRef] [PubMed]
- Ritzi-Lehnert, M. Development of chip-compatible sample preparation for diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 2012, 12, 189–206. [Google Scholar] [CrossRef]
- Neely, L.A.; Audeh, M.; Phung, N.A.; Min, M.; Suchocki, A.; Plourde, D.; Blanco, M.; Demas, V.; Skewis, L.R.; Anagnostou, T.; et al. T2 Magnetic resonance enables nanoparticle-mediated rapid detection of candidemia in whole blood. Sci. Transl. Med. 2013, 5, ra54–ra182. [Google Scholar] [CrossRef] [PubMed]
- Achyuthan, K.; Whitten, D. Design considerations for high throughput screening and in vitro diagnostic assays. Comb. Chem. High Throughput Screen. 2007, 10, 399–412. [Google Scholar] [CrossRef]
- Dolci, A.; Giavarina, D.; Pasqualetti, S.; Szőke, D.; Panteghini, M. Total laboratory automation: Do stat tests still matter? Clin. Biochem. 2017, 50, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Elpa, D.P.; Prabhu, G.R.D.; Wu, S.-P.; Tay, K.S.; Urban, P.L. Automation of mass spectrometric detection of analytes and related workflows: A review. Talanta 2020, 208, 120304. [Google Scholar] [CrossRef]
- Mafra, G.; Vieira, A.A.; Merib, J.; Anderson, J.L.; Carasek, E. Single drop microextraction in a 96-well plate format: A step toward automated and high-throughput analysis. Anal. Chim. Acta 2019, 1063, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Li, W.; Entcheva, E.; Li, Z. Microfluidics-enabled 96-well perfusion system for high-throughput tissue engineering and long-term all-optical electrophysiology. Lab Chip 2020, 20, 4031–4042. [Google Scholar] [CrossRef]
- Mottaz-Brewer, H.M.; Norbeck, A.D.; Adkins, J.N.; Manes, N.P.; Ansong, C.; Shi, L.; Rikihisa, Y.; Kikuchi, T.; Wong, S.W.; Estep, R.D.; et al. Optimization of Proteomic sample preparation procedures for comprehensive protein characterization of pathogenic systems. J. Biomol. Tech. 2008, 19, 285. [Google Scholar]
- Hess, J.F.; Kohl, T.A.; Kotrová, M.; Rönsch, K.; Paprotka, T.; Mohr, V.; Hutzenlaub, T.; Brüggemann, M.; Zengerle, R.; Niemann, S.; et al. Library preparation for next generation sequencing: A review of automation strategies. Biotechnol. Adv. 2020, 41, 107537. [Google Scholar] [CrossRef]
- Ye, X.; Tang, J.; Mao, Y.; Lu, X.; Yang, Y.; Chen, W.; Zhang, X.; Xu, R.; Tian, R. Integrated proteomics sample preparation and fractionation: Method development and applications. TrAC Trends Anal. Chem. 2019, 120, 115667. [Google Scholar] [CrossRef]
- Berger, S.T.; Ahmed, S.; Muntel, J.; Cuevas Polo, N.; Bachur, R.; Kentsis, A.; Steen, J.; Steen, H. MStern blotting–high throughput polyvinylidene fluoride (PVDF) membrane-based proteomic sample preparation for 96-well plates. Mol. Cell. Proteom. 2015, 14, 2814–2823. [Google Scholar] [CrossRef] [PubMed]
- Switzar, L.; van Angeren, J.; Pinkse, M.; Kool, J.; Niessen, W.M.A. A high-throughput sample preparation method for cellular proteomics using 96-well filter plates. Proteomics 2013, 13, 2980–2983. [Google Scholar] [CrossRef]
- Yu, Y.; Suh, M.-J.; Sikorski, P.; Kwon, K.; Nelson, K.E.; Pieper, R. Urine sample preparation in 96-well filter plates for quantitative clinical proteomics. Anal. Chem. 2014, 86, 5470–5477. [Google Scholar] [CrossRef]
- Wiśniewski, J.R. Filter aided sample preparation—A tutorial. Anal. Chim. Acta 2019, 1090, 23–30. [Google Scholar] [CrossRef]
- Solovjev, A.M.; Kurzeev, S.A.; Sakharov, I.Y. Chemiluminescent microplate-based assay of DNA based on Isothermal Circular Strand-Displacement Polymerization Reaction (ICSDPR). Talanta 2020, 215, 120895. [Google Scholar] [CrossRef]
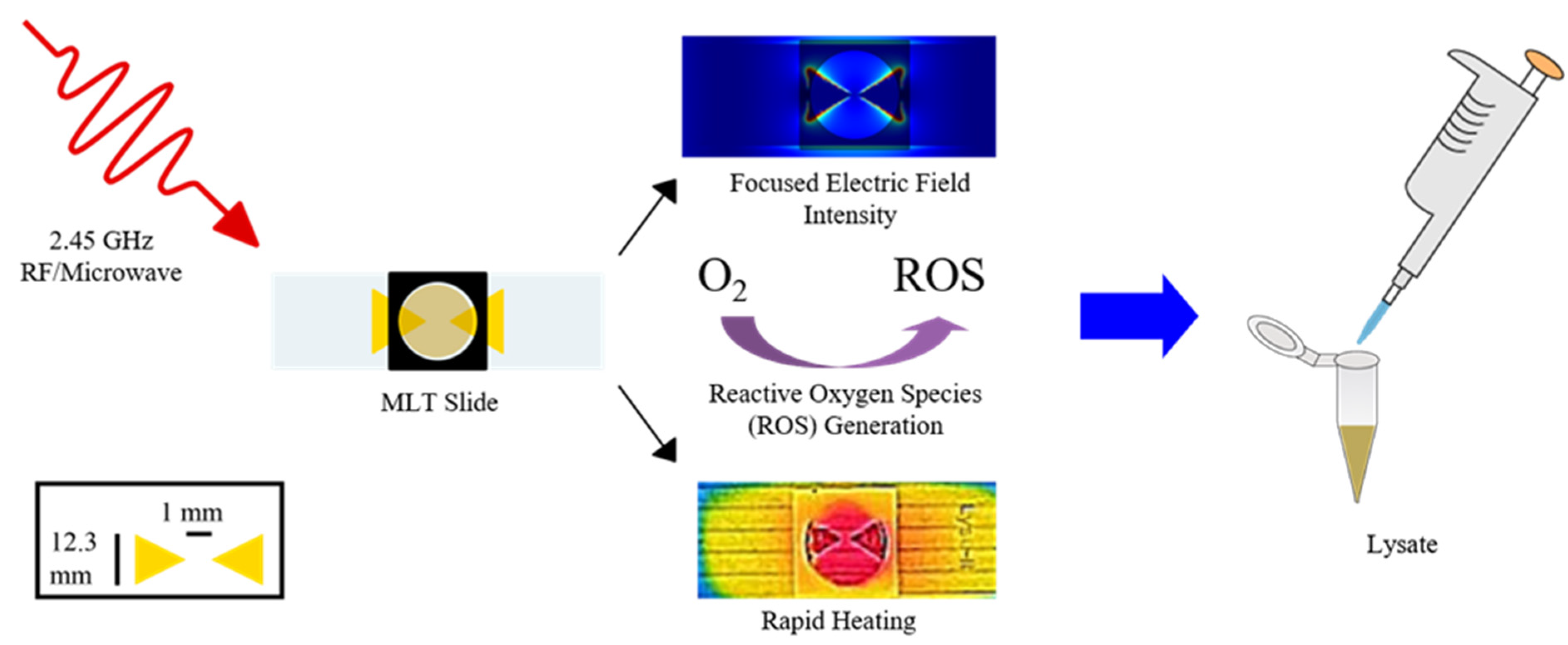
- Nichols, Z.E.; Saha, L.; Knoblauch, R.; Santaus, T.M.; Geddes, C.D. Development of a microplate platform for high-throughput sample preparation based on microwave metasurfaces. IEEE Access 2021, 9, 37823–37833. [Google Scholar] [CrossRef]
- Wang, L.-J.; Naudé, N.; Demissie, M.; Crivaro, A.; Kamoun, M.; Wang, P.; Li, L. Analytical validation of an ultra low-cost mobile phone microplate reader for infectious disease testing. Clin. Chim. Acta 2018, 482, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Clark, D.; Liu, Y.; Li, S.; Zhang, H. High-Throughput analysis of n-glycans using autotip via glycoprotein immobilization. Sci. Rep. 2017, 7, 10216. [Google Scholar] [CrossRef]
- Mishra, R.; Zapatero-Rodríguez, J.; Sharma, S.; Kelly, D.; McAuley, D.; Gilgunn, S.; O’Kennedy, R.; Ducrée, J. Automation of multi-analyte prostate cancer biomarker immunoassay panel from whole blood by minimum-instrumentation rotational flow control. Sens. Actuators B Chem. 2018, 263, 668–675. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nichols, Z.E.; Geddes, C.D. Sample Preparation and Diagnostic Methods for a Variety of Settings: A Comprehensive Review. Molecules 2021, 26, 5666. https://doi.org/10.3390/molecules26185666
Nichols ZE, Geddes CD. Sample Preparation and Diagnostic Methods for a Variety of Settings: A Comprehensive Review. Molecules. 2021; 26(18):5666. https://doi.org/10.3390/molecules26185666
Chicago/Turabian StyleNichols, Zach E., and Chris D. Geddes. 2021. "Sample Preparation and Diagnostic Methods for a Variety of Settings: A Comprehensive Review" Molecules 26, no. 18: 5666. https://doi.org/10.3390/molecules26185666
APA StyleNichols, Z. E., & Geddes, C. D. (2021). Sample Preparation and Diagnostic Methods for a Variety of Settings: A Comprehensive Review. Molecules, 26(18), 5666. https://doi.org/10.3390/molecules26185666





