Abstract
We report the design and synthesis of a series of new 5-chloropyridinyl esters of salicylic acid, ibuprofen, indomethacin, and related aromatic carboxylic acids for evaluation against SARS-CoV-2 3CL protease enzyme. These ester derivatives were synthesized using EDC in the presence of DMAP to provide various esters in good to excellent yields. Compounds are stable and purified by silica gel chromatography and characterized using 1H-NMR, 13C-NMR, and mass spectral analysis. These synthetic derivatives were evaluated in our in vitro SARS-CoV-2 3CLpro inhibition assay using authentic SARS-CoV-2 3CLpro enzyme. Compounds were also evaluated in our in vitro antiviral assay using quantitative VeroE6 cell-based assay with RNAqPCR. A number of compounds exhibited potent SARS-CoV-2 3CLpro inhibitory activity and antiviral activity. Compound 9a was the most potent inhibitor, with an enzyme IC50 value of 160 nM. Compound 13b exhibited an enzyme IC50 value of 4.9 µM. However, it exhibited a potent antiviral EC50 value of 24 µM in VeroE6 cells. Remdesivir, an RdRp inhibitor, exhibited an antiviral EC50 value of 2.4 µM in the same assay. We assessed the mode of inhibition using mass spectral analysis which suggested the formation of a covalent bond with the enzyme. To obtain molecular insight, we have created a model of compound 9a bound to SARS-CoV-2 3CLpro in the active site.
1. Introduction
Novel Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for the ongoing COVID-19 pandemic [1,2]. The first cases of the disease were reported in Wuhan, China and then rapidly spread worldwide, overwhelming health care systems, disrupting economies, and leading to extensive loss of human lives [3,4,5]. At present, there is no specific efficacious treatment for COVID-19, except remdesivir, which shows only modest benefits to patients with COVID-19 infection [6,7]. Current vaccination efforts are showing benefits, but the hope that COVID-19 will be under control with ‘herd immunity’ is quite uncertain due to the emergence of COVID-19 variants [8,9]. Therefore, it is critical to develop effective antiviral drugs that mitigate the lethal cytokine storm in COVID-19 patients.
SARS-CoV-2 belongs to a family of beta-coronaviruses, including SARS-CoV and MERS-CoV, which were responsible for earlier outbreaks of SARS and MERS in 2003 and 2012, respectively [10,11,12]. SARS-CoV-2 encodes two proteases, a 3-chymotrypsin-like cysteine protease (3CLpro) also known as main protease (Mpro) and a papain-like protease (PLpro) for proteolytic processing of viral replication and maturation. [13,14] Both of these proteases are essential for replication of SARS-CoV-2 and other coronaviruses. The SARS-CoV-2 genome shares 80% nucleotide identity with SARS-CoV. Structurally, 3CLpro protease of SARS-CoV-2 shares more than 90% amino acid sequence identity with highly conserved substrate-binding sites with in the active site. [15,16] Thus, previous medicinal chemistry efforts leading to the development of 3CL protease inhibitors of SARS-CoV and MERS-CoV provided important groundwork for drug design efforts against COVID-19 [17,18,19].
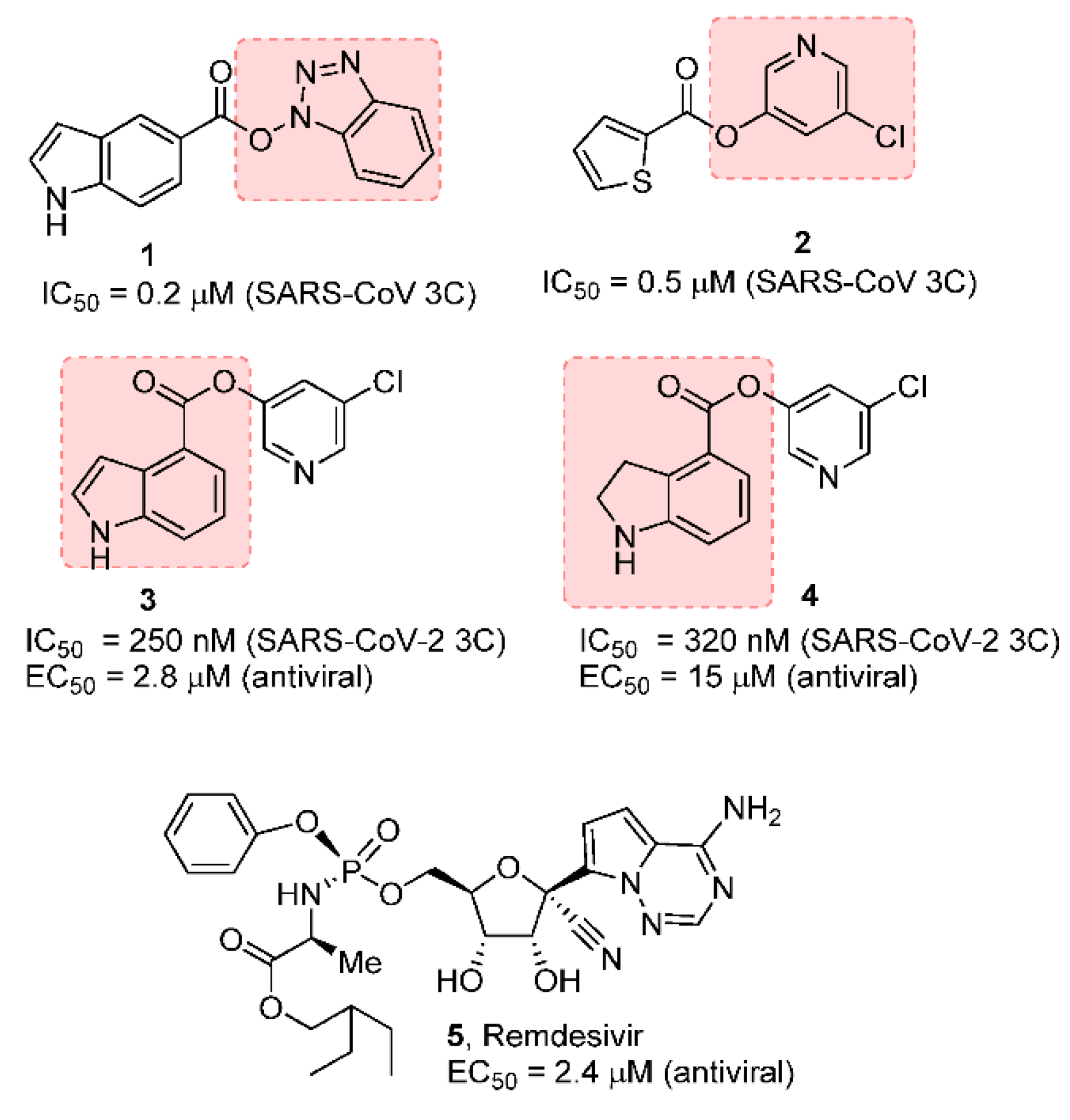
Our laboratories previously designed, synthesized, and performed X-ray structural studies of a variety of peptidomimetic and nonpeptide, covalent, noncovalent MERS-CoV 3CLpro inhibitors that show potent enzyme inhibitory activity and exerted significant antiviral activity against SARS-CoV. [17,20] In particular, we and others have designed a new class of potent and small-molecule covalent SARS-CoV inhibitors that involve acylation of the active site Cys145, forming a covalent bond with the small-molecule inhibitors [21,22,23,24]. As shown in Figure 1, the indole carboxylic acid-derived benzotriazole ester 1 and thiophene carboxylic acid-derived 5-chloropyridinyl ester 2 are potent against SARS-CoV 3CL protease. However, these compounds did not show antiviral activity.
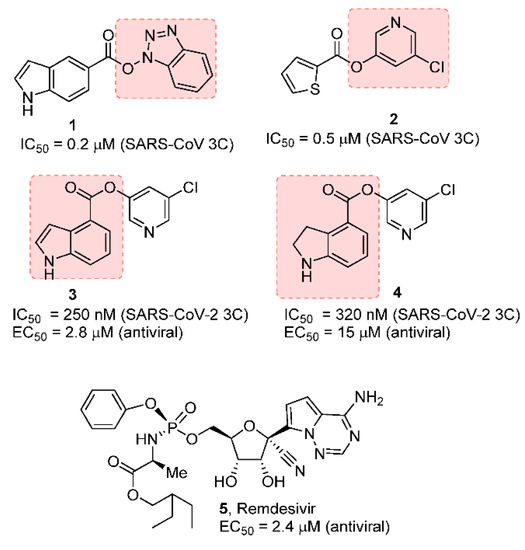
Figure 1.
Covalent SARS-CoV and SARS-CoV-2 3CL inhibitors 1–4 and remdesivir.
We subsequently designed a series of 5-chloropyridin-3-yl esters and demonstrated both enzyme inhibitory and antiviral activity. Compound 3 exhibited a SARS-CoV 3CL protease inhibitory IC50 value of 250 nM and an antiviral EC50 value of 2.8 µM in VeroE6 cells [25]. Based upon our previous results, we recently developed indole chloropyridin-3-yl ester-derived SARS-CoV-2 3CL protease inhibitors and demonstrated that prototype compounds, such as inhibitor 3, are potent inhibitors of SARS-CoV-2 3CL protease. It exerts comparable antiviral activity to remdesivir (5), an RNA-dependent RNA-polymerase inhibitor [26,27]. Furthermore, we have shown that compound 3 blocked the infectivity and cytopathic effect of SARS-CoV-2wk-521 in VeroE6 cells in our immunocytochemistry assay.
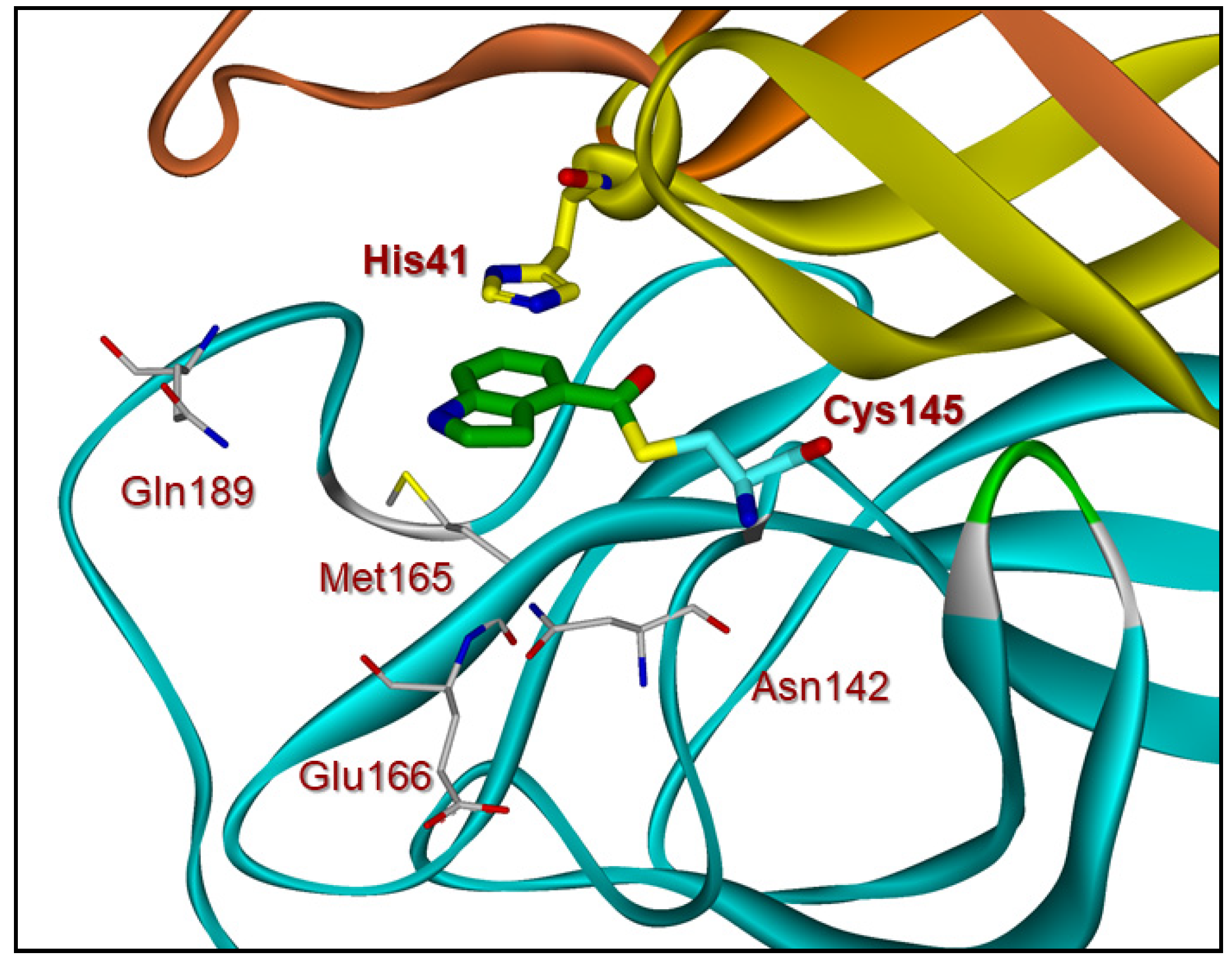
Compound 4 has also shown potent enzyme inhibitory and antiviral activity in our immunocytochemistry assays [28]. Our X-ray structural analysis of inhibitor 4 and SARS-CoV-2 3CL protease complex demonstrates that the mode of inhibition involved the formation of a covalent bond with the inhibitor carbonyl group and catalytic Cys145 in the active site as shown in Figure 2. Furthermore, our recent structure–activity relationship (SAR) suggested that the position of the carboxylic acid on the indole scaffold is critical for the enzyme activity. Based upon our X-ray structural studies and mode of inhibition, we have further investigated other aromatic and heteroaromatic scaffolds and their ability to block SARS-CoV-2 3CL protease activity as well as antiviral activity in VeroE6 cells [29]. In particular, we plan to synthesize 5-chloropyridin-3-yl esters of widely used nonsteroidal anti-inflammatory agents (NSAIDs) [30,31] and evaluate their potential as irreversible inhibitors of SARS-CoV-2 3CLpro enzyme. Presumably, such acylated thioesters of these NSAIDs would hydrolyze slowly over time and release parent NSAIDs in the cell [32]. Interestingly, this would lead to inhibition of cyclooxygenase in the cell, leading to analgesic and anti-inflammatory effects. In the present studies, we report a series of 5-chloropyridinyl ester derivatives of salicylic acid, ibuprofen, naproxane, indomethacin, and related interesting derivatives. A number of compounds exhibited potent enzyme inhibitory and antiviral activity.
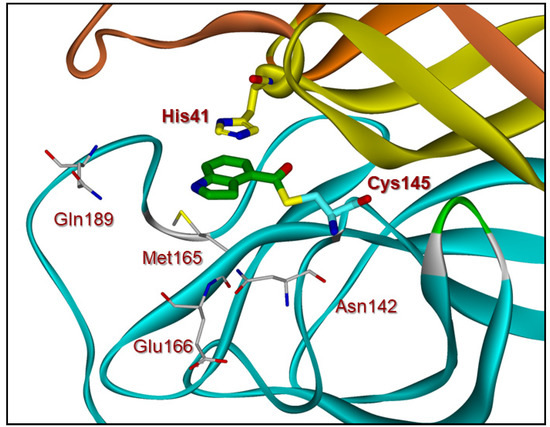
Figure 2.
X-ray crystal structure of inhibitor 4-bound SARS-CoV-2 3CL protease (PDB code:7RBZ). The thioester bond between Cys145 and compound 4 (green) is shown.
2. Results and Discussion
2.1. Chemistry
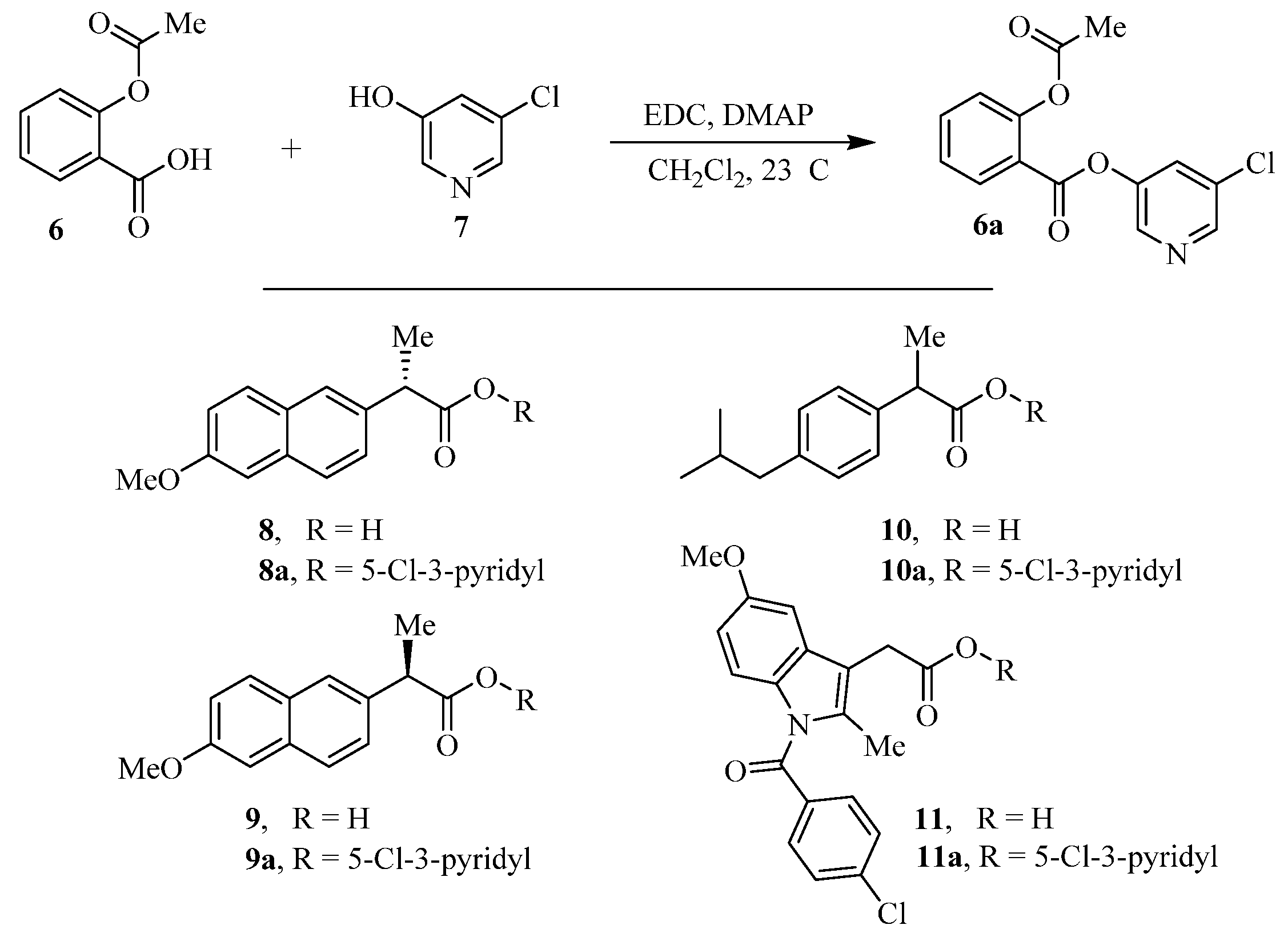
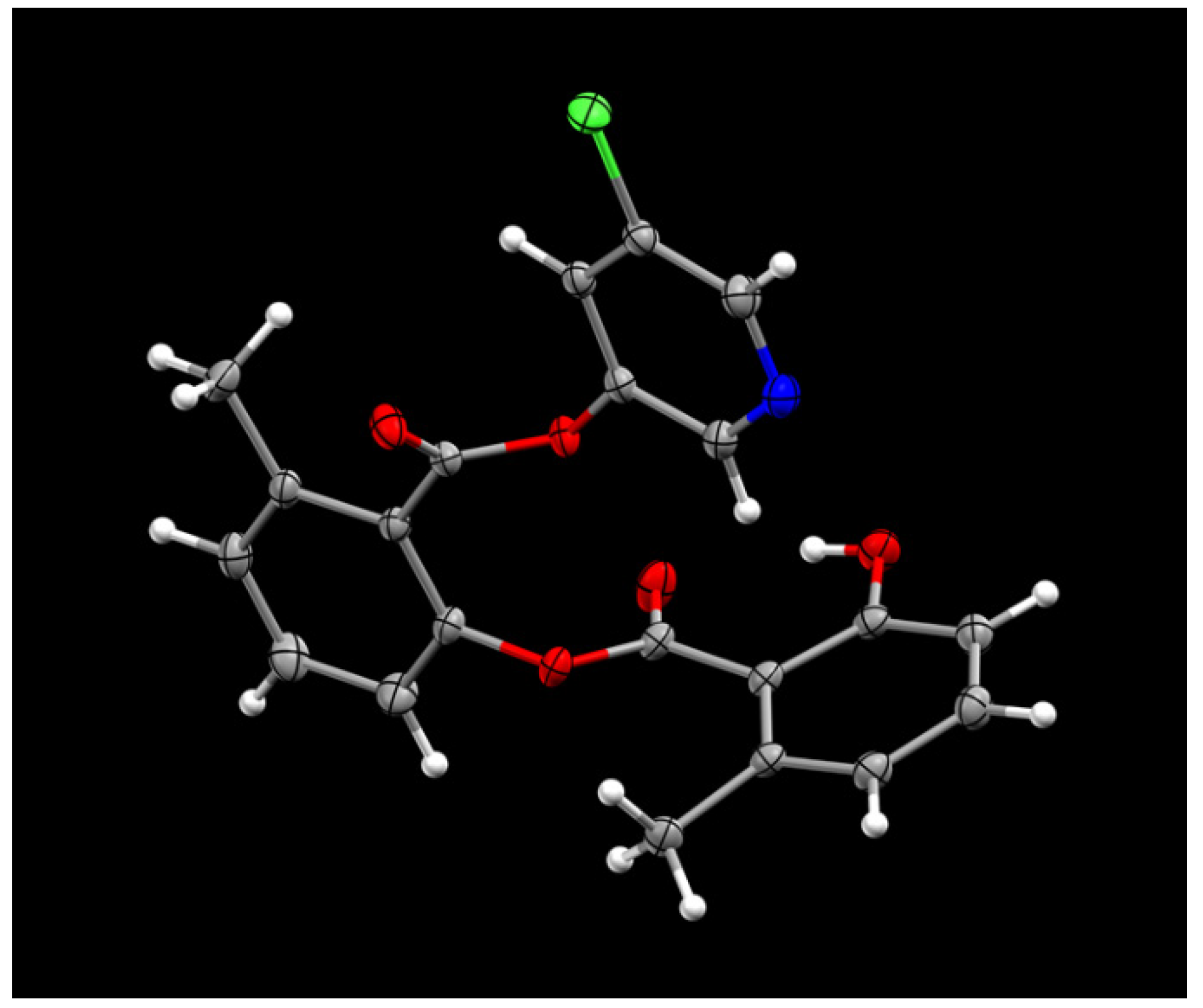
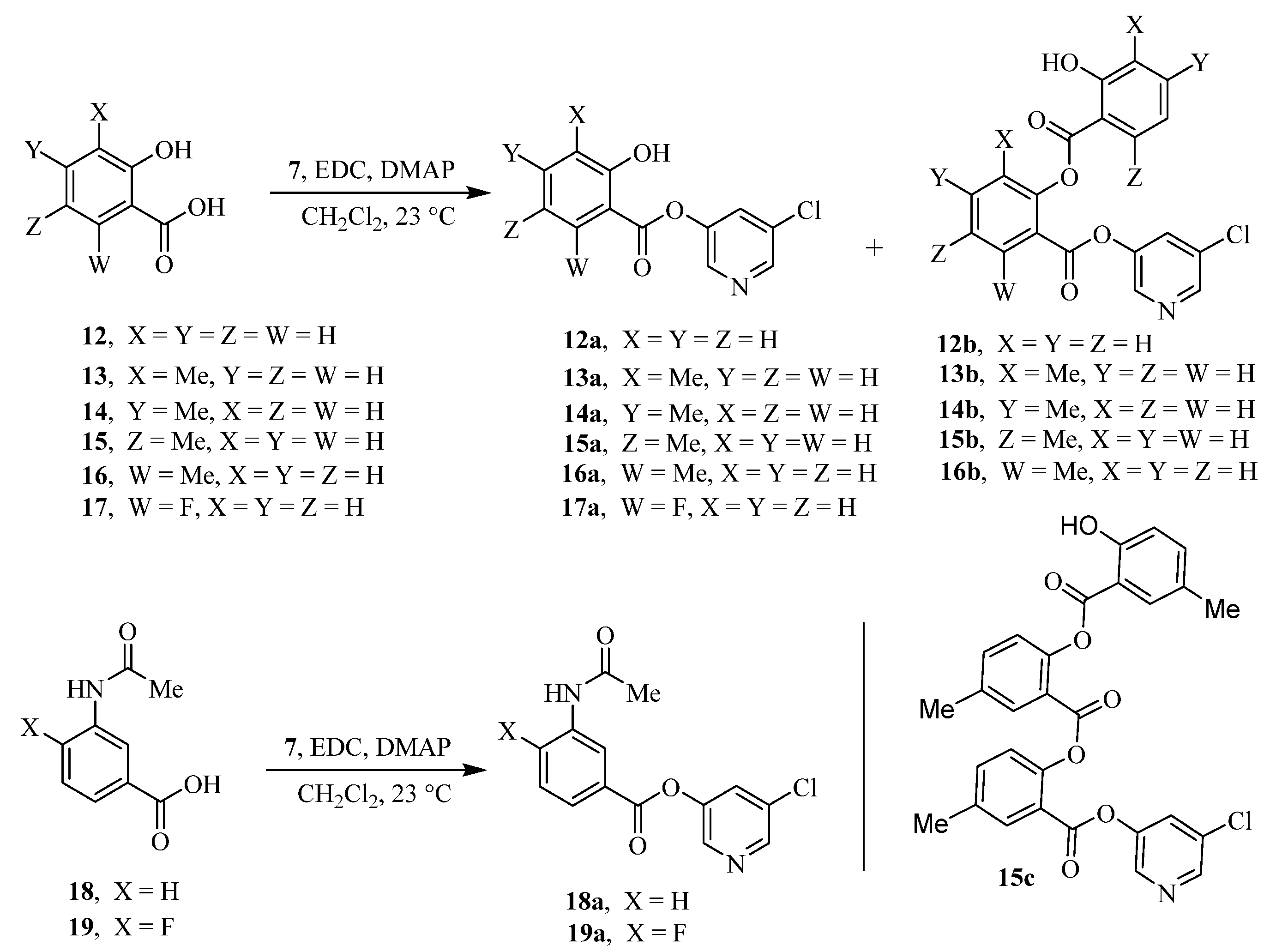
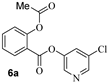
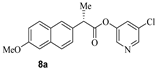
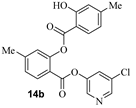
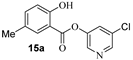
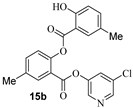
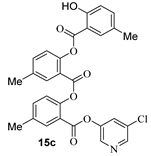
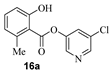
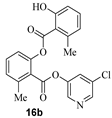
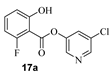
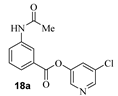
The synthesis of various 5-chloropyridinyl esters of common nonsteroidal anti-inflammatory agents [30,31] is shown in Scheme 1. Commercially available aspirin 6 was esterified with 1.2 equivalents of 5-chloro-3-pyridinol using 1.5 equivalents of EDC in the presence of 1 equivalent of DMAP in CH2Cl2 at 23 °C for 12 h. This condition has provided 6a in 12% yield after silica gel chromatography, we have then exposed (S)- and (R)-naproxen, racemic ibuprofen and indomethacin under these esterification conditions for the synthesis of other 5-chloropyridinyl esters 8a–11a (46–53% yield). The structures of these ester derivatives are shown in Table 1. We have also prepared chloropyridinyl esters derived from salicylic acid and its methyl-substituted derivatives as shown in Scheme 2. Initially, we attempted the synthesis of salicylic acid derivatives by using conditions mentioned above. However, the above conditions provided variable results and provided mixture of monomeric and dimeric ester along with small amounts of higher oligomers. The monomeric products were variable. We then carried out the esterification reaction of salicylic acid and its methyl-substituted derivatives by first exposing 0.25 equivalent of acid, 1.2 equivalent of 3-hydroxy-5-chloro-pyridine and 1.5 equivalent of EDC in the presence of 1.0 equivalent DMAP at 23 °C. The mixture was stirred for 2 h and then 0.25 equivalent of acid was added at 23 °C every 2 h interval. The resulting mixture was stirred at 23 °C for 12 h. This condition provided a mixture of monomeric esters (12a–17a) and dimeric esters (12b–16b) respectively, in good yields. In the case of 2-hydroxy-5-methyl benzoic acid 15, in addition to monomer 15a and dimer derivative 15b, we have also obtained triester derivative 15c. 2-Hydroxy-6-methyl benzoic acid 16 provided monoester 16a and diester 16b. The diester 16b was crystalized in CH2Cl2 solution and the identity of the structure was unambiguously determined by X-ray crystallography as depicted in Figure 3 [33,34]. Please see Supporting Information for further details. For our structure–activity relationship studies, we have also prepared 3-acetamido-benzoic acid ester derivatives 18a and 19a in good yield.
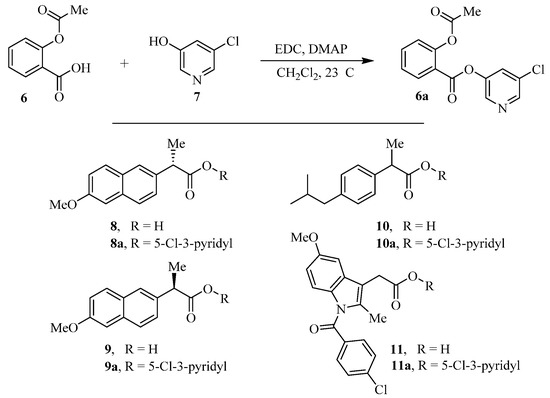
Scheme 1.
Synthesis of 5-chloropyridin-3-yl esters of common NSAIDs.

Table 1.
Structure and activity of NSAIDs-derived chloropyridinyl esters.
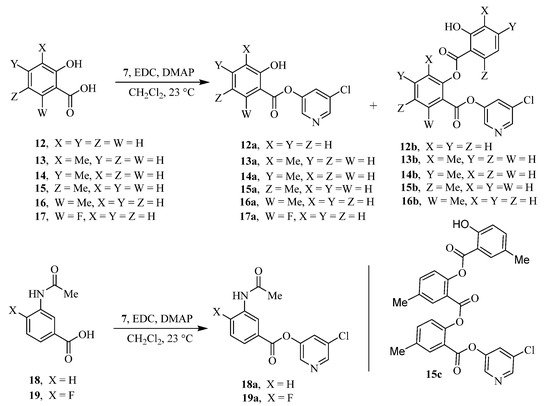
Scheme 2.
Synthesis of chloropyridinyl esters of salicylic acid derivatives.
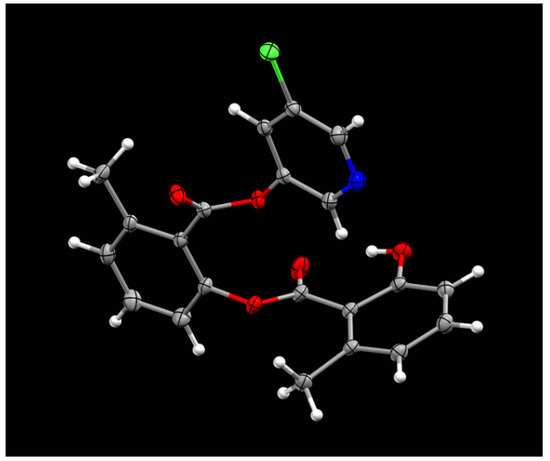
Figure 3.
ORTEP drawing of compound 16b at 50% probability level.
2.2. Biological Evaluation
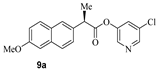
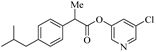
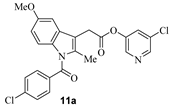
We have carried out SARS-CoV-2 3CLpro inhibition assays using the authentic SARS-CoV-2 3CLpro enzyme as described recently [35]. The enzyme inhibitory activity (IC50 values) of synthetic active esters was assessed using a continuous fluorescence assay and the FRET-based substrate UIVT3 (HiLyteFluor488TM-EATLQSGLRKAK-QXL520-NH2 (HPLC > 90%); Anaspec, Fremont, CA, USA) described by us previously [20,36]. The antiviral activity (EC50 value) of compounds was evaluated using quantitative VeroE6 cell-based assay with RNA-qPCR as described by us recently [28]. The structures and activity of synthetic ester derivatives are shown in Table 1 and Table 2. We first assessed common NSAIDs-derived chloropyridinyl esters shown in Table 1. As can be seen, acetoxysalicylic acid-derived ester 6a exhibited SARS-CoV-2 3CL protease IC50 value of 360 nM (entry 1). This compound exhibited an antiviral EC50 value > 100 µM. (S)-Naproxen-derived ester 8a exhibited an enzyme inhibitory activity value of 670 nM (entry 2), while the (R)-naproxen derivative 9a exhibited an enzyme IC50 value of 160 nM (entry 3), an over 4-fold improvement. Racemic ibuprofen-derived ester 10a exhibited an IC50 value of 810 nM (entry 4). Indomethacin-derived ester 11a exhibited a significant reduction in enzyme activity while structurally related indole derivatives exhibited excellent enzyme inhibitory activity (entry 5) [29]. Interestingly, compound 11a exhibited an antiviral EC50 value of 30 µM while aspirin, ibuprofen, and naproxen-derived esters did not show appreciable antiviral activity. We presume that the mode of inhibition involves covalent bond formation with catalytic Cys145 as observed in our previous X-ray structural analysis of inhibitor-bound SARS-CoV-2 3CL protease as well as mass spectral analysis. [20,29] The irreversible enzyme acylation of the NSAIDs-based inhibitor was examined by using MALDI-TOF. Authentic SARS-CoV-2 3CLpro was incubated with inhibitor 8a and then analyzed with untreated enzyme. As expected, we were able to see a signal for enzyme-bound compound 8a on the LC–MS spectrum that corresponds to acylation of 3CL protease with a mass shift of +212 Daltons.

Table 2.
Structure and activity of salicylic acid and its substituted chloropyridinyl esters.
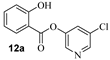
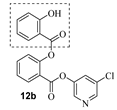
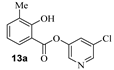
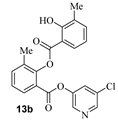
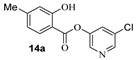
Based upon the encouraging enzyme inhibitory activity of NSAIDS derivatives, we then prepared a range of salicylic acid derivatives and evaluated their activity. As shown in Table 2, salicylic acid-derived pyridinyl ester 12a exhibited an IC50 value of 3.47 µM. The corresponding diester derivative 12b exhibited a 5-fold reduction in enzyme activity. However, compound 12b exhibited an antiviral EC50 value of 64 µM (entry 2). In an effort to modulate activity, we incorporated the methyl group on the aromatic ring. Methyl substitution at C3 resulted in monoester 13a, which exhibited a significant improvement in enzyme activity (IC50 650 nM) over its unsubstituted derivative 12a. Diester derivative 13b exhibited a reduction in the enzyme IC50 value but some improvement in antiviral activity (entries 3 and 4). Incorporation of methyl group at C4 provided slight improvement in enzyme activity for both mono- and di-ester derivatives 14a and 14b (entries 5 and 6). Substitution of methyl group at C5 led to the syntheses of mono-ester 15a, diester 15b, and tri-ester 15c. Both mono-ester 15a and diester 15b exhibited improvement in enzyme activity over other substituted derivatives (entries 7–9). Interestingly, substitution of methyl group at C6 resulted in significant loss of enzyme activity (entries 10 and 11). Incorporation of fluorine at C6 also resulted in a further reduction in enzyme inhibitory activity (entry 12). We have also investigated amide derivatives 18a and 19a. Both compounds exhibited enzyme inhibitory activity in low micromolar range (entries 13 and 14). All compounds in Table 1 and Table 2 exhibited a cytotoxicity (CC50) value >100 µM. While a number of chloropyridinyl esters exhibited low nanomolar 3CLpro inhibitory activity, the majority of these ester derivatives did not show appreciable antiviral activity, except compounds 11a, 12b, 13b, 15a and 15b, which exhibited antiviral EC50 values of 24–64 µM. Such high ratios of antiviral EC50 and enzyme IC50 values may be due to the expression of the efflux transporter P-glycoprotein in VeroE6 cells. [37,38] We, therefore, examined the antiviral activity of selected compounds (6a, 9a, 11a, 14a, and 15a) in the presence of P-glycoprotein inhibitor, CP-100356 [39]. Interestingly, none of these compounds exhibited any significant antiviral activity in the presence of the P-glycoprotein inhibitor.
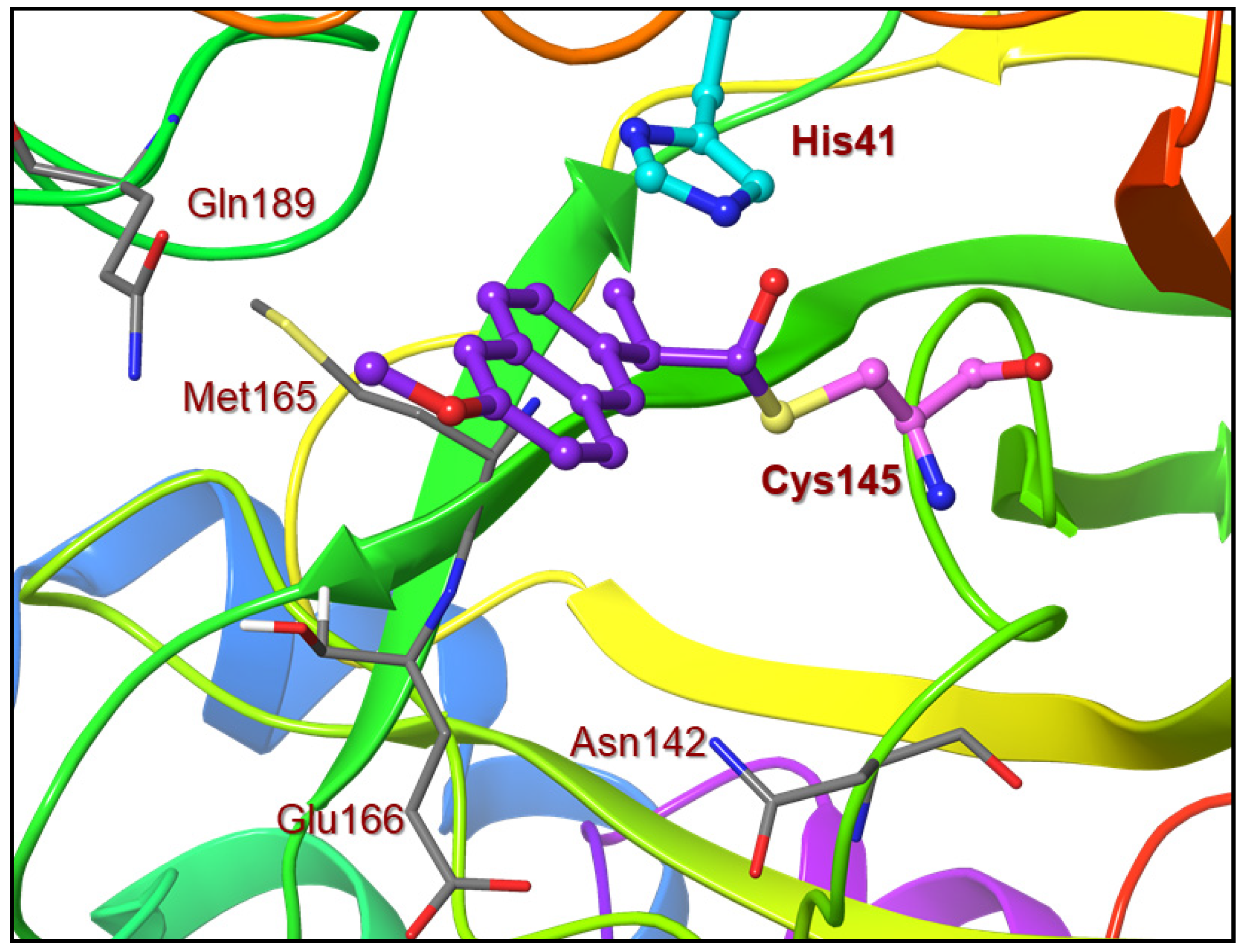
Based upon the X-ray structure of an irreversible inhibitor (GRL-017-20) bound to SARS-CoV-2 3CL protease (PDB code: 7RBZ), we modeled the complex of inhibitor 9a with SARS-CoV-2 3CL protease [29]. The model of inhibitor 9a bound to the catalytic Cys 145 residue in the active site of the 3CL protease is shown in Figure 4. The sulfur atom of Cys145 forms a covalent bond to the carbonyl and the chloropyridinyl group acts as a leaving group. The ligand sits in the binding pocket formed from Asn142, Met165, Glu166 and Gln 189. The model shows a similar π–π stacking of the aromatic ring with the imidazole ring of the His 41 residue [29]. Unfortunately, we did not observe the anticipated hydrogen bond between the methoxy group oxygen atom of the ligand and the side chain of the Gln 189 residue. The current study can provide a foundation to design new irreversible inhibitors to target SARS-CoV-2 3CL protease. Our laboratory is actively working on the design and synthesis of potent COVID-19 inhibitors.
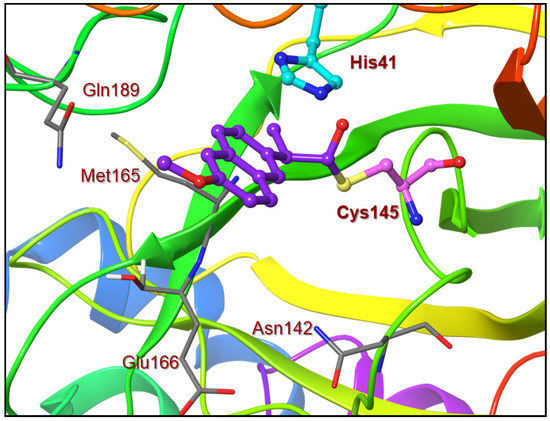
Figure 4.
An optimized model of inhibitor 9a (violet) in the active site of SARS-CoV-2 3CL protease. The model was created based upon the X-ray structure of inhibitor 4 bound to SARS-CoV-2 3CL protease (PDB code:7RBZ).
3. Materials and Methods
3.1. Chemistry
All compounds were purified by column chromatography. Column chromatography was performed using silica gel 230–400 mesh, with a 60 Å pore diameter. Proton Nuclear Magnetic Resonance NMR (1H NMR) spectra and carbon nuclear magnetic resonance (13C NMR) spectra were recorded on Bruker AV-III-400HD and Bruker AVIII-800 spectrometers. Optical rotations were measured on a Rudolph’s AUTOPOL-III automatic digital polarimeter with a sodium lamp and are reported as follows: [α]λ T °C (c = g/100 mL, solvent). High-resolution mass spectrometry (HRMS) spectra were recorded under positive electron spray ionization (ESI+) using a LTQ Orbitrap Mass Spectrometer at the Purdue University Department of Chemistry Mass Spectrometry Center and an Agilent 6550 Q-TOF LC/MS instrument at the Purdue University Analytical Mass Spectrometry Facility.
X-ray diffraction data for the single crystal of compound 16b were collected on a Bruker Quest diffractometer. Examination and data collection were performed with Mo Kα radiation (λ = 0.71073 Å) at 150 K. Data were collected, reflections were indexed and processed, and the files scaled and corrected for absorption using APEX3 [40]. The space groups were assigned, and the structures were solved by direct methods using XPREP within the SHELXTL suite of programs [41,42] and refined by full-matrix least squares against F2 with all reflections using Shelxl2018 using the graphical interface Shelxle [43]. Complete crystallographic data, in CIF format, have been deposited with the Cambridge Crystallographic Data Centre. The details of the structure are provided in the Supporting Information Section.
- Synthesis of 6-chloropyridin-2-yl 2-acetoxybenzoate (6a). To a stirred solution of 2-acetoxybenzoic acid (150 mg, 0.83 mmol) in DCM (3 mL), 5-chloropyridin-3-ol (129.4 mg, 0.10 mmol), EDC (240 mg, 1.25 mmol) and DMAP (102 mg, 0.83 mmol) were added. The resulting reaction mixture was stirred at 23 °C for 10 h. After this period, the reaction mixture was washed with saturated aqueous NaHCO3. The organic layer was dried over anhydrous Na2SO4 and concentrated under reduced pressure to give a crude residue. The residue was purified via silica gel column chromatography (30% ethyl acetate in hexanes) to afford the title ester 6a (30 mg, 12%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.51 (d, J = 2.1 Hz, 1H), 8.42 (d, J = 2.4 Hz, 1H), 8.20 (dd, J = 7.9, 1.7 Hz, 1H), 7.68 (ddd, J = 8.1, 7.5, 1.7 Hz, 1H), 7.64 (t, J = 2.2 Hz, 1H), 7.41 (td, J = 7.7, 1.2 Hz, 1H), 7.20 (dd, J = 8.1, 1.2 Hz, 1H), 2.32 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 169.5, 161.9, 151.4, 146.1, 145.7, 141.3, 135.2, 132.0, 129.6, 129.4, 126.2, 124.1, 121.3, 20.9; LRMS-ESI (m/z): 292.0 [M + H]+. HRMS (ESI/LTQ) m/z. [M + H]+ calcd for C14H11ClNO4 292.03711; found 292.03630.
- 5-chloropyridin-3-yl (S)-2-(6-methoxynaphthalen-2-yl)propanoate (8a). Commercially available (S)-2-(6-methoxynaphthalen-2-yl)propanoic acid (50 mg, 0.22 mmol) was esterified with 5-chloropyridin-3-ol (34 mg, 0.26 mmol) by following the procedure for ester 6a to provide the title ester 8a (36 mg, 49%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.42–8.40 (m, 1H), 8.25–8.23 (m, 1H), 7.78–7.72 (m, 3H), 7.48–7.41 (m, 2H), 7.20–7.13 (m, 2H), 4.11 (q, J = 7.1 Hz, 1H), 3.93 (s, 3H), 1.70 (d, J = 7.1 Hz, 3H); 13C NMR (100 MHz, CDCl3) δ 172.25, 157.8, 147.2, 145.6, 140.9, 134.1, 133.8, 131.6, 129.2, 128.8, 127.5, 126.1, 125.7, 119.2, 105.5, 55.2, 45.4, 18.2; LRMS-ESI (m/z): 342.0 [M + H]+. HRMS (ESI/LTQ) m/z. [M + H]+ calcd for C19H17ClNO3 342.08915; found 342.08907.
- 5-chloropyridin-3-yl (R)-2-(6-methoxynaphthalen-2-yl)propanoate (9a). Commercially available (R)-2-(6-methoxynaphthalen-2-yl)propanoic acid (60 mg, 0.26 mmol) was esterified with 5-chloropyridin-3-ol (40 mg, 0.31 mmol) by following the procedure for ester 6a to provide the title ester 9a (45 mg, 50%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.42 (d, J = 2.1 Hz, 1H), 8.24 (d, J = 2.3 Hz, 1H), 7.79–7.72 (m, 3H), 7.49–7.42 (m, 2H), 7.18 (dd, J = 8.9, 2.5 Hz, 1H), 7.14 (d, J = 2.5 Hz, 1H), 4.12 (q, J = 7.1 Hz, 1H), 3.92 (s, 3H), 1.70 (d, J = 7.1 Hz, 3H); 13C NMR (100 MHz, CDCl3) δ 172.2, 157.8, 147.2, 145.8, 141.1, 134.2, 133.9, 131.5, 129.2, 129.1, 128.9, 127.5, 126.1, 125.7, 119.3, 105.5, 55.2, 45.4, 18.3; LRMS-ESI (m/z): 342.1 [M + H]+.
- 5-chloropyridin-3-yl 2-(4-isobutylphenyl)propanoate (10a). Commercially available 2-(4-isobutylphenyl)propanoic acid (50 mg, 0.24 mmol) was esterified with 5-chloropyridin-3-ol (47 mg, 0.36 mmol) by following the procedure for ester 6a to provide the title ester 10a (35 mg, 46%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.43–8.41 (m, 1H), 8.25–8.22 (m, 1H), 7.44 (t, J = 2.2 Hz, 1H), 7.29–7.25 (m, 2H), 7.15 (d, J = 8.1 Hz, 2H), 3.95 (q, J = 7.1 Hz, 1H), 2.47 (d, J = 7.2 Hz, 2H), 1.93–1.81 (m, 1H), 1.61 (d, J = 7.1 Hz, 3H), 0.91 (d, J = 6.6 Hz, 6H); 13C NMR (100 MHz, CDCl3) δ 172.3, 147.3, 145.7, 141.1, 141.1, 136.3, 131.6, 129.6, 129.2, 127.1, 45.1, 44.9, 30.1, 22.3, 18.3; LRMS-ESI (m/z): 318.1 [M + H]+. HRMS (ESI/LTQ) m/z. [M + H]+ calcd for C18H21ClNO2 318.1255; found 318.1251.
- 5-chloropyridin-3-yl 2-(1-(4-chlorobenzoyl)-5-methoxy-2-methyl-1H-indol-3-yl)acetate (11a). Commercially available 2-(1-(4-chlorobenzoyl)-5-methoxy-2-methyl-1H-indol-3-yl)acetic acid (50 mg, 0.14 mmol) was esterified with 5-chloropyridin-3-ol (22 mg, 0.17mmol) by following the procedure for ester 6a to provide the title ester 11a (36 mg, 53%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.39 (d, J = 53.8 Hz, 2H), 7.68 (d, J = 8.2 Hz, 2H), 7.56–7.43 (m, 3H), 7.01 (d, J = 2.6 Hz, 1H), 6.87 (d, J = 9.0 Hz, 1H), 6.70 (dd, J = 9.0, 2.5 Hz, 1H), 3.94 (s, 2H), 3.84 (s, 3H), 2.46 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 168.3, 168.2, 156.1, 145.9, 141.0, 139.4, 136.4, 133.6, 131.1, 130.7, 130.1, 129.2, 129.1, 115.0, 111.7, 111.0, 101.1, 55.7, 30.3, 13.3; LRMS-ESI (m/z): 470.1 [M + H]+. HRMS (ESI/LTQ) m/z. [M + H]+ calcd for C24H19Cl2N2O4 469.07164; found 469.07131.
- 5-chloropyridin-3-yl 2-hydroxybenzoate (12a) and 5-chloropyridin-3-yl 2-((2-hydroxybenzoyl)oxy)benzoate (12b). Commercially available 2-hydroxybenzoic acid (50 mg, 0.36 mmol) was esterified with 5-chloropyridin-3-ol (56.3 mg, 0.43mmol) by following the procedure for ester 6a to provide the title monoester 12a (10 mg, 11%), and diester 12b (13 mg, 10%) as amorphous solid.
- 12a: 1H NMR (400 MHz, CDCl3) δ 10.17 (d, J = 0.5 Hz, 1H), 8.54 (dd, J = 2.0, 0.5 Hz, 1H), 8.48 (dd, J = 2.3, 0.5 Hz, 1H), 8.04 (ddd, J = 8.0, 1.8, 0.5 Hz, 1H), 7.68 (t, J = 2.2 Hz, 1H), 7.58 (dddd, J = 8.4, 7.2, 1.7, 0.5 Hz, 1H), 7.07 (ddd, J = 8.5, 1.1, 0.5 Hz, 1H), 7.03–6.98 (m, 1H); 13C NMR (100 MHz, CDCl3) δ 167.9, 162.4, 146.4, 141.3, 137.2 (×2), 131.9, 130.2, 129.5, 119.7, 118.1, 110.8; LRMS-ESI (m/z): 250.0 [M + H]+. HRMS (ESI/LTQ) m/z. [M + H]+ calcd for C12H9ClNO3 250.02655; found 250.02579.
- 12b:1H NMR (400 MHz, CDCl3) δ 10.24 (d, J = 0.4 Hz, 1H), 8.44 (dd, J = 2.1, 0.5 Hz, 1H), 8.32–8.26 (m, 2H), 8.09 (ddd, J = 8.0, 1.8, 0.4 Hz, 1H), 7.76 (ddd, J = 8.2, 7.5, 1.7 Hz, 1H), 7.55–7.48 (m, 3H), 7.35 (dd, J = 8.2, 1.1 Hz, 1H), 7.02 (ddd, J = 8.5, 1.1, 0.4 Hz, 1H), 6.96 (ddd, J = 8.3, 7.3, 1.1 Hz, 1H); 13C NMR (200 MHz, CDCl3) δ 168.7, 162.1, 161.7, 150.5, 146.9, 146.1, 141.1, 136.7, 135.3, 132.4, 131.8, 130.4, 129.4, 126.8, 124.3, 121.7, 119.7, 117.9, 111.5; LRMS-ESI (m/z): 370.0 [M + H]+. HRMS (ESI/LTQ) m/z. [M + H]+ calcd for C19H13ClNO5 370.04768; found 370.04688.
- 5-chloropyridin-3-yl 2-hydroxy-3-methylbenzoate (13a) and 5-chloropyridin-3-yl 2-((2-hydroxy-3-methylbenzoyl)oxy)-3-methylbenzoate (13b). Commercially available 2-hydroxy-3-methylbenzoic acid (30 mg, 0.20 mmol) was esterified with 5-chloropyridin-3-ol (31.0 mg, 0.24 mmol) by following the procedure for ester 6a to provide the title monoester 13a (10 mg, 19%), and diester 13b (17 mg, 22%) as amorphous solid.
- 13a: 1H NMR (400 MHz, CDCl3) δ 10.41 (s, 1H), 8.51 (d, J = 27.8 Hz, 2H), 7.89 (d, J = 8.0 Hz, 1H), 7.71–7.63 (m, 1H), 7.44 (d, J = 7.2 Hz, 1H), 6.89 (t, J = 7.7 Hz, 1H), 1.56 (s, 3H).; 13C NMR (200 MHz, CDCl3) δ 168.3, 160.8, 146.2, 141.2, 137.9 (2), 129.7, 127.7, 127.2, 119.1 (2), 110.0, 15.6; LRMS-ESI (m/z): 264.0 [M + H]+. HRMS (ESI/LTQ) m/z. [M + H]+ calcd for C13H11ClNO3 264.04220; found 264.04152.
- 13b: 1H NMR (400 MHz, CDCl3) δ 10.55 (s, 1H), 8.42 (s, 1H), 8.30 (s, 1H), 8.13–8.05 (m, 1H), 8.00–7.92 (m, 1H), 7.61 (d, J = 7.5 Hz, 1H), 7.54–7.47 (m, 1H), 7.44–7.33 (m, 2H), 6.86 (t, J = 7.7 Hz, 1H), 2.32 (s, 3H), 2.28 (s, 3H).; 13C NMR (200 MHz, CDCl3) δ 168.8, 162.0, 160.5, 149.1, 145.9, 141.1, 137.5, 136.9, 132.9, 130.1, 129.5, 127.9 (2), 127.0, 126.4 (2), 121.7, 119.1, 110.6, 16.2, 15.6; LRMS-ESI (m/z): 398.0 [M + H]+. HRMS (ESI/LTQ) m/z. [M + H]+ calcd for C21H17ClNO5 398.07898; found 398.07824.
- 5-chloropyridin-3-yl 2-hydroxy-4-methylbenzoate (14a) and 5-chloropyridin-3-yl 2-((2-hydroxy-4-methylbenzoyl)oxy)-4-methylbenzoate (14b). Commercially available 2-hydroxy-4-methylbenzoic acid (200 mg, 1.31 mmol) was esterified with 5-chloropyridin-3-ol (204.3 mg, 1.58 mmol) by following the procedure for ester 6a to provide the title monoester 14a (52 mg, 15%), and diester 14b (15 mg, 3%) as amorphous solid.
- 14a: 1H NMR (400 MHz, CDCl3) δ 10.11 (s, 1H), 8.52 (d, J = 2.1 Hz, 1H), 8.46 (d, J = 2.4 Hz, 1H), 7.89 (d, J = 8.2 Hz, 1H), 7.66 (t, J = 2.2 Hz, 1H), 6.86 (dd, J = 1.8, 0.9 Hz, 1H), 6.83–6.76 (m, 1H), 2.39 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 167.7, 162.3, 148.9, 146.7, 146.2, 141.3, 131.8, 130.0, 129.5, 121.0, 118.1, 108.1, 22.0; LRMS-ESI (m/z): 264.0 [M + H]+. HRMS (ESI/LTQ) m/z. [M + H]+ calcd for C13H11ClNO3 264.04220; found 264.04148.
- 14b: 1H NMR (400 MHz, CDCl3) δ 10.21 (s, 1H), 8.42 (d, J = 2.1 Hz, 1H), 8.30 (d, J = 2.3 Hz, 1H), 8.14 (d, J = 8.0 Hz, 1H), 7.95 (d, J = 8.1 Hz, 1H), 7.51 (t, J = 2.2 Hz, 1H), 7.28 (ddd, J = 8.1, 1.7, 0.8 Hz, 1H), 7.14 (d, J = 0.8 Hz, 1H), 6.82 (dd, J = 1.8, 0.9 Hz, 1H), 6.76 (ddd, J = 8.1, 1.6, 0.6 Hz, 1H), 2.50 (s, 3H), 2.36 (s, 3H); 13C NMR (200 MHz, CDCl3) δ 168.8, 162.0, 161.8, 150.5, 148.3, 147.0, 147.0, 145.9, 141.1, 132.3, 131.7, 130.2, 129.5, 127.6, 124.8, 121.0, 118.8, 117.9, 109.0, 21.9, 21.6; LRMS-ESI (m/z): 398.0 [M + H]+. HRMS (ESI/LTQ) m/z. [M + H]+ calcd for C21H17ClNO5 398.07898; found 398.07865.
- 5-chloropyridin-3-yl 2-hydroxy-5-methylbenzoate (15a) and 5-chloropyridin-3-yl 2-((2-hydroxy-5-methylbenzoyl)oxy)-5-methylbenzoate (15b) and 6-chloropyridin-2-yl 2-((2-((2-hydroxy-5-methylbenzoyl)oxy)-5-methylbenzoyl)oxy)-5-methylbenzoate (15c). Commercially available 2-hydroxy-5-methylbenzoic acid (200 mg, 1.31 mmol) was esterified with 5-chloropyridin-3-ol (204.3 mg, 1.58 mmol) by following the procedure for ester 6a to provide the title monoester 15a (72 mg, 21%), diester 15b (29 mg, 5%), and triester 15c (11 mg, 5%) as amorphous solid.
- 15a: 1H NMR (400 MHz, CDCl3) δ 9.97 (s, 1H), 8.49 (dd, J = 24.3, 2.3 Hz, 2H), 7.81 (d, J = 2.4 Hz, 1H), 7.66 (t, J = 2.2 Hz, 1H), 7.37 (dd, J = 8.6, 2.3 Hz, 1H), 6.95 (d, J = 8.5 Hz, 1H), 2.33 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 167.8, 160.3, 146.7, 146.3, 141.3, 138.2, 131.8, 129.7, 129.5, 129.0, 117.8, 110.3, 20.3; LRMS-ESI (m/z): 264.0 [M + H]+. HRMS (ESI/LTQ) m/z. [M + H]+ calcd for C13H11ClNO3 264.04220; found 264.04128.
- 15b:1H NMR (400 MHz, CDCl3) δ 10.09 (s, 1H), 8.43 (s, 1H), 8.31 (s, 1H), 8.05 (d, J = 2.4 Hz, 1H), 7.87 (d, J = 2.4 Hz, 1H), 7.53 (dt, J = 4.4, 2.0 Hz, 2H), 7.35–7.19 (m, 2H), 6.91 (d, J = 8.5 Hz, 1H), 2.48 (s, 3H), 2.30 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 168.9, 161.9, 160.0, 148.2, 145.9, 141.0, 137.7, 136.8 (2), 135.92, 132.66, 129.92, 129.53, 128.80, 123.91, 121.18, 117.59 (2), 111.02, 20.77, 20.32.; LRMS-ESI (m/z): 398.0 [M + H]+. HRMS (ESI/LTQ) m/z. [M + H]+ calcd for C21H17ClNO5 398.07898; found 398.07802.
- 15c:1H NMR (400 MHz, CDCl3) δ 10.10 (d, J = 7.8 Hz, 1H), 8.44 (t, J = 3.3 Hz, 1H), 8.32 (d, J = 2.6 Hz, 1H), 8.09 (dd, J = 2.3, 0.8 Hz, 1H), 7.96 (dd, J = 2.2, 0.9 Hz, 1H), 7.84 (dd, J = 2.2, 1.0 Hz, 1H), 7.57–7.51 (m, 2H), 7.46 (dddd, J = 11.6, 8.3, 2.3, 0.8 Hz, 2H), 7.16 (d, J = 8.2 Hz, 1H), 7.08 (d, J = 8.2 Hz, 1H), 6.87 (d, J = 8.5 Hz, 1H), 2.43 (s, 3H), 2.41 (s, 3H), 2.25 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 168.8, 162.9, 159.8, 148.6, 148.2, 145.8, 137.3 (2), 136.6, 136.4, 135.9, 135.4 (2), 132.6, 132.5 (2), 130.1 (2), 129.7, 128.6, 123.8, 123.7, 121.1, 117.3 (2), 111.3, 20.8, 20.7, 20.3; LRMS-ESI (m/z): 532.0 [M + H]+. HRMS (ESI/LTQ) m/z. [M + H]+ calcd for C29H23ClNO7 532.11576; found 532.11478.
- 5-chloropyridin-3-yl 2-hydroxy-6-methylbenzoate (16a) and 5-chloropyridin-3-yl 2-((2-hydroxy-6-methylbenzoyl)oxy)-6-methylbenzoate (16b). Commercially available 2-hydroxy-6-methylbenzoic acid (70 mg, 0.46 mmol) was esterified with 5-chloropyridin-3-ol (72 mg, 0.55 mmol) by following the procedure for ester 6a to provide the title monoester 16a (14 mg, 12%), and diester 16b (15 mg, 8%) as amorphous solid.
- 16a: 1H NMR (400 MHz, CDCl3) δ 10.67 (s, 1H), 8.55 (d, J = 2.1 Hz, 1H), 8.46 (d, J = 2.4 Hz, 1H), 7.67 (t, J = 2.2 Hz, 1H), 7.39 (dd, J = 8.4, 7.5 Hz, 1H), 6.92 (ddd, J = 8.4, 1.3, 0.6 Hz, 1H), 6.84–6.81 (m, 1H), 2.69 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 169.4, 163.7, 146.4 (2), 141.4, 141.2, 135.7, 131.9, 129.6, 123.5, 116.1, 110.7, 24.2; LRMS-ESI (m/z): 264.0 [M + H]+. HRMS (ESI/LTQ) m/z. [M + H]+ calcd for C13H11ClNO3 264.04220; found 264.04109.
- 16b:1H NMR (400 MHz, CDCl3) δ 10.78 (s, 1H), 8.42 (d, J = 2.1 Hz, 1H), 8.21 (d, J = 2.4 Hz, 1H), 7.53 (t, J = 7.9 Hz, 1H), 7.40–7.27 (m, 3H), 7.16 (ddd, J = 8.2, 1.1, 0.6 Hz, 1H), 6.91–6.86 (m, 1H), 6.79 (ddd, J = 7.5, 1.3, 0.7 Hz, 1H), 2.66 (s, 3H), 2.59 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 170.1, 163.6, 163.4, 148.0, 146.1, 141.4, 140.7, 139.5, 135.5, 132.2, 129.1, 129.1, 124.3, 123.4 (2), 120.7 (2), 116.0, 111.0, 24.1, 20.5; LRMS-ESI (m/z): 398.0 [M + H]+. HRMS (ESI/LTQ) m/z. [M + H]+ calcd for C21H17ClNO5 398.07898; found 398.07808.
- 5-chloropyridin-3-yl 2-fluoro-6-hydroxybenzoate (17a). Commercially available 2-fluoro-6-hydroxybenzoic acid (50 mg, 0.32 mmol) was esterified with 5-chloropyridin-3-ol (50 mg, 0.38 mmol) by following the procedure for ester 6a to provide the title ester 17a (11 mg, 13%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 10.64 (s, 1H), 8.52 (dd, J = 21.6, 2.2 Hz, 2H), 7.68 (t, J = 2.2 Hz, 1H), 7.50 (td, J = 8.4, 6.0 Hz, 1H), 6.87 (dt, J = 8.5, 1.1 Hz, 1H), 6.72 (ddd, J = 11.0, 8.3, 1.1 Hz, 1H); 13C NMR (100 MHz, CDCl3) δ 163.5, 146.5, 146.4, 141.2, 137.0, 136.8, 131.8, 129.5, 113.8, 107.5, 107.3, 101.4; LRMS-ESI (m/z): 268.0 [M + H]+.
- 5-chloropyridin-3-yl 3-acetamidobenzoate (18a). Commercially available 3-acetamidobenzoic acid (50 mg, 0.28 mmol) was esterified with 5-chloropyridin-3-ol (43.4 mg, 0.33 mmol) by following the procedure for ester 6a to provide the title ester 18a (21 mg, 26%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.50 (d, J = 2.1 Hz, 1H), 8.45 (d, J = 2.3 Hz, 1H), 8.20 (t, J = 2.0 Hz, 1H), 7.96 (dd, J = 8.2, 2.1 Hz, 1H), 7.91 (dt, J = 7.9, 1.4 Hz, 1H), 7.66 (t, J = 2.2 Hz, 1H), 7.58 (s, 1H), 7.48 (t, J = 8.0 Hz, 1H), 2.21 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 169.6, 164.1, 145.6, 141.1, 138.9, 131.9, 129.8 (2), 129.3, 128.6, 125.5 (2), 121.1, 23.9; LRMS-ESI (m/z): 291.0 [M + H]+. HRMS (ESI/LTQ) m/z. [M + H]+ calcd for C14H12ClN2O3 291.05310; found 291.05263.
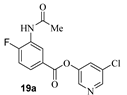
- 5-chloropyridin-3-yl 3-acetamido-4-fluorobenzoate (19a). Commercially available 3-acetamido-4-fluorobenzoic acid (50 mg, 0.25 mmol) was esterified with 5-chloropyridin-3-ol (39.4 mg, 0.30 mmol) by following the procedure for ester 6a to provide the title ester 19a (70 mg, 90%) as an amorphous solid. 1H NMR (400 MHz, MeOD) δ 8.88 (dd, J = 7.4, 2.3 Hz, 1H), 8.41 (dd, J = 10.6, 2.2 Hz, 2H), 7.91 (ddd, J = 8.6, 4.8, 2.3 Hz, 1H), 7.72 (s, 1H), 7.22 (dd, J = 10.3, 8.6 Hz, 1H), 2.19 (s, 3H); 13C NMR (100 MHz, MeOD) δ 170.4, 163.3, 147.6, 145.5, 141.0, 132.0, 130.1, 127.6, 127.5, 125.7, 124.4, 115.8, 115.6, 23.2; LRMS-ESI (m/z): 309.0 [M + H]+. HRMS (ESI/LTQ) m/z. [M + H]+ calcd for C14H11ClFN2O3 309.04367; found 309.04413.
3.2. IC50 Value Determination
IC50 values were determined for compounds that covalently inhibit SARS-CoV-2 3CLpro using our recently described assay [28] and data fitting methods that were derived from our previous work on SARS-CoV 3CLpro and inhibition by chloropyridyl esters [22]. The only differences were that pre-incubation of the enzyme with the compounds was 10 min instead of 20 min. In addition, the Morrison Equation was only used to determine the IC50 values when they were below 1 µM.
3.3. Mass Analysis of Enzyme-Inhibitor Complex
Purified SARS-CoV-2 3CLpro was injected onto a Superdex™ 200 Increase 10/300 GL gel filtration column (GE Healthcare) equilibrated in 20 mM HEPES pH 7.5. Fractions containing pure, active protein were pooled for further analysis. Protein was diluted to a final concentration of 2 μM using 20 mM HEPES pH 7.5 and incubated at room temperature with a final concentration of 20 μM compound 8a. The protein and ligand were incubated together for ten minutes before analysis.
Analysis of the proteins was performed on a 6550 iFunnel Q-TOF LC/MS (Agilent Technologies, Santa Clara, CA, USA). A sample (6 ul) was injected on to a Zorbax Extend C18 column (Agilent Technologies) kept at 60 degrees C. The mobile phase consisted of B = acetonitrile and A = 0.1% aqueous formic acid. The flow rate was 0.4 mL/min with a gradient as follows: 0–2 min 3% B; 2–7 min 95% B; 7–9 min 95% B; 9–11 min 3% B. For the first 2 min of the analysis, the column flow was diverted off to waste. TOF MS conditions: drying gas temperature 290 degrees C; drying gas flow 14 L/min; sheath gas temperature 400 degrees C; nebulizer 20 psi; capillary voltage 5000 V; nozzle 2000 V.
Positive ion spectra obtained were analyzed using Agilent MassHunter Qualitative Analysis Software B.06 and the deconvolution spectra were generated using Agilent MassHunter BioConfirm software B.06.
3.4. Cells, Viruses, and Antiviral Activity
VeroE6 cells and TMPRSS2-overexpressing VeroE6 (VeroE6TMPRSS2) cells were obtained from the Japanese Collection of Research Bioresources (JCRB) Cell Bank (Osaka, Japan). VeroE6 cells were maintained in Dulbecco’s modified Eagle’s medium (d-MEM) supplemented with 10% fetal bovine serum (FCS), 100 µg/mL of penicillin, and 100 µg/mL of streptomycin. VeroE6TMPRSS2 cells were maintained in d-MEM as reported [25] in the presence of 1 mg/mL of G418. SARS-CoV-2 strain JPN/TY/WK-521 (SARS-CoV-2WK-521) was obtained from the National Institute of Infectious Diseases (Tokyo, Japan).
Antiviral assay was carried out as described recently [28]. Cells were seeded in a 96-well plate (2 × 104 cells/well) and incubated. After 24 h, virus was inoculated into cells at multiplicity of infection (MOI) of 0.05. After an additional 72 h, cell culture supernatants were harvested and viral RNA was extracted using a QIAamp viral RNA minikit (Qiagen, Hilden, Germany), and quantitative RT-PCR (RT-qPCR) was then performed using One Step PrimeScript III RT-qPCR mix (TaKaRa Bio, Shiga, Japan) following the instructions of the manufacturers. The primers and probe used for detecting SARS-CoV-2 envelope (6) were 5′-ACT TCT TTT TCT TGC TTT CGT GGT-3′ (forward), 5′-GCA GCA GTA CGC ACA CAA TC-3′ (reverse), and 5′-FAM-CTA GTT ACA CTA GCC ATC CTT ACT GC-black hole quencher 1 (BHQ1)-3′ (probe). To determine the cytotoxicity of each compound, cells were seeded in a 96-well plate (2 × 104 cells/well). One day later, various concentrations of each compound were added, and cells were incubated for an additional 3 days. The 50% cytotoxic concentrations (CC50) values were determined using the WST-8 assay and Cell Counting Kit-8 (Dojindo, Kumamoto, Japan).
4. Conclusions
In conclusion, we have designed a series of 5-chloropyridinyl esters of common nonsteroidal anti-inflammatory agents. These ester derivatives were synthesized conveniently using EDC in the presence of DMAP to provide stable ester derivatives which were purified by silica gel chromatography. These compounds were evaluated in our in vitro SARS-CoV-2 3CL protease inhibitory assay. A number of compounds exhibited potent low nanomolar enzyme inhibitory activity. Presumably, the mode of inhibition involves the formation of a covalent bond with catalytic Cys145 in the active site. We expanded our studies with salicylic acid-derived derivatives. Methyl group substitution on the aromatic resulted in the synthesis of monomeric, dimeric, and trimeric ester derivatives. Interestingly, methyl substitution led to modulation of enzyme inhibitory activity. A number of these active ester derivatives also exhibited antiviral activity in VeroE6 cell-based assays with RNA-qPCR and immunocytochemistry assays. These compounds did not show cytotoxicity up to 100 µM. To obtain molecular insight, we have created an active model of compound 9a-bound SARS-CoV-2 3CL protease. The model shows formation of a covalent bond with Cys145 and a strong hydrogen bond with Gln189. Active site His41 forms a nice π–π stacking interaction with the napthyl ring of (R)-naproxen. Similar π–π stacking interactions were observed in our X-ray crystallographic studies of indol-derived active ester derivatives. Further modification of structures to improve antiviral activity is in progress.
Supplementary Materials
The following materials are available online. Characterization data, X-ray structural information, and 1H-NMR, 13C NMR and HRMS data.
Author Contributions
Conceptualization, A.K.G.; investigation, A.K.G., D.S. (Dana Shahabi), M.Y., S.K., B.J.A., E.K.L., S.-i.H., D.S.(Devika Sirohi), C.A.B.-S., R.K., H.M. and A.D.M.; software, A.K.G., D.S. (Dana Shahabi) and M.Y.; methodology, D.S. (Dana Shahabi), M.Y., B.J.A., E.K.L., A.K.G., H.M. and A.D.M.; original draft writing, A.K.G. and D.S. (Dana Shahabi); writing and editing, all authors; funding acquisition, A.K.G., S.-i.H., A.D.M. and H.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by NIH-NAID; A.K.G.—AI158649 and A.D.M—A1150466. Contract No. HHSN272201700060C.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The research was supported in part by grants from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (A.K.G., AI158649 and AI085089, and A.D.M., contract No. HHSN272201700060C). The present work was also supported by a grant for Development of Novel Drugs for Treating COVID-19 from the Intramural Research Program of National Center for Global Health and Medicine (H.M., 19A3001 and S.-i.H., 20A2001D), in part by Japan Agency for Medical Research and Development (AMED) (H.M., 20fk0108257) and by the Intramural Research Program of Center for Cancer Research, National Cancer Institute, National Institutes of Health (H.M.). The authors acknowledge support from the Purdue Center for Cancer Research, NIH grant P30 CA023168 for use of the shared NMR and mass spectrometry facilities. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817).
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are not available from the authors.
References
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef]
- Fauci, A.S.; Lane, H.C.; Redfield, R.R. Covid-19—Navigating the uncharted. N. Engl. Med. 2020, 382, 1268–1269. [Google Scholar] [CrossRef]
- World Health Organization. Coronavirus Disease (COVID-19) Situation Report. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 23 September 2021).
- Siemieniuk, R.A.; Bartoszko, J.J.; Ge, L.; Zeraatkar, D.; Izcovich, A.; Kum, E.; Pardo-Hernandez, H.; Qasim, A.; Martinez, J.P.D.; Rochwerg, B.; et al. Drug treatments for covid-19: Living systematic review and network meta-analysis. BMJ 2020, 370, m2980. [Google Scholar] [CrossRef]
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of Covid-19—Final Report. N. Engl. J. Med. 2020, 383, 1813–1826. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.P.; Gupta, V. COVID-19 vaccine: A comprehensive status report. Virus Res. 2020, 288, 198114. [Google Scholar] [CrossRef]
- Poland, G.A.; Ovsyannikova, I.G.; Kennedy, R.B. SARS-CoV-2 immunity: Review and applications to phase 3 vaccine candidates. Lancet 2020, 396, 1595–1606. [Google Scholar] [CrossRef]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [Green Version]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef]
- Forni, D.; Cagliani, R.; Clerici, M.; Sironi, M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017, 25, 35–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajpoot, S.; Alagumuthu, M.; Baig, M.S. Dual targeting of 3CLpro and PLpro of SARS-CoV-2: A novel structure-based design approach to treat COVID-19. Curr. Res. Struct. Biol. 2021, 3, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, A.K.; Brindisi, M.; Shahabi, D.; Chapman, M.E.; Mesecar, A.D. Drug development and medicinal chemistry efforts toward SARS-coronavirus and Covid-19 therapeutics. ChemMedChem 2020, 15, 907–932. [Google Scholar] [CrossRef]
- Pillaiyar, T.; Manickam, M.; Namasivayam, V.; Hayashi, Y.; Jung, S.-H. An overview of severe acute respiratory syndrome–coronavirus (SARS-CoV) 3CL protease inhibitors: Peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016, 59, 6595–6628. [Google Scholar] [CrossRef]
- Pillaiyar, T.; Meenakshisundaram, S.; Manickam, M. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov. Today 2020, 25, 668–688. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Xi, K.; Johnson, M.E.; Baker, S.C.; Mesecar, A.D. Progress in anti-SARS coronavirus chemistry, biology and chemotherapy. Annu. Rep. Med. Chem. 2007, 41, 183–196. [Google Scholar]
- Wu, C.Y.; King, K.Y.; Kuo, C.J.; Fang, J.M.; Wu, Y.T.; Ho, M.Y.; Liao, C.L.; Shie, J.J.; Liang, P.H.; Wong, C.H. Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL protease. Chem. Biol. 2006, 13, 261–268. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, A.K.; Gong, G.; Grum-Tokars, V.; Mulhearn, D.C.; Baker, S.C.; Coughlin, M.; Prabhakar, B.S.; Sleeman, K.; Johnson, M.E.; Mesecar, A.D. Design, synthesis and antiviral efficacy of a series of potent chloropyridyl ester-derived SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2008, 18, 5684–5688. [Google Scholar] [CrossRef]
- Blanchard, J.E.; Elowe, N.H.; Huitema, C.; Fortin, P.D.; Cechetto, J.D.; Eltis, L.D.; Brown, E.D. High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chem. Biol. 2004, 11, 1445–1453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Pettersson, H.I.; Huitema, C.; Niu, C.; Yin, J.; James, M.N.; Eltis, L.D.; Vederas, J.C. Design, synthesis, and evaluation of inhibitors for severe acute respiratory syndrome 3C-like protease based on phthalhydrazide ketones or heteroaromatic esters. J. Med. Chem. 2007, 50, 1850–1864. [Google Scholar] [CrossRef]
- Hattori, S.-i.; Higshi-Kuwata, N.; Raghavaiah, J.; Das, D.; Bulut, H.; Davis, D.A.; Takamatsu, Y.; Matsuda, K.; Takamune, N.; Kishimoto, N.; et al. GRL-0920, an indole chloropyridinyl ester, completely blocks SARS-CoV-2 infection. MBio 2020, 11, e01833-20. [Google Scholar] [CrossRef]
- Spinner, C.D.; Gottlieb, R.L.; Criner, G.J.; López, J.R.A.; Cattelan, A.M.; Viladomiu, A.S.; Ogbuagu, O.; Malhotra, P.; Mullane, K.M.; Castagna, A.; et al. Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19. A randomized clinical trial. JAMA 2020, 324, 1048–1057. [Google Scholar] [CrossRef] [PubMed]
- McCreary, E.K.; Angus, D.C. Efficacy of remdesivir in COVID-19. JAMA 2020, 324, 1041–1042. [Google Scholar] [CrossRef] [PubMed]
- Hattori, S.-I.; Higashi-Kuwata, N.; Hayashi, H.; Allu, S.R.; Raghavaiah, J.; Bulut, H.; Das, D.; Anson, B.J.; Lendy, E.K.; Takamatsu, Y.; et al. A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2021, 12, 668. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Raghavaiah, J.; Shahabi, D.; Yadav, M.; Anson, B.J.; Lendy, E.K.; Hattori, S.-I.; Higashi-Kuwata, N.; Mitsuya, H.; Mesecar, A.D. Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme inhibition, Antiviral Efficacy, Structure-Activity and X-ray Structural Studies. J. Med. Chem. 2021, in press. [Google Scholar] [CrossRef]
- Wongrakpanich, S.; Wongrakpanich, A.; Melhado, K.; Rangaswami, J. A comprehensive review of non-steroidal anti-inflammatory drug use in the elderly. Aging Dis. 2018, 9, 143–150. [Google Scholar] [CrossRef] [Green Version]
- Peesa, J.P.; Yalavarthi, P.R.; Rasheed, A.; Mandava, V.B.R. A perspective review on role of novel NSAID prodrugs in the management of acute inflammation. J. Acute Dis. 2016, 5, 364–381. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; StJohn, S.E.; Osswald, H.L.; O’Brien, A.; Banach, B.S.; Sleeman, K.; Ghosh, A.K.; Mesecar, A.D.; Baker, S.C. Coronaviruses resistant to a 3C-like protease inhibitor are attenuated for replication and pathogenesis, revealing a low genetic barrier but high fitness cost of resistance. J. Virol. 2014, 88, 11886–11898. [Google Scholar] [CrossRef] [Green Version]
- Zeller, M. Single-Crystal X-Ray Analysis was Performed in Our X-ray Crystallography Laboratory; Department of Chemistry, Purdue University: West Lafayette, IN, USA.
- CCDC 2105273 Contains the Supplementary Crystallographic Data for Compound 16b. This Data Can be Obtained Free of Charge from The Cambridge Crystallographic Data Centre via. Available online: www.ccdc.cam.ac.uk/data_request/cif (accessed on 23 September 2021).
- Anson, B.J.; Chapman, M.E.; Lendy, E.K.; Pshenychnyi, S.; D’Aquila, R.T.; Satchell, K.J.F.; Mesecar, A.D. Broad-spectrum inhibition of coronavirus main and papain-like proteases by HCV drugs. Res. Sq. 2020. preprint. [Google Scholar] [CrossRef]
- Grum-Tokars, V.; Ratia, K.; Begaye, A.; Baker, S.C.; Mesecar, A.D. Evaluating the 3C-like protease activity of SARS-Coronavirus: Recommendations for standardized assays for drug discovery. Virus. Res. 2008, 133, 63–73. [Google Scholar] [CrossRef]
- Hoffman, R.L.; Kania, R.S.; Brothers, M.A.; Davies, J.F.; Ferre, R.A.; Gajiwala, K.S.; He, M.; Hogan, R.J.; Kozminski, K.; Li, L.Y.; et al. Discovery of ketone-based covalent inhibitors of coronavirus 3CL proteases for the potential therapeutic treatment of COVID-19. J. Med. Chem. 2020, 63, 12725–12747. [Google Scholar] [CrossRef] [PubMed]
- de Vries, M.; Mohamed, A.S.; Prescott, R.A.; Valero-Jimenez, A.M.; Desvignes, L.; O’Connor, R.; Steppan, C.; Devlin, J.C.; Ivanova, E.; Herrera, A.; et al. A comparative analysis of SARS-CoV-2 antivirals characterizes 3CLpro inhibitor PF-00835231 as a potential new treatment for COVID-19. J. Virol. 2021, 95, e01819-20. [Google Scholar] [CrossRef] [PubMed]
- Kalgutkar, A.S.; Frederick, K.S.; Chupka, J.; Feng, B.; Kempshall, S.; Mireles, R.J.; Fenner, K.S.; Trouman, M.D. N-(3,4-dimethoxyphenethyl)-4-(6,7-dimethoxy-3,4-dihydroisoquinolin-2[1H]-yl)-6,7-dimethoxyquinazolin-2-amine (CP-100,356) as a “chemical knock-out equivalent” to assess the impact of efflux transporters on oral drug absorption in the rat. J. Pharm. Sci. 2009, 98, 4914–4927. [Google Scholar] [CrossRef] [PubMed]
- Bruker (2016). Apex3 v2016.9-0, Saint V8.34A, SAINT V8.37A; Bruker AXS Inc.: Madison, WI, USA, 2013/2014. [Google Scholar]
- SHELXTL Suite of Programs, Version 6.14, 2000–2003, Bruker Advanced X-ray Solutions; Bruker AXS Inc.: Madison, WI, USA, 2000–2003.
- Sheldrick, G. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008, 64, 112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheldrick, G. Acta. Crystallogr. Sect. C. Struct. Chem. 2015, 71, 3. [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).