A Pillar-Based High-Throughput Myogenic Differentiation Assay to Assess Drug Safety
Abstract
:1. Introduction
2. Results
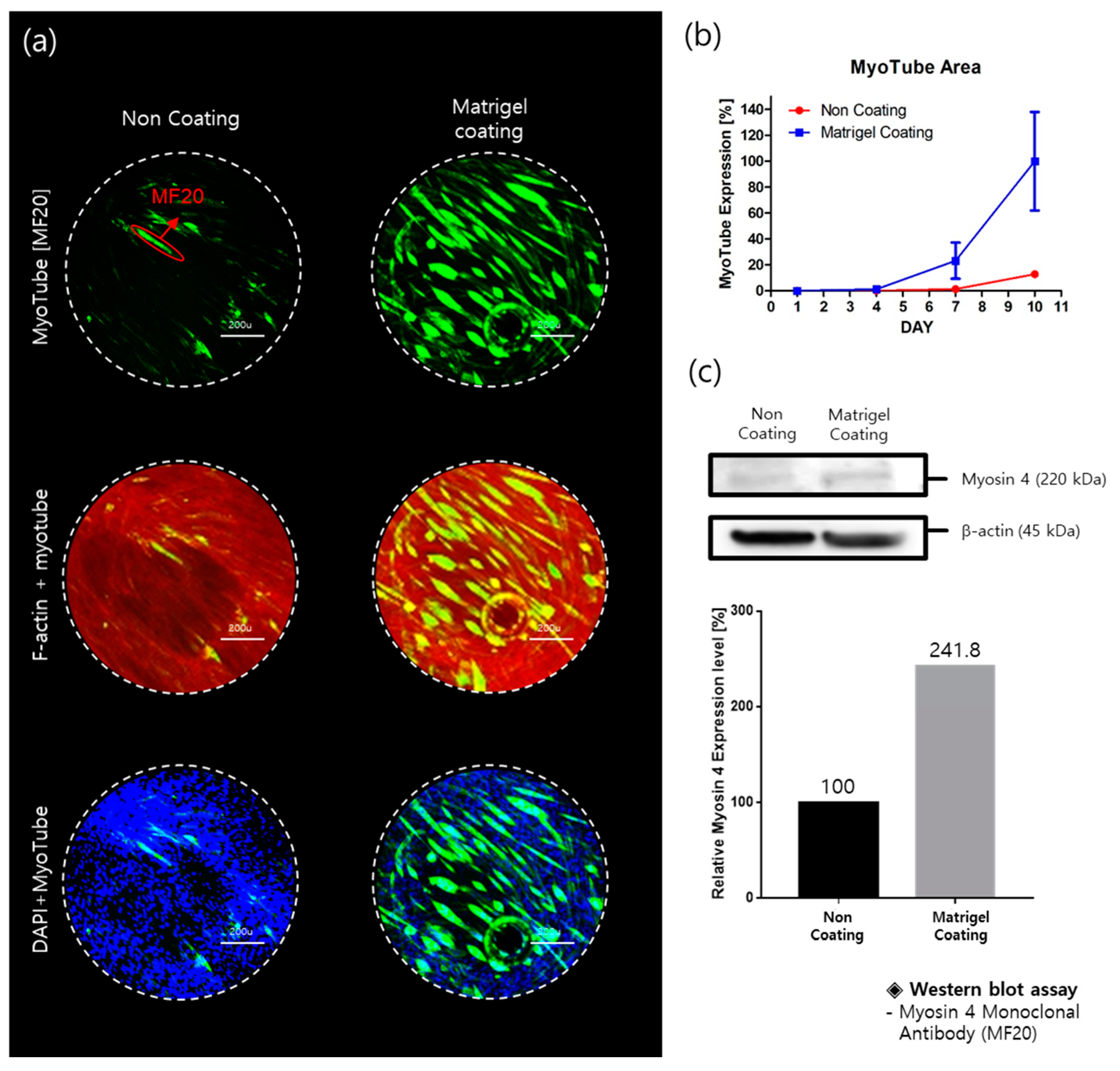
2.1. Myotube Expression
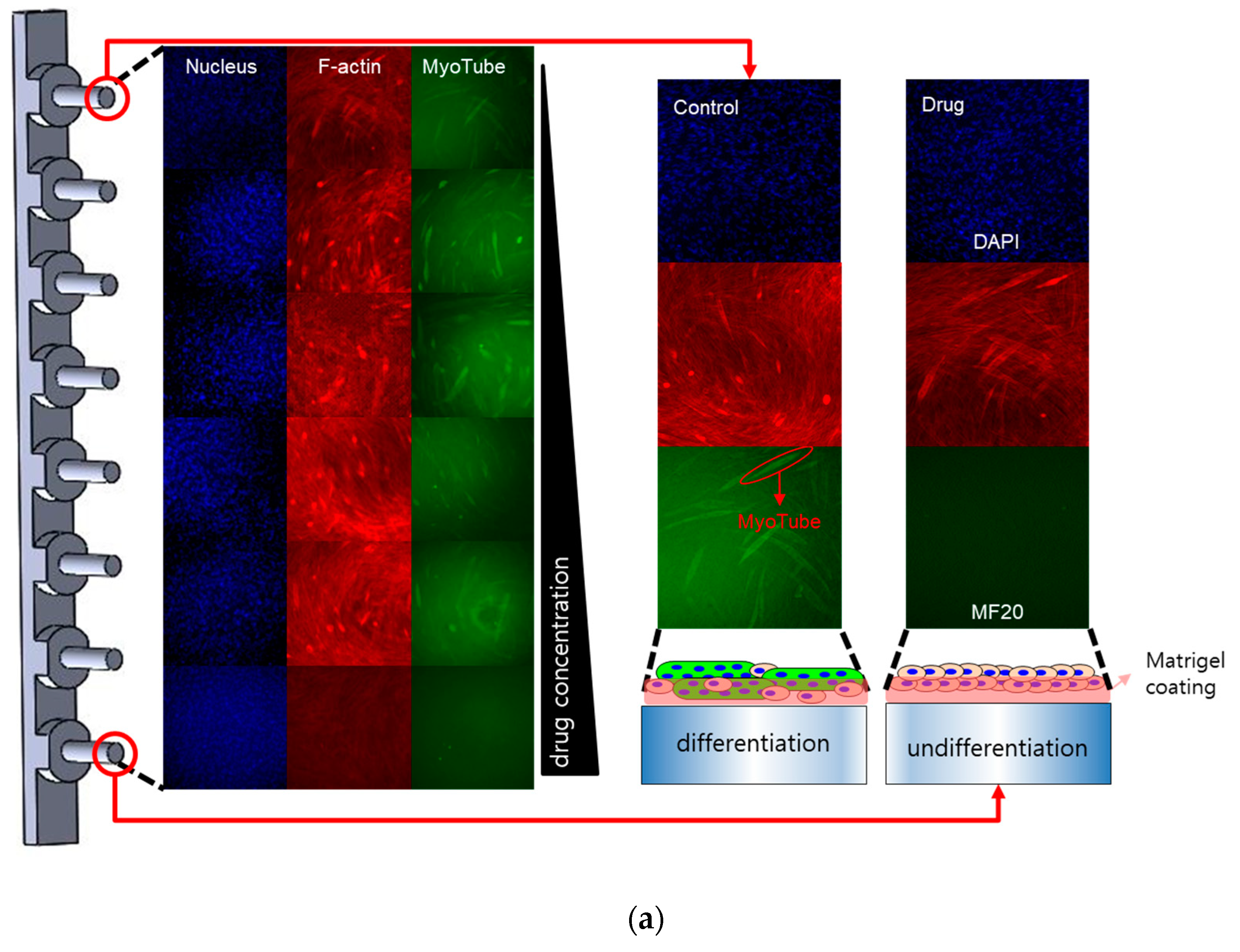
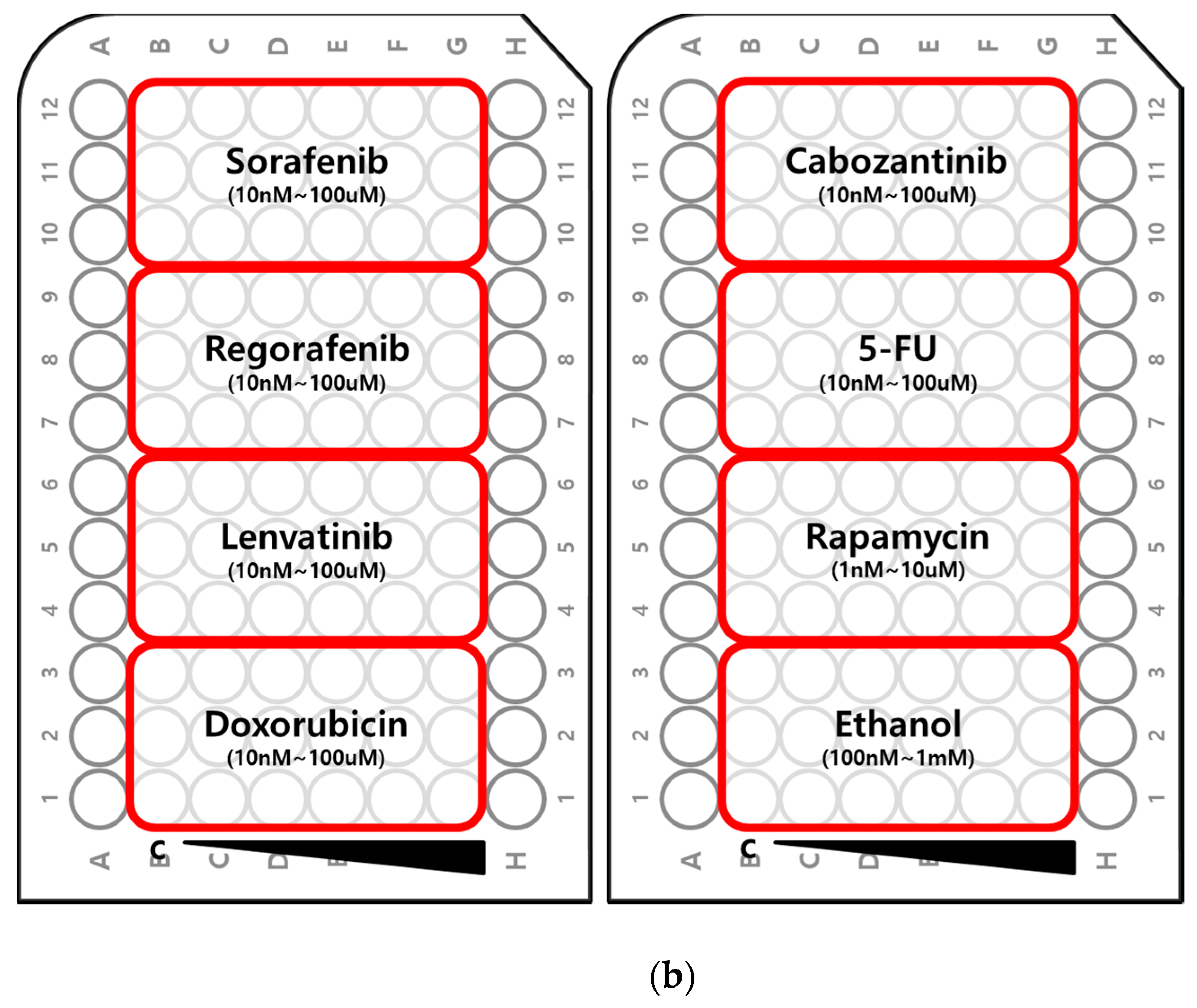
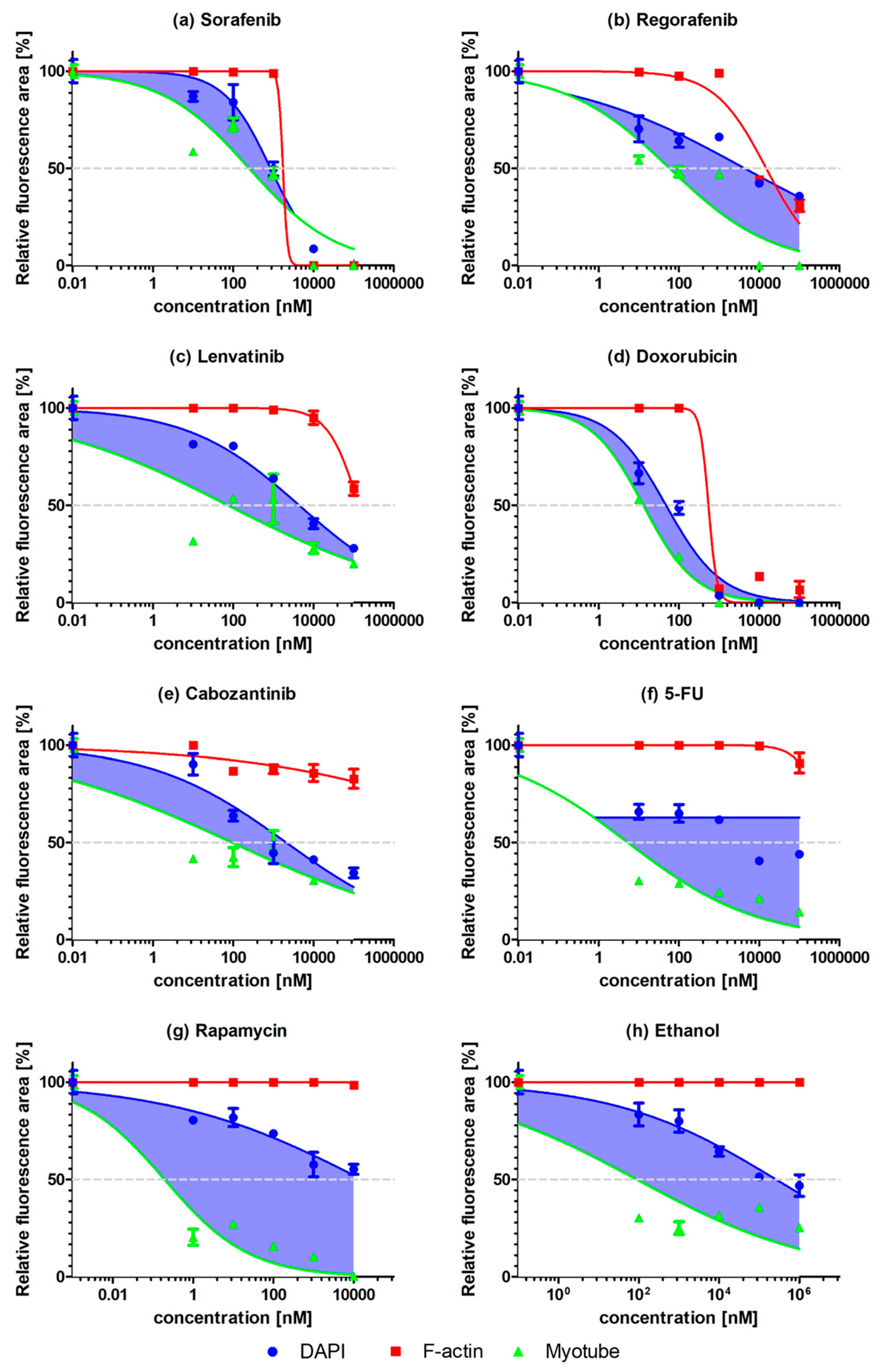
2.2. Myogenic Differentiation Assay
3. Discussion
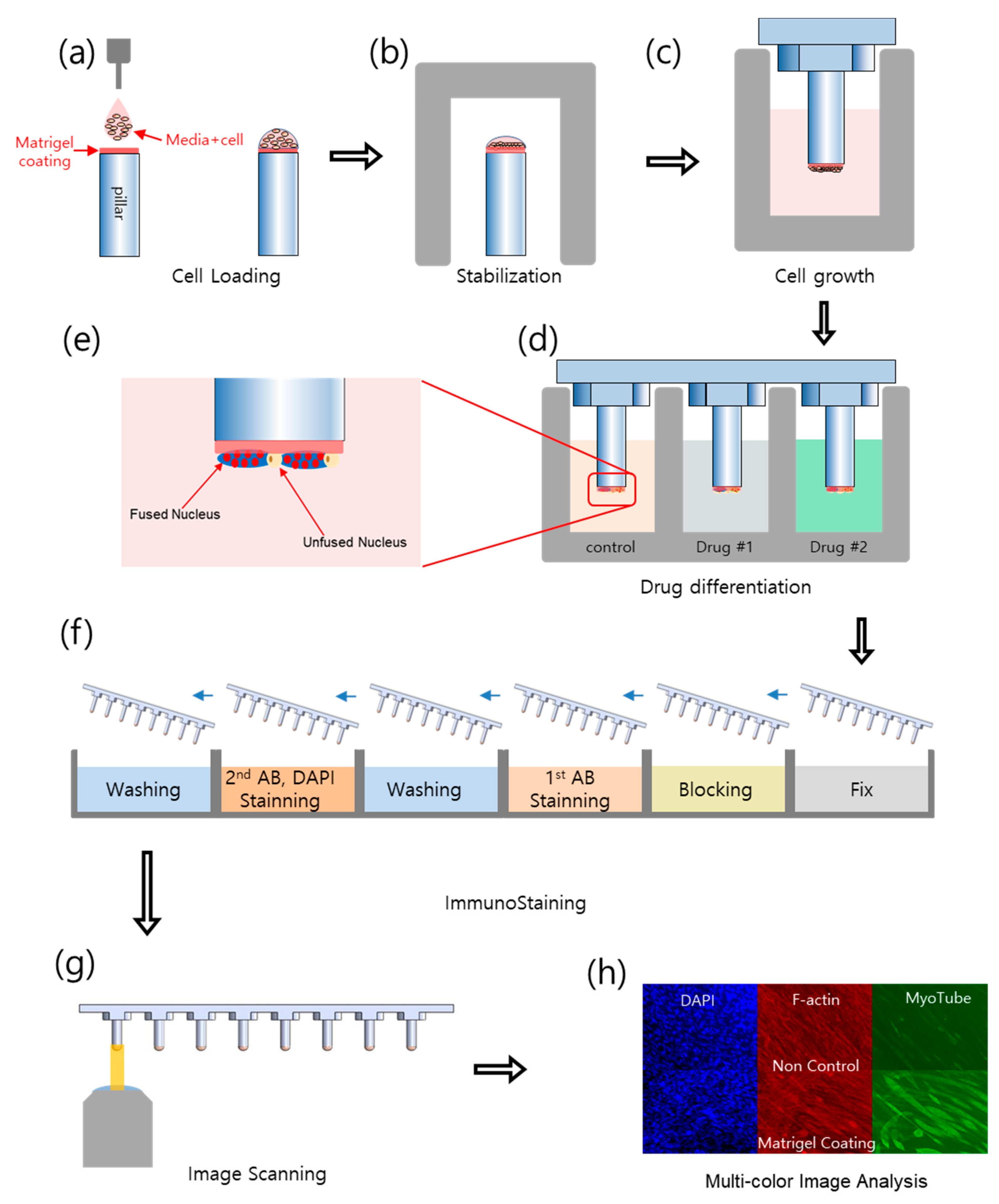
4. Materials and Methods
4.1. Cell Culture
4.2. Experiential Procedure
4.3. Immunofluorescence Staining
4.4. Image Processing and Data Analysis
4.5. Western Blot Assay
4.6. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Höfer, T.; Gerner, I.; Gundert-Remy, U.; Liebsch, M.; Schulte, A.; Spielmann, H.; Vogel, R.; Wettig, K. Animal Testing and Alternative Approaches for the Human Health Risk Assessment Under the Proposed New European Chemicals Regulation. Arch. Toxicol. 2004, 78, 549–564. [Google Scholar] [CrossRef]
- Hong, E.; Jeung, E. Assessment of Developmental Toxicants using Human Embryonic Stem Cells. Toxicol. Res. 2013, 29, 221–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Jong, E.; Louisse, J.; Verwei, M.; Blaauboer, B.J.; van de Sandt, J.J.M.; Woutersen, R.A.; Rietjens, I.M.C.M.; Piersma, A.H. Relative Developmental Toxicity of Glycol Ether Alkoxy Acid Metabolites in the Embryonic Stem Cell Test as Compared with the in Vivo Potency of their Parent Compounds. Toxicol. Sci. 2009, 110, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Flick, B.; Rietjens, I.M.; Louisse, J.; Schneider, S.; van Ravenzwaay, B. Extended Evaluation on the ES-D3 Cell Differentiation Assay Combined with the BeWo Transport Model, to Predict Relative Developmental Toxicity of Triazole Compounds. Arch. Toxicol. 2015, 90, 1225–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louisse, J.; Gönen, S.; Rietjens, I.M.C.M.; Verwei, M. Relative Developmental Toxicity Potencies of Retinoids in the Embryonic Stem Cell Test Compared with their Relative Potencies in in Vivo and Two Other in Vitro Assays for Developmental Toxicity. Toxicol. Lett. 2011, 203, 1–8. [Google Scholar] [CrossRef]
- Strikwold, M.; Woutersen, R.A.; Spenkelink, B.; Punt, A.; Rietjens, I.M. Relative Embryotoxic Potency of P-Substituted Phenols in the Embryonic Stem Cell Test (EST) and Comparison to their Toxic Potency in Vivo and in the Whole Embryo Culture (WEC) Assay. Toxicol. Lett. 2012, 213, 235–242. [Google Scholar] [CrossRef]
- Tang, J.; He, A.; Yan, H.; Jia, G.; Liu, G.; Chen, X.; Cai, J.; Tian, G.; Shang, H.; Zhao, H. Damage to the Myogenic Differentiation of C2C12 Cells by Heat Stress is Associated with Up-Regulation of several Selenoproteins. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Burattini, S.; Ferri, P.; Battistelli, M.; Curci, R.; Luchetti, F.; Falcieri, E. C2C12 Murine Myoblasts as a Model of Skeletal Muscle Development: Morpho-Functional Characterization. Eur. J. Histochem. 2004, 223–234. [Google Scholar]
- Denes, L.T.; Riley, L.A.; Mijares, J.R.; Arboleda, J.D.; McKee, K.; Esser, K.A.; Wang, E.T. Culturing C2C12 Myotubes on Micromolded Gelatin Hydrogels Accelerates Myotube Maturation. Skeletal Muscle 2019, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- McMahon, D.K.; Anderson, P.A.; Nassar, R.; Bunting, J.B.; Saba, Z.; Oakeley, A.E.; Malouf, N.N. C2C12 Cells: Biophysical, Biochemical, and Immunocytochemical Properties. Am. J. Physiol.-Cell Physiol. 1994, 266, C1795–C1802. [Google Scholar] [CrossRef]
- Blau, H.M.; Chiu, C.; Webster, C. Cytoplasmic Activation of Human Nuclear Genes in Stable Heterocaryons. Cell 1983, 32, 1171–1180. [Google Scholar] [CrossRef]
- Ikeda, K.; Ito, A.; Imada, R.; Sato, M.; Kawabe, Y.; Kamihira, M. In Vitro Drug Testing Based on Contractile Activity of C2C12 Cells in an Epigenetic Drug Model. Sci. Rep. 2017, 7, 44570. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Huang, J.; Tibbits, G.F.; Li, P.C.H. Real-Time Monitoring of Intracellular Calcium Dynamic Mobilization of a Single Cardiomyocyte in a Microfluidic Chip Pertaining to Drug Discovery. Electrophoresis 2007, 28, 4723–4733. [Google Scholar] [CrossRef]
- Hoppeler, H.; Flück, M. Normal Mammalian Skeletal Muscle and its Phenotypic Plasticity. J. Exp. Biol. 2002, 205, 2143–2152. [Google Scholar] [CrossRef] [PubMed]
- Zurlo, F.; Larson, K.; Bogardus, C.; Ravussin, E. Skeletal Muscle Metabolism is a Major Determinant of Resting Energy Expenditure. J. Clin. Investig. 1990, 86, 1423–1427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.S.J.; Zhang, M.; Evans, D.J.R. Changes in the Proportion and Number of Pax(7 + ve) and MF20(+ve) Myoblasts during Chick Myogenesis in the Head and Limb. Int. J. Dev. Biol. 2004, 48, 31–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krueger, C.; Hoffmann, F.M. Identification of Retinoic Acid in a High Content Screen for Agents that Overcome the Anti-Myogenic Effect of TGF-Beta-1. PLoS ONE 2010, 5, e15511. [Google Scholar] [CrossRef] [PubMed]
- Sin, J.; Andres, A.M.; Taylor, D.J.R.; Weston, T.; Hiraumi, Y.; Stotland, A.; Kim, B.J.; Huang, C.; Doran, K.S.; Gottlieb, R.A. Mitophagy is Required for Mitochondrial Biogenesis and Myogenic Differentiation of C2C12 Myoblasts. Autophagy 2015, 12, 369–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Shi, L.; Liang, T.; Wang, B.; Wu, W.; Su, G.; Wei, J.; Li, P.; Huang, R. MiR-696 Regulates C2C12 Cell Proliferation and Differentiation by Targeting CNTFRα. Int. J. Biol. Sci. 2017, 13, 413–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langen, R.C.; Schols, A.M.; Kelders, M.C.; Wouters, E.F.; Janssen-Heininger, Y.M. Enhanced Myogenic Differentiation by Extracellular Matrix is Regulated at the Early Stages of Myogenesis. In Vitro Cell. Dev. Biol.-Animal. 2003, 39, 163–169. [Google Scholar] [CrossRef]
- Erbay, E.; Chen, J. The Mammalian Target of Rapamycin Regulates C2C12 Myogenesis Via a Kinase-Independent Mechanism. J. Biol. Chem. 2001, 276, 36079–36082. [Google Scholar] [CrossRef] [Green Version]
- Damaraju, V.L.; Kuzma, M.; Cass, C.E.; Putman, C.T.; Sawyer, M.B. Multitargeted Kinase Inhibitors Imatinib, Sorafenib and Sunitinib Perturb Energy Metabolism and Cause Cytotoxicity to Cultured C2C12 Skeletal Muscle Derived Myotubes. Biochem. Pharmacol. 2018, 155, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Thakkar, N.S.; Potten, C.S. Inhibition of Doxorubicin-Induced Apoptosis in Vivo by 2-Deoxy-D-Glucose. Cancer Res. 1993, 53, 2057–2060. [Google Scholar]
- Takigawa, H.; Kitadai, Y.; Shinagawa, K.; Yuge, R.; Higashi, Y.; Tanaka, S.; Yasui, W.; Chayama, K. Multikinase Inhibitor Regorafenib Inhibits the Growth and Metastasis of Colon Cancer with Abundant Stroma. Cancer Sci. 2016, 107, 601–608. [Google Scholar] [CrossRef]
- Schlumberger, M.; Tahara, M.; Wirth, L.J.; Robinson, B.; Brose, M.S.; Elisei, R.; Habra, M.A.; Newbold, K.; Shah, M.H.; Hoff, A.O.; et al. Lenvatinib Versus Placebo in Radioiodine-Refractory Thyroid Cancer. N. Engl. J. Med. 2015, 372, 621–630. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhu, M.; Xie, L.; Sun, X.; Xu, J.; Guo, Y.; Liu, D.; Shi, Y.; Xu, X.; Song, E. Cabozantinib, a Multityrosine Kinase Inhibitor of MET and VEGF Receptors which Suppresses Mouse Laser-Induced Choroidal Neovascularization. J. Ophthalmol. 2020, 2020, e5905269. [Google Scholar] [CrossRef] [PubMed]
- 5-Fluorouracil Inhibits Neural Differentiation Via Mfn1/2 Reduction in Human Induced Pluripotent Stem Cells. J. Toxicol. Sci. 2018, 43, 727–734. [CrossRef] [PubMed] [Green Version]
- Arya, M.A.; Tai, A.K.; Wooten, E.C.; Parkin, C.D.; Kudryavtseva, E.; Huggins, G.S. Notch Pathway Activation Contributes to Inhibition of C2C12 Myoblast Differentiation by Ethanol. PLoS ONE 2013, 8, e71632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
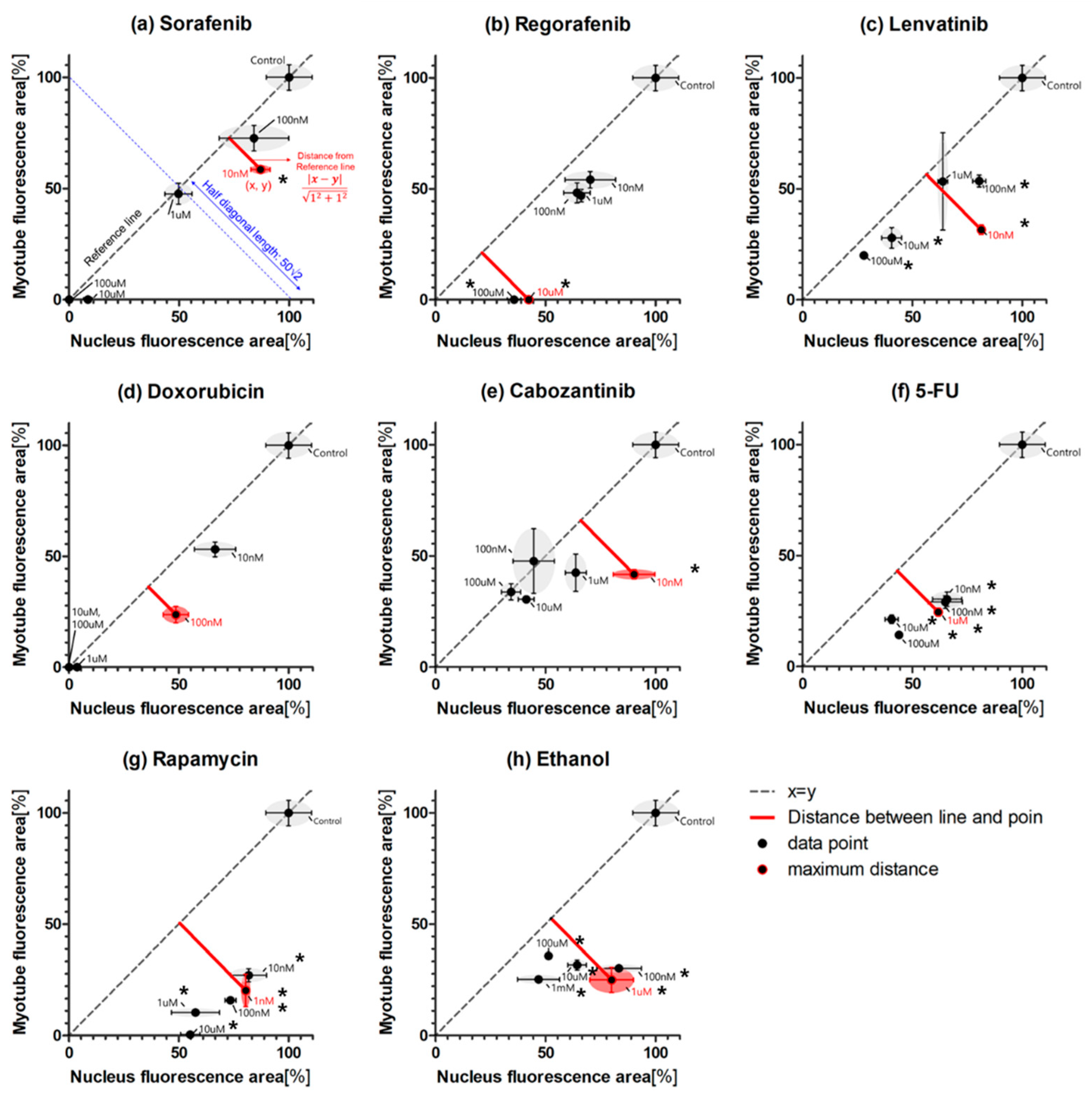
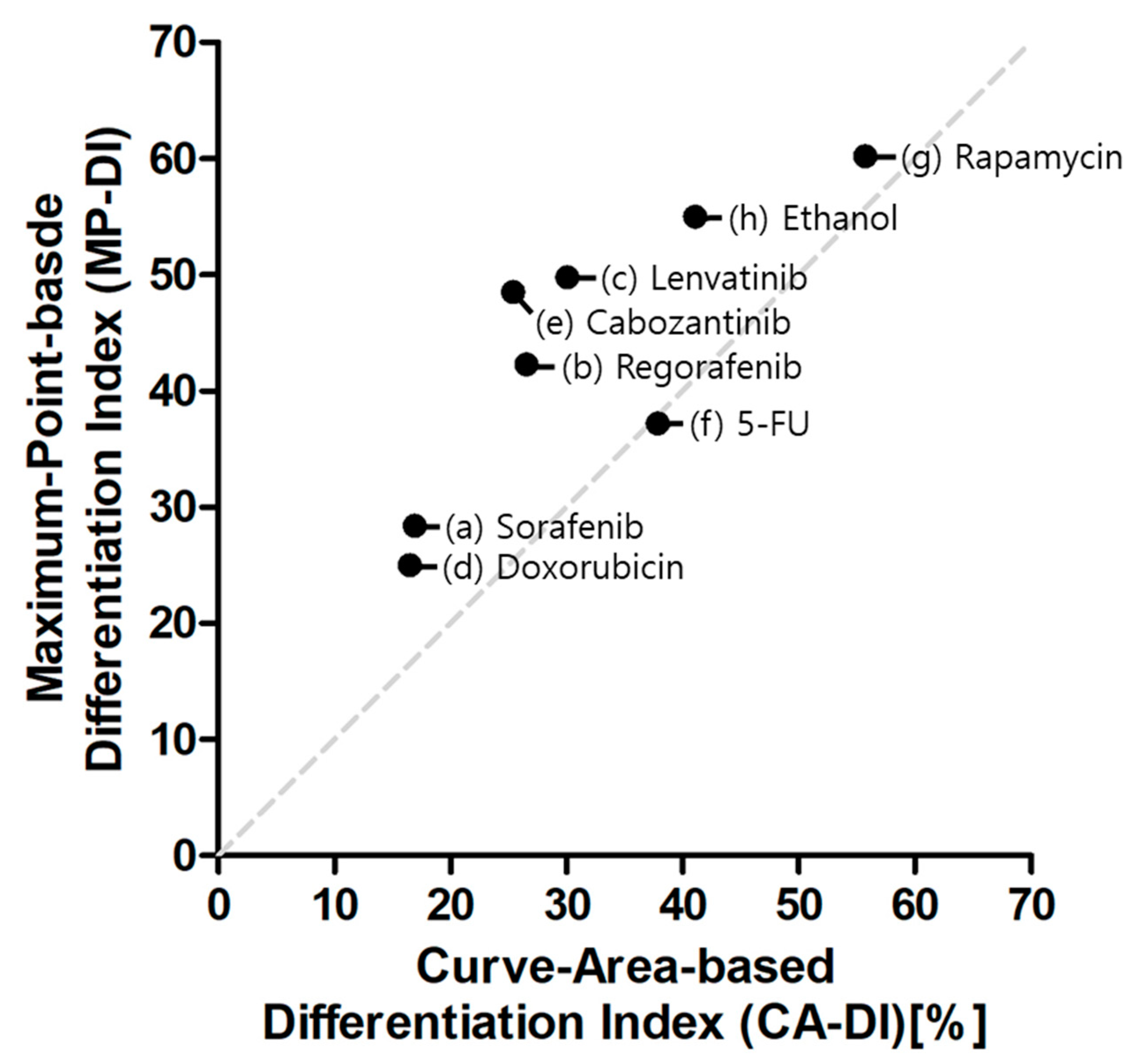
| CA-DI | MP-DI | ||||||
|---|---|---|---|---|---|---|---|
| Myotube [A.U.] | Nucleus [A.U.] | Differentiation Index [%] | Drug Concentration | Nucleus [%] | Myotube [%] | Differentiation Index [%] | |
| 1. Sorafenib [22] | 387.4 | 466.4 | 16.93 | 10 nM | 87.0 | 58.5 | 28.4 |
| 2. Regorafenib [24] | 353.5 | 481.1 | 26.52 | 10 µM | 42.3 | 0 | 42.3 |
| 3. Lenvatinib [25] | 357.8 | 511.5 | 30.04 | 10 nM | 81.4 | 31.6 | 49.8 |
| 4. Doxorubicin [23] | 280 | 335.4 | 16.51 | 100 nM | 48.6 | 23.7 | 25.0 |
| 5. Cabozantinib [26] | 370.9 | 497.2 | 25.40 | 10 nM | 90.2 | 41.7 | 48.5 |
| 6. 5-FU [27] | 292.7 | 470.8 | 37.82 | 1 µM | 61.7 | 24.5 | 37.2 |
| 7. Rapamycin [21] | 244.2 | 551.5 | 55.72 | 10 µM | 80.4 | 20.3 | 60.2 |
| 8. Ethanol [28] | 315.7 | 535.8 | 41.07 | 100 µM | 79.9 | 25.0 | 55.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahn, K.H.; Kim, S.; Yang, M.; Lee, D.W. A Pillar-Based High-Throughput Myogenic Differentiation Assay to Assess Drug Safety. Molecules 2021, 26, 5805. https://doi.org/10.3390/molecules26195805
Ahn KH, Kim S, Yang M, Lee DW. A Pillar-Based High-Throughput Myogenic Differentiation Assay to Assess Drug Safety. Molecules. 2021; 26(19):5805. https://doi.org/10.3390/molecules26195805
Chicago/Turabian StyleAhn, Kyeong Hwan, Sooil Kim, Mihi Yang, and Dong Woo Lee. 2021. "A Pillar-Based High-Throughput Myogenic Differentiation Assay to Assess Drug Safety" Molecules 26, no. 19: 5805. https://doi.org/10.3390/molecules26195805
APA StyleAhn, K. H., Kim, S., Yang, M., & Lee, D. W. (2021). A Pillar-Based High-Throughput Myogenic Differentiation Assay to Assess Drug Safety. Molecules, 26(19), 5805. https://doi.org/10.3390/molecules26195805







