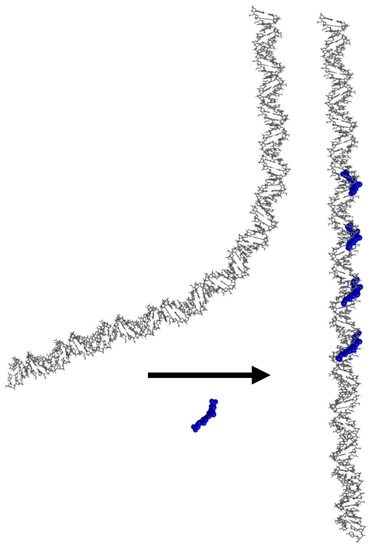
Examining the Effects of Netropsin on the Curvature of DNA A-Tracts Using Electrophoresis
Abstract
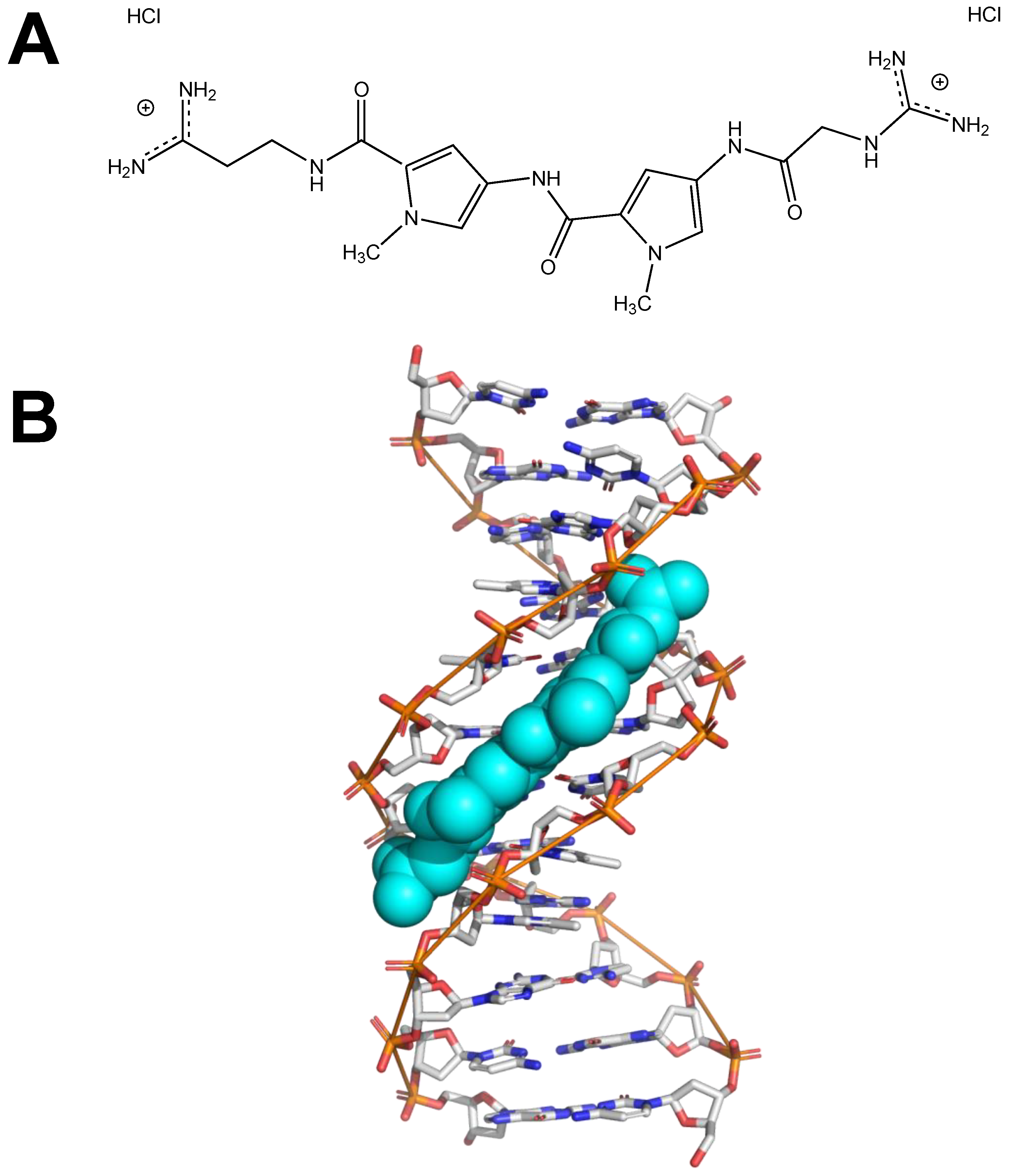
:1. Introduction
2. Results and Discussion
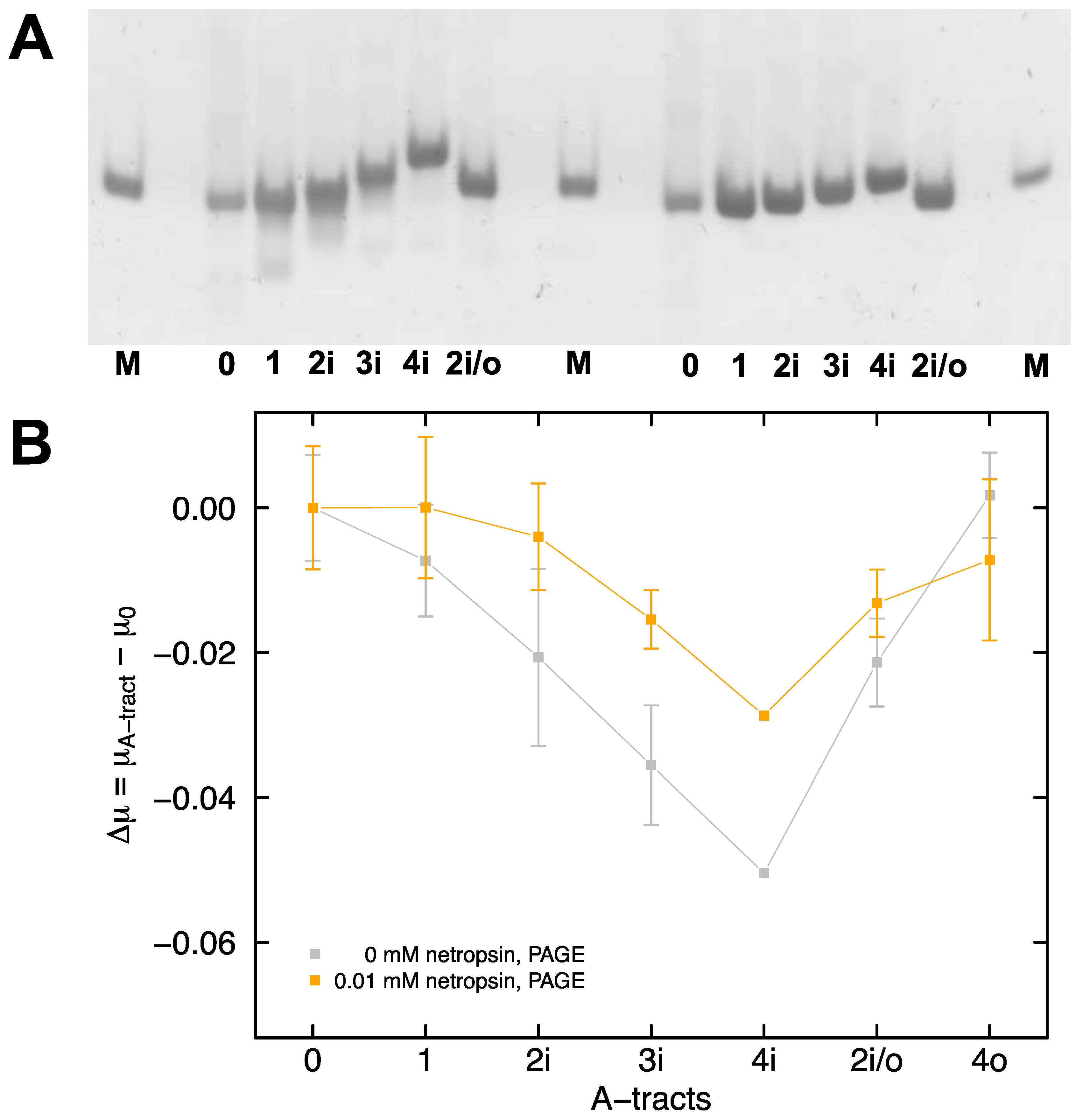
2.1. Electrophoretic Mobility of DNA in Polyacrylamide Gels
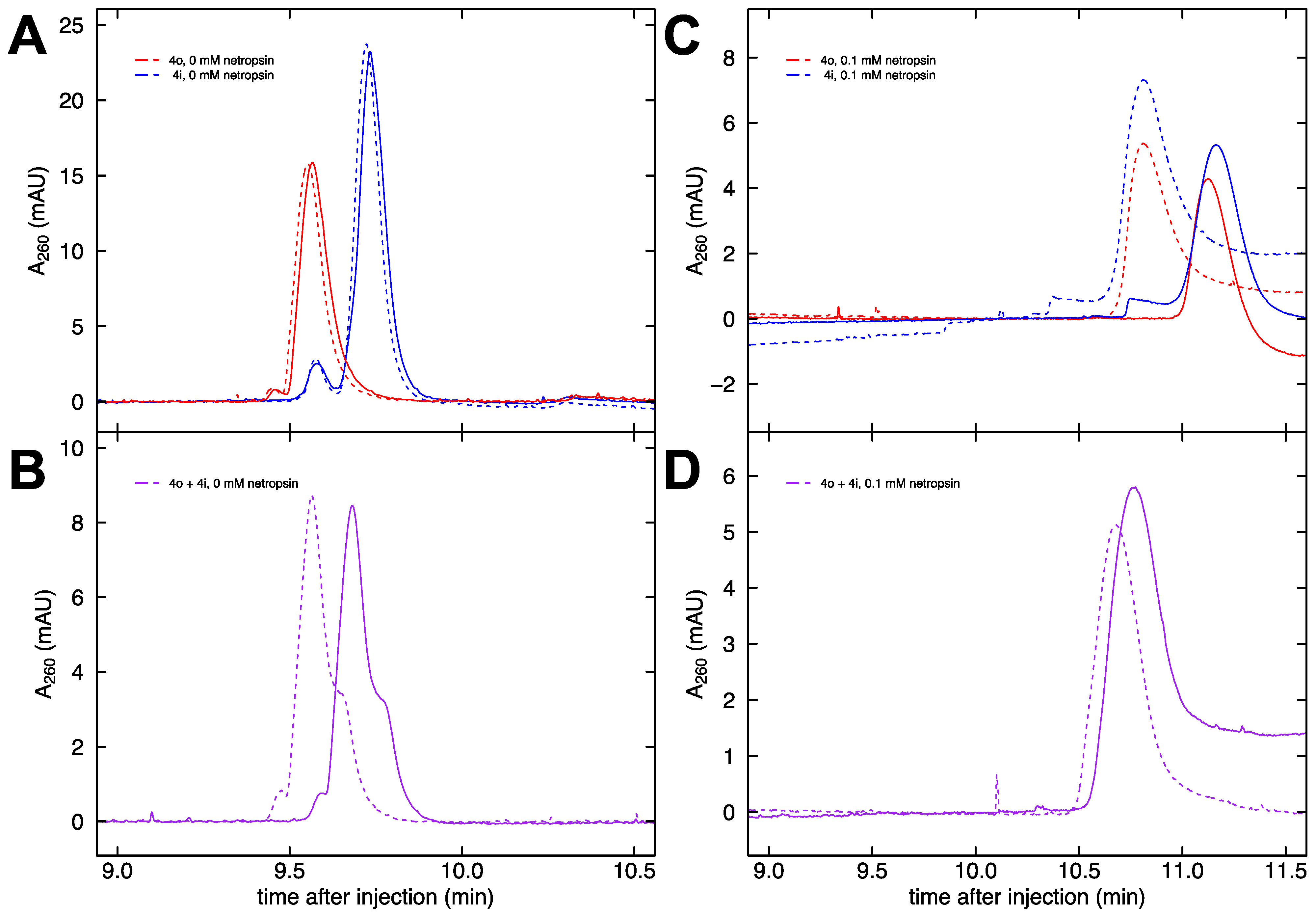
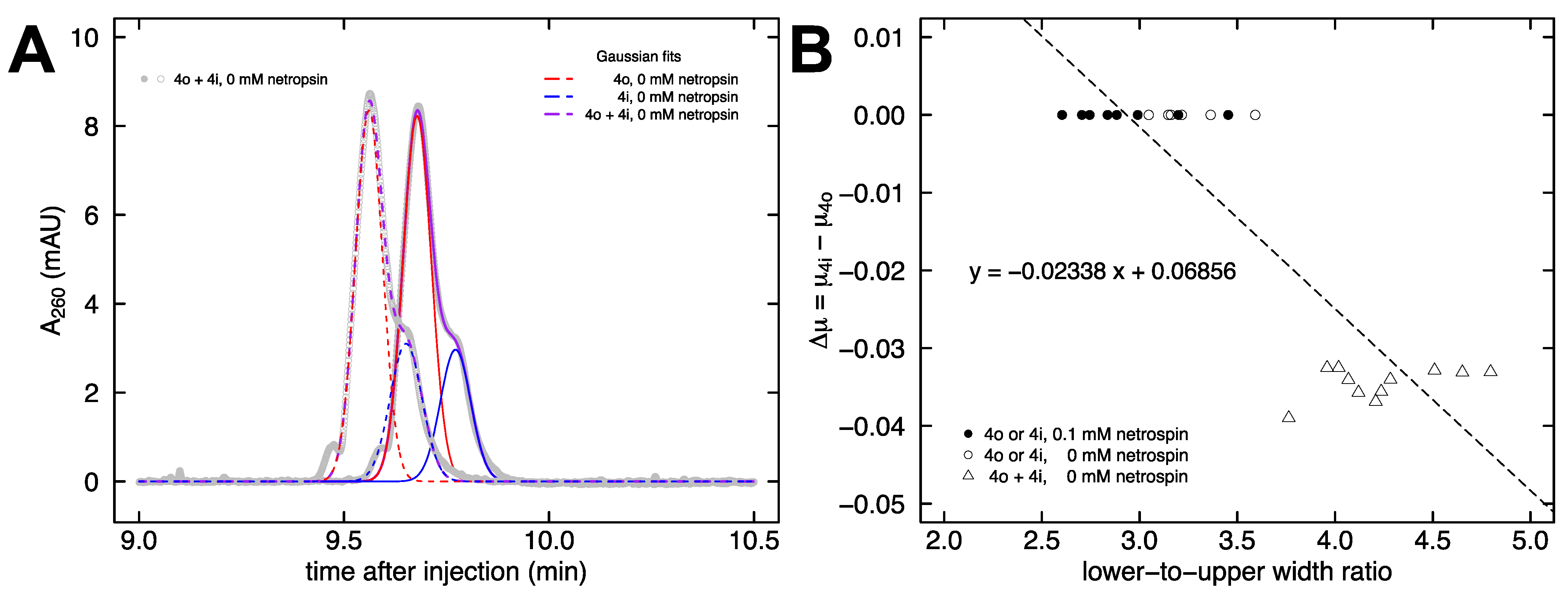
2.2. Electrophoretic Mobility of DNA in Free Solution
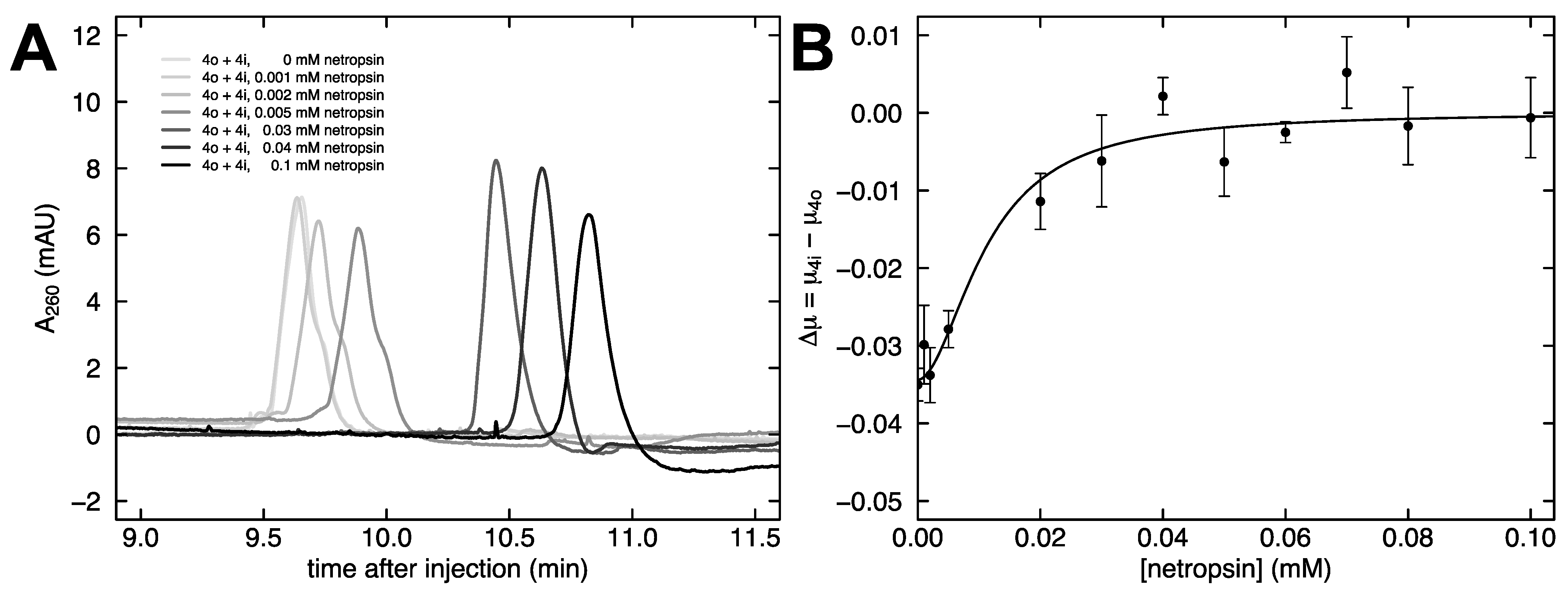
2.3. Dependence of Free Solution Mobility Difference on Netropsin Concentration
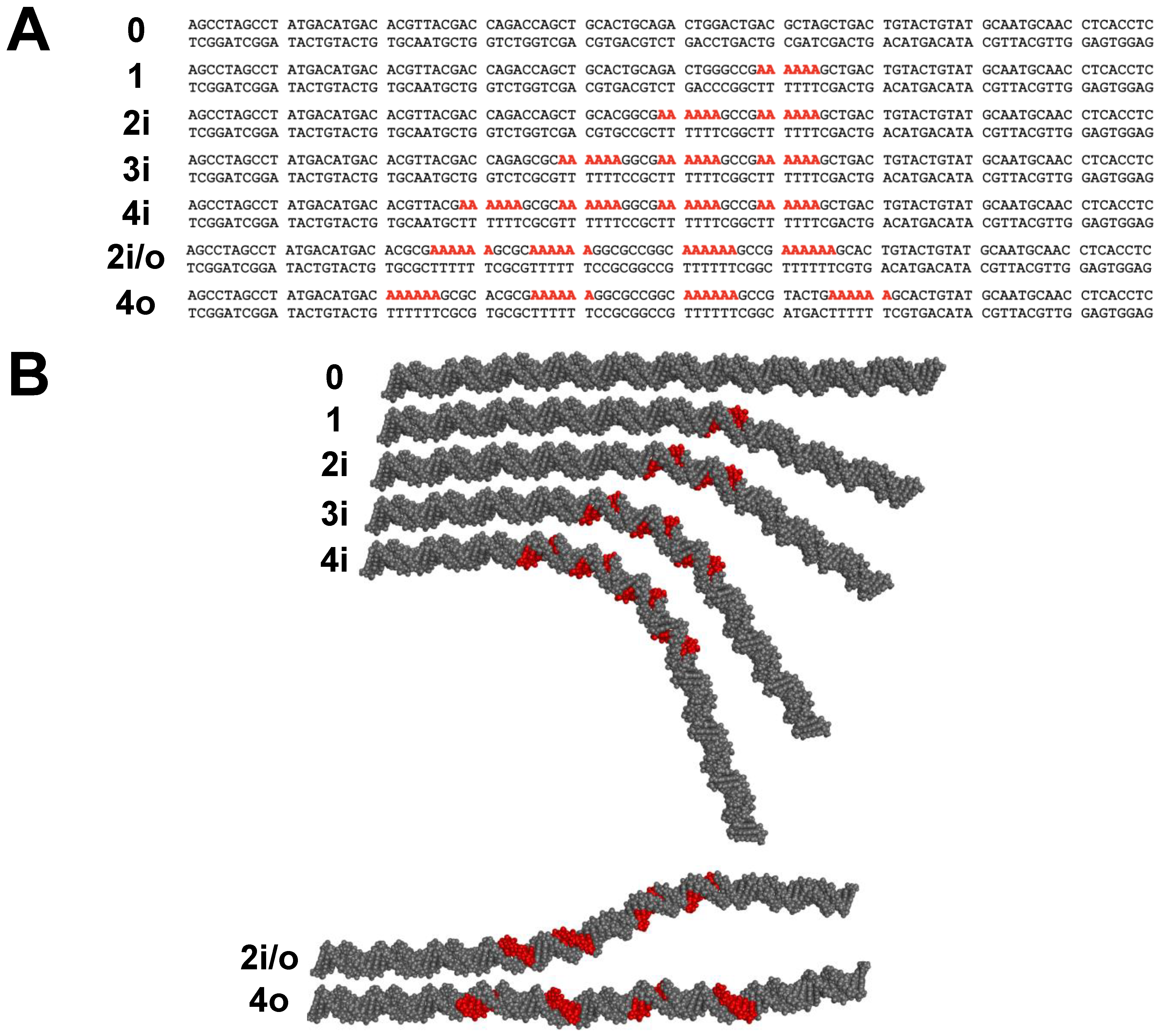
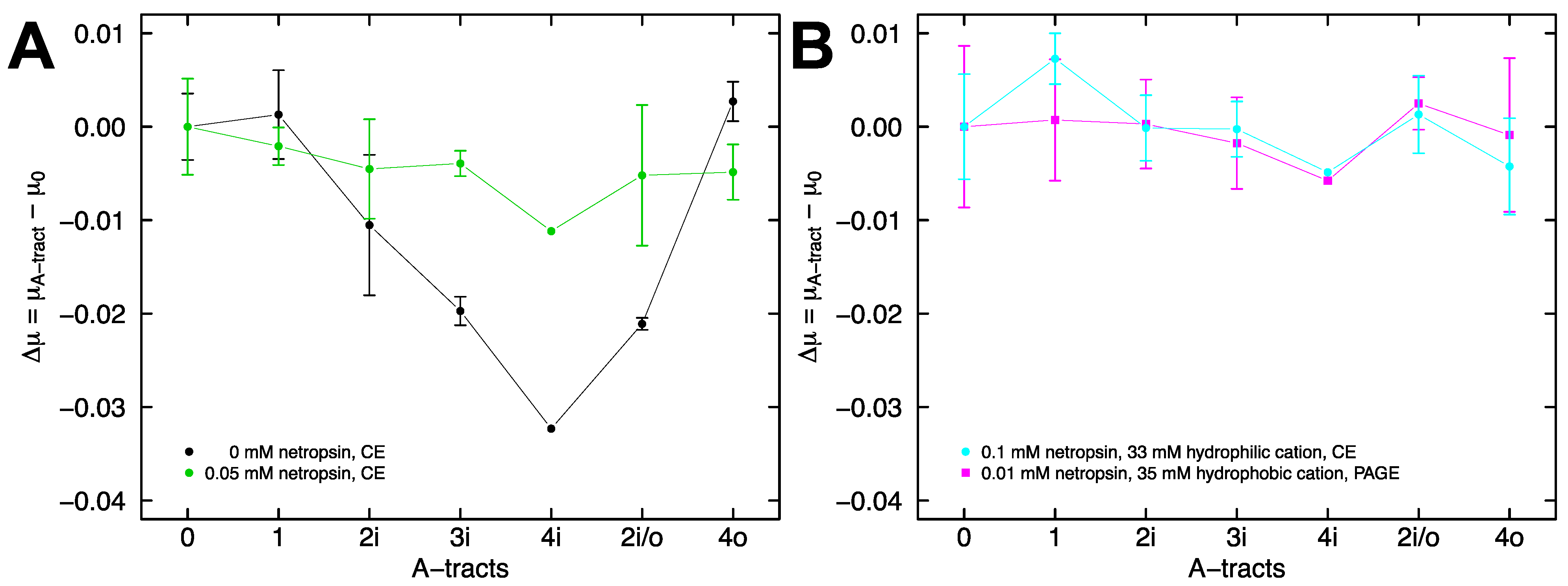
2.4. Electrophoretic Mobility of A-Tract Constructs
3. Materials and Methods
3.1. DNA Fragments
3.2. Buffers
3.3. Polyacrylamide Gel Electrophoresis (PAGE)
3.4. Capillary Electrophoresis (CE)
3.5. Calculation of Mobility
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Marini, J.C.; Levene, S.D.; Crothers, D.M.; Englund, P.T. Bent Helical Structure in Kinetoplast DNA. Proc. Natl. Acad. Sci. USA 1982, 79, 7664–7668. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.-M.; Crothers, D.M. The Locus of Sequence-Directed and Protein-Induced DNA Bending. Nature 1984, 308, 509–513. [Google Scholar] [CrossRef]
- Levene, S.D.; Wu, H.M.; Crothers, D.M. Bending and Flexibility of Kinetoplast DNA. Biochemistry 1986, 25, 3988–3995. [Google Scholar] [CrossRef]
- Beutel, B.A.; Gold, L. In Vitro Evolution of Intrinsically Bent DNA. J. Mol. Biol. 1992, 228, 803–812. [Google Scholar] [CrossRef]
- Stellwagen, N.C. Curved DNA Molecules Migrate Anomalously Slowly in Polyacrylamide Gels Even at Zero Gel Concentration. Electrophoresis 2006, 27, 1163–1168. [Google Scholar] [CrossRef]
- Stellwagen, N.C.; Gelfi, C.; Righetti, P.G. The Free Solution Mobility of DNA. Biopolym. Orig. Res. Biomol. 1997, 42, 687–703. [Google Scholar] [CrossRef]
- Stellwagen, E. Curved DNA Molecules Migrate Anomalously Slowly in Free Solution. Nucleic Acids Res. 2005, 33, 4425–4432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koo, H.-S.; Wu, H.-M.; Crothers, D.M. DNA Bending at Adenine· Thymine Tracts. Nature 1986, 320, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.S.; Drak, J.; Rice, J.A.; Crothers, D.M. Determination of the Extent of DNA Bending by an Adenine-Thymine Tract. Biochemistry 1990, 29, 4227–4234. [Google Scholar] [CrossRef] [PubMed]
- DiGabriele, A.D.; Sanderson, M.R.; Steitz, T.A. Crystal Lattice Packing Is Important in Determining the Bend of a DNA Dodecamer Containing an Adenine Tract. Proc. Natl. Acad. Sci. USA 1989, 86, 1816–1820. [Google Scholar] [CrossRef] [Green Version]
- Dickerson, R.E.; Goodsell, D.; Kopka, M.L. MPD and DNA Bending in Crystals and in Solution. J. Mol. Biol. 1996, 256, 108–125. [Google Scholar] [CrossRef] [PubMed]
- Strahs, D.; Schlick, T. A-Tract Bending: Insights into Experimental Structures by Computational Models. J. Mol. Biol. 2000, 301, 643–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McConnell, K.J.; Beveridge, D.L. Molecular Dynamics Simulations of B′-DNA: Sequence Effects on A-Tract-Induced Bending and Flexibility. J. Mol. Biol. 2001, 314, 23–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balasubramanian, C.; Ojha, R.P.; Maiti, S. Hydration Pattern of A4T4 and T4A4 DNA: A Molecular Dynamics Study. Biochem. Biophys. Res. Commun. 2007, 355, 1081–1086. [Google Scholar] [CrossRef]
- MacDonald, D.; Herbert, K.; Zhang, X.; Polgruto, T.; Lu, P. Solution Structure of an A-Tract DNA Bend. J. Mol. Biol. 2001, 306, 1081–1098. [Google Scholar] [CrossRef]
- Barbič, A.; Zimmer, D.P.; Crothers, D.M. Structural Origins of Adenine-Tract Bending. Proc. Natl. Acad. Sci. USA 2003, 100, 2369–2373. [Google Scholar] [CrossRef] [Green Version]
- Stellwagen, N.C. Anomalous Electrophoresis of Deoxyribonucleic Acid Restriction Fragments on Polyacrylamide Gels. Biochemistry 1983, 22, 6186–6193. [Google Scholar] [CrossRef]
- Diekmann, S. Temperature and Salt Dependence of the Gel Migration Anomaly of Curved DNA Fragments. Nucleic Acids Res. 1987, 15, 247–265. [Google Scholar] [CrossRef] [Green Version]
- Augustyn, K.E.; Wojtuszewski, K.; Hawkins, M.E.; Knutson, J.R.; Mukerji, I. Examination of the Premelting Transition of DNA A-Tracts Using a Fluorescent Adenosine Analogue. Biochemistry 2006, 45, 5039–5047. [Google Scholar] [CrossRef]
- Stellwagen, E.; Peters, J.P.; Maher, L.J.; Stellwagen, N.C. DNA A-Tracts Are Not Curved in Solutions Containing High Concentrations of Monovalent Cations. Biochemistry 2013, 52, 4138–4148. [Google Scholar] [CrossRef] [Green Version]
- Stefl, R.; Wu, H.; Ravindranathan, S.; Sklenář, V.; Feigon, J. DNA A-Tract Bending in Three Dimensions: Solving the dA4T4 vs. dT4A4 Conundrum. Proc. Natl. Acad. Sci. USA 2004, 101, 1177–1182. [Google Scholar] [CrossRef] [Green Version]
- Snoussi, K.; Leroy, J.-L. Alteration of A•T Base-Pair Opening Kinetics by the Ammonium Cation in DNA A-Tracts. Biochemistry 2002, 41, 12467–12474. [Google Scholar] [CrossRef]
- Diekmann, S. DNA Methylation Can Enhance or Induce DNA Curvature. EMBO J. 1987, 6, 4213–4217. [Google Scholar] [CrossRef]
- Hagerman, P.J. Pyrimidine 5-Methyl Groups Influence the Magnitude of DNA Curvature. Biochemistry 1990, 29, 1980–1983. [Google Scholar] [CrossRef]
- Zhurkin, V.B.; Ulyanov, N.B.; Gorin, A.A.; Jernigan, R.L. Static and Statistical Bending of DNA Evaluated by Monte Carlo Simulations. Proc. Natl. Acad. Sci. USA 1991, 88, 7046–7050. [Google Scholar] [CrossRef] [Green Version]
- Hagerman, P.J.; Ramadevi, V. Application of the Method of Phage T4 DNA Ligase-Catalyzed Ring-Closure to the Study of DNA Structure: I. Computational Analysis. J. Mol. Biol. 1990, 212, 351–362. [Google Scholar] [CrossRef]
- Lankaš, F.; Cheatham, T.E.; Hobza, P.; Langowski, J.; Šponer, J. Others Critical Effect of the N2 Amino Group on Structure, Dynamics, and Elasticity of DNA Polypurine Tracts. Biophys. J. 2002, 82, 2592–2609. [Google Scholar] [CrossRef] [Green Version]
- Tereshko, V.; Minasov, G.; Egli, M. A “Hydrat-Ion” Spine in a B-DNA Minor Groove. J. Am. Chem. Soc. 1999, 121, 3590–3595. [Google Scholar] [CrossRef]
- Hamelberg, D. Effect of a Neutralized Phosphate Backbone on the Minor Groove of B-DNA: Molecular Dynamics Simulation Studies. Nucleic Acids Res. 2002, 30, 3615–3623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madhumalar, A.; Bansal, M. Structural Insights into the Effect of Hydration and Ions on A-Tract DNA: A Molecular Dynamics Study. Biophys. J. 2003, 85, 1805–1816. [Google Scholar] [CrossRef] [Green Version]
- Subirana, J.A.; Soler-López, M. Cations as Hydrogen Bond Donors: A View of Electrostatic Interactions in DNA. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 27–45. [Google Scholar] [CrossRef]
- Yonetani, Y.; Kono, H. Sequence Dependencies of DNA Deformability and Hydration in the Minor Groove. Biophys. J. 2009, 97, 1138–1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Schatz, G.C. Molecular Dynamics Study of the Role of the Spine of Hydration in DNA A-Tracts in Determining Nucleosome Occupancy. J. Phys. Chem. B 2012, 116, 13672–13681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woods, K.K. The Role of Minor Groove Functional Groups in DNA Hydration. Nucleic Acids Res. 2003, 31, 1536–1540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harvey, S.C.; Dlakic, M.; Griffith, J.; Harrington, R.; Park, K.; Sprous, D.; Zacharias, W. What Is the Basis of Sequence-Directed Curvature in DNAs Containing A Tracts? J. Biomol. Struct. Dyn. 1995, 13, 301–307. [Google Scholar] [CrossRef]
- Sprous, D.; Zacharias, W.; Wood, Z.A.; Harvey, S.C. Dehydrating Agents Sharply Reduce Curvature in DNAs Containing A Tracts. Nucleic Acids Res. 1995, 23, 1816–1821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dlakic, M.; Park, K.; Griffith, J.D.; Harvey, S.C.; Harrington, R.E. The Organic Crystallizing Agent 2-Methyl-2, 4-Pentanediol Reduces DNA Curvature by Means of Structural Changes in A-Tracts. J. Biol. Chem. 1996, 271, 17911–17919. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Stellwagen, E.; Stellwagen, N.C. Effect of Organic Cosolvents on the Free Solution Mobility of Curved and Normal DNA Molecules. Electrophoresis 2006, 27, 1462–1470. [Google Scholar] [CrossRef]
- Haran, T.E.; Cohen, I.; Spasic, A.; Yang, K.; Mohanty, U. Dynamics of Curved DNA Molecules: Prediction and Experiment. J. Am. Chem. Soc. 2003, 125, 11160–11161. [Google Scholar] [CrossRef]
- Haran, T.E.; Cohen, I.; Spasic, A.; Yang, K.; Mohanty, U. Characteristics of Migration Patterns of DNA Oligomers in Gels and the Relationship to the Question of Intrinsic DNA Bending. J. Am. Chem. Soc. 2004, 126, 2372–2377. [Google Scholar] [CrossRef] [PubMed]
- Crothers, D.M.; Haran, T.E.; Nadeau, J.G. Intrinsically Bent DNA. J. Biol. Chem. 1990, 265, 7093–7096. [Google Scholar] [CrossRef]
- McCarthy, J.G.; Frederick, C.A.; Nicolas, A. A Structural Analysis of the Bent Kinetoplast DNA from Crithidia Fasciculata by High Resolution Chemical Probing. Nucleic Acids Res. 1993, 21, 3309–3317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodsell, D.S.; Dickerson, R.E. Bending and Curvature Calculations in B-DNA. Nucleic Acids Res. 1994, 22, 5497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dršata, T.; Špačková, N.; Jurečka, P.; Zgarbová, M.; Šponer, J.; Lankaš, F. Mechanical Properties of Symmetric and Asymmetric DNA A-Tracts: Implications for Looping and Nucleosome Positioning. Nucleic Acids Res. 2014, 42, 7383–7394. [Google Scholar] [CrossRef] [Green Version]
- Mack, D.R.; Chiu, T.K.; Dickerson, R.E. Intrinsic Bending and Deformability at the T-A Step of CCTTTAAAGG: A Comparative Analysis of T-A and A-T Steps within A-Tracts. J. Mol. Biol. 2001, 312, 1037–1049. [Google Scholar] [CrossRef]
- Lankas, F.; Spackova, N.; Moakher, M.; Enkhbayar, P.; Sponer, J. A Measure of Bending in Nucleic Acids Structures Applied to A-Tract DNA. Nucleic Acids Res. 2010, 38, 3414–3422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Møllegaard, N.E.; Lindemose, S.; Nielsen, P.E. Uranyl Photoprobing of Nonbent A/T-and Bent A-Tracts. A Difference of Flexibility? Biochemistry 2005, 44, 7855–7863. [Google Scholar] [CrossRef]
- Woods, K.K.; McFail-Isom, L.; Sines, C.C.; Howerton, S.B.; Stephens, R.K.; Williams, L.D. Monovalent Cations Sequester within the A-Tract Minor Groove of [d(CGCGAATTCGCG)]2. J. Am. Chem. Soc. 2000, 122, 1546–1547. [Google Scholar] [CrossRef]
- Lindemose, S.; Nielsen, P.E.; Møllegaard, N.E. Polyamines Preferentially Interact with Bent Adenine Tracts in Double-Stranded DNA. Nucleic Acids Res. 2005, 33, 1790–1803. [Google Scholar] [CrossRef] [Green Version]
- Hud, N.V.; Polak, M. DNA–Cation Interactions: The Major and Minor Grooves Are Flexible Ionophores. Curr. Opin. Struct. Biol. 2001, 11, 293–301. [Google Scholar] [CrossRef]
- Egli, M. DNA-Cation Interactions: Quo Vadis? Chem. Biol. 2002, 9, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Tereshko, V.; Minasov, G.; Egli, M. The Dickerson-Drew B-DNA Dodecamer Revisited at Atomic Resolution. J. Am. Chem. Soc. 1999, 121, 470–471. [Google Scholar] [CrossRef]
- Ballin, J.D.; Wilson, G.M. Role and Applications of Electrostatic Effects on Nucleic Acid Conformational Transitions and Binding Processes. Appl. Thermodyn. Biol. Mater. Sci. 2011, 129–174. [Google Scholar]
- Brukner, I.; Dlakic, M.; Savic, A.; Susic, S.; Pongor, S.; Suck, D. Evidence for Opposite Groove-Directed Curvature of GGGCCC and AAAAA Sequence Elements. Nucleic Acids Res. 1993, 21, 1025–1029. [Google Scholar] [CrossRef] [Green Version]
- Hud, N.V.; Plavec, J. A Unified Model for the Origin of DNA Sequence-Directed Curvature. Biopolymers 2003, 69, 144–158. [Google Scholar] [CrossRef]
- Lu, Y.; Stellwagen, N.C. Monovalent Cation Binding by Curved DNA Molecules Containing Variable Numbers of A-Tracts. Biophys. J. 2008, 94, 1719–1725. [Google Scholar] [CrossRef] [Green Version]
- Dong, Q.; Stellwagen, E.; Stellwagen, N.C. Monovalent Cation Binding in the Minor Groove of DNA A-Tracts. Biochemistry 2009, 48, 1047–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watkins, D.; Mohan, S.; Koudelka, G.B.; Williams, L.D. Sequence Recognition of DNA by Protein-Induced Conformational Transitions. J. Mol. Biol. 2010, 396, 1145–1164. [Google Scholar] [CrossRef] [PubMed]
- Watkins, D.; Harris, L.-A.; Koudelka, G.B.; Williams, L.D. Protein: Feeling the Groove of DNA. In Frontiers in Nucleic Acids; Sheardy, R.D., Winkle, S.A., Eds.; American Chemical Society: Washington, DC, USA, 2011; Volume 1082, pp. 133–145. ISBN 978-0-8412-2623-4. [Google Scholar]
- Poot, M.; Kausch, K.; Köhler, J.; Haaf, T.; Hoehn, H. The Minor-Groove Binding DNA-Ligands Netropsin, Distamycin A and Berenil Cause Polyploidisation via Impairment of the G2 Phase of the Cell Cycle. Cell Struct. Funct. 1990, 15, 151–157. [Google Scholar] [CrossRef] [Green Version]
- Jeon, H.; Nam, H.; Lee, J.B. Sustained Release of Minor-Groove-Binding Antibiotic Netropsin from Calcium-Coated Groove-Rich DNA Particles. Pharmaceutics 2019, 11, 387. [Google Scholar] [CrossRef] [Green Version]
- Tabernero, L.; Verdaguer, N.; Coll, M.; Fita, I.; Van Der Marel, G.A.; Van Boom, J.H.; Rich, A.; Aymami, J. Molecular Structure of the A-Tract DNA Dodecamer d(CGCAAATTTGCG) Complexed with the Minor Groove Binding Drug Netropsin. Biochemistry 1993, 32, 8403–8410. [Google Scholar] [CrossRef]
- Goodsell, D.S.; Kopka, M.L.; Dickerson, R.E. Refinement of Netropsin Bound to DNA: Bias and Feedback in Electron Density Map Interpretation. Biochemistry 1995, 34, 4983–4993. [Google Scholar] [CrossRef]
- Nunn, C.M.; Garman, E.; Neidle, S. Crystal Structure of the DNA Decamer d(CGCAATTGCG) Complexed with the Minor Groove Binding Drug Netropsin. Biochemistry 1997, 36, 4792–4799. [Google Scholar] [CrossRef] [PubMed]
- Barcelo, F.; Muzard, G.; Mendoza, R.; Revet, B.; Roques, B.P.; Le Pecq, J.B. Removal of DNA Curving by DNA Ligands: Gel Electrophoresis Study. Biochemistry 1991, 30, 4863–4873. [Google Scholar] [CrossRef] [PubMed]
- Freyer, M.W.; Buscaglia, R.; Nguyen, B.; Wilson, W.D.; Lewis, E.A. Binding of Netropsin and 4, 6-Diamidino-2-Phenylindole to an A2T2 DNA Hairpin: A Comparison of Biophysical Techniques. Anal. Biochem. 2006, 355, 259–266. [Google Scholar] [CrossRef]
- Lewis, E.A.; Munde, M.; Wang, S.; Rettig, M.; Le, V.; Machha, V.; Wilson, W.D. Complexity in the Binding of Minor Groove Agents: Netropsin Has Two Thermodynamically Different DNA Binding Modes at a Single Site. Nucleic Acids Res. 2011, 39, 9649–9658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrera, J.E.; Chaires, J.B. A Premelting Conformational Transition in Poly (DA)-Poly (DT) Coupled to Daunomycin Binding. Biochemistry 1989, 28, 1993–2000. [Google Scholar] [CrossRef]
- Alniss, H.Y. Thermodynamics of DNA Minor Groove Binders: Perspective. J. Med. Chem. 2019, 62, 385–402. [Google Scholar] [CrossRef]
- Degtyareva, N.N.; Fresia, M.J.; Petty, J.T. DNA Conformational Effects on the Interaction of Netropsin with A-Tract Sequences. Biochemistry 2007, 46, 15136–15143. [Google Scholar] [CrossRef]
- Bailly, C.; Chaires, J.B. Sequence-Specific DNA Minor Groove Binders. Design and Synthesis of Netropsin and Distamycin Analogues. Bioconjugate Chem. 1998, 9, 513–538. [Google Scholar] [CrossRef]
- Koo, H.-S.; Crothers, D.M. Calibration of DNA Curvature and a Unified Description of Sequence-Directed Bending. Proc. Natl. Acad. Sci. USA 1988, 85, 1763–1767. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Liu, J.; Li, W. Resolution of Overlapping Spectra by Curve-Fitting. Anal. Chim. Acta 2005, 538, 383–389. [Google Scholar] [CrossRef]
- Durney, B.C.; Crihfield, C.L.; Holland, L.A. Capillary Electrophoresis Applied to DNA: Determining and Harnessing Sequence and Structure to Advance Bioanalyses (2009–2014). Anal. Bioanal. Chem. 2015, 407, 6923–6938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowak, P.M.; Woźniakiewicz, M.; Gładysz, M.; Janus, M.; Kościelniak, P. Improving Repeatability of Capillary Electrophoresis—A Critical Comparison of Ten Different Capillary Inner Surfaces and Three Criteria of Peak Identification. Anal. Bioanal. Chem. 2017, 409, 4383–4393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, B.A.; Vigh, G. Fast, Accurate Mobility Determination Method for Capillary Electrophoresis. Anal. Chem. 1996, 68, 1174–1180. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miller, J.; Peters, J.P. Examining the Effects of Netropsin on the Curvature of DNA A-Tracts Using Electrophoresis. Molecules 2021, 26, 5871. https://doi.org/10.3390/molecules26195871
Miller J, Peters JP. Examining the Effects of Netropsin on the Curvature of DNA A-Tracts Using Electrophoresis. Molecules. 2021; 26(19):5871. https://doi.org/10.3390/molecules26195871
Chicago/Turabian StyleMiller, Jillian, and Justin P. Peters. 2021. "Examining the Effects of Netropsin on the Curvature of DNA A-Tracts Using Electrophoresis" Molecules 26, no. 19: 5871. https://doi.org/10.3390/molecules26195871
APA StyleMiller, J., & Peters, J. P. (2021). Examining the Effects of Netropsin on the Curvature of DNA A-Tracts Using Electrophoresis. Molecules, 26(19), 5871. https://doi.org/10.3390/molecules26195871