Photodynamic Therapy with Tumor Cell Discrimination through RNA-Targeting Ability of Photosensitizer
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis of QICY and Intermediates
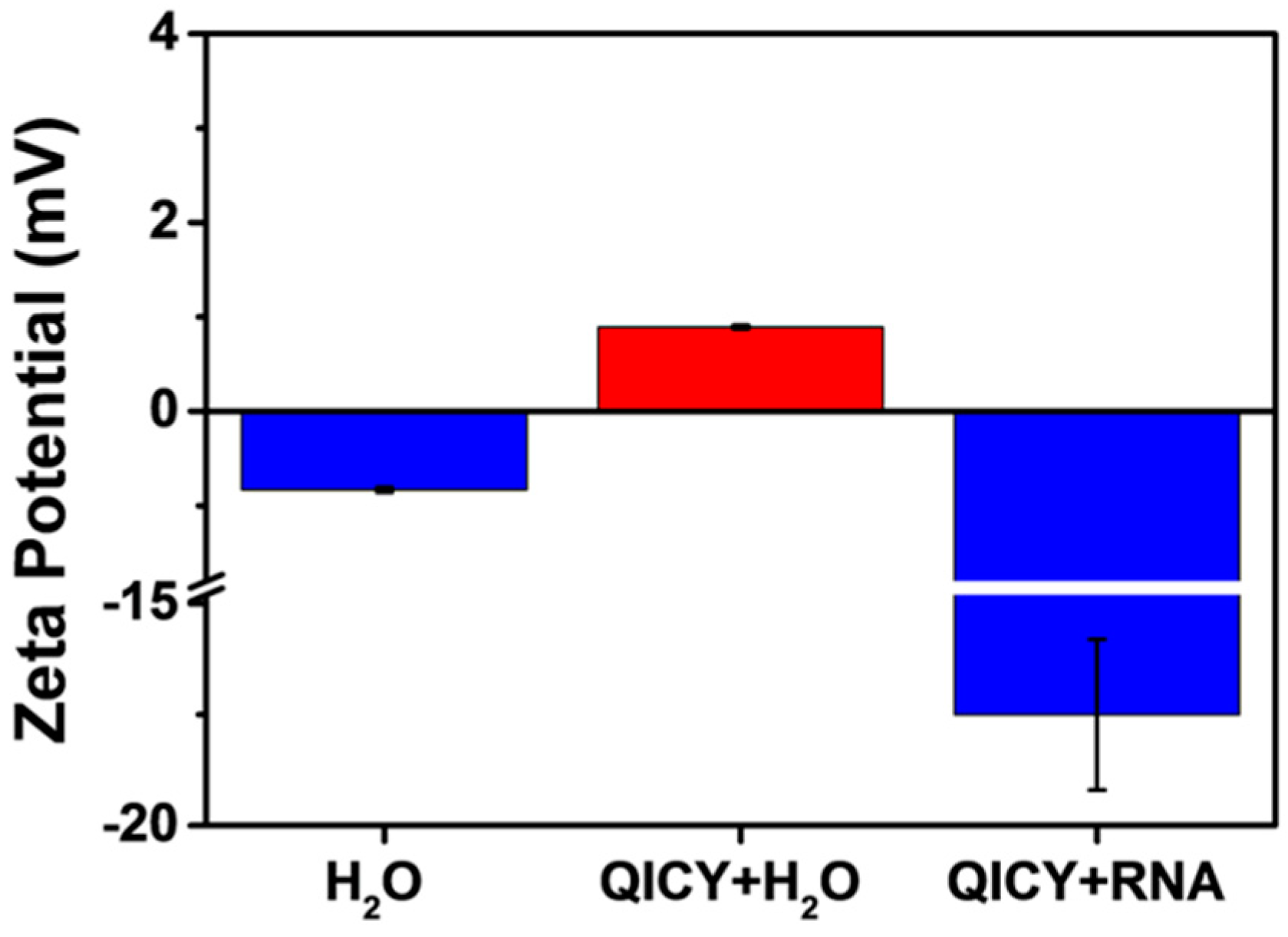
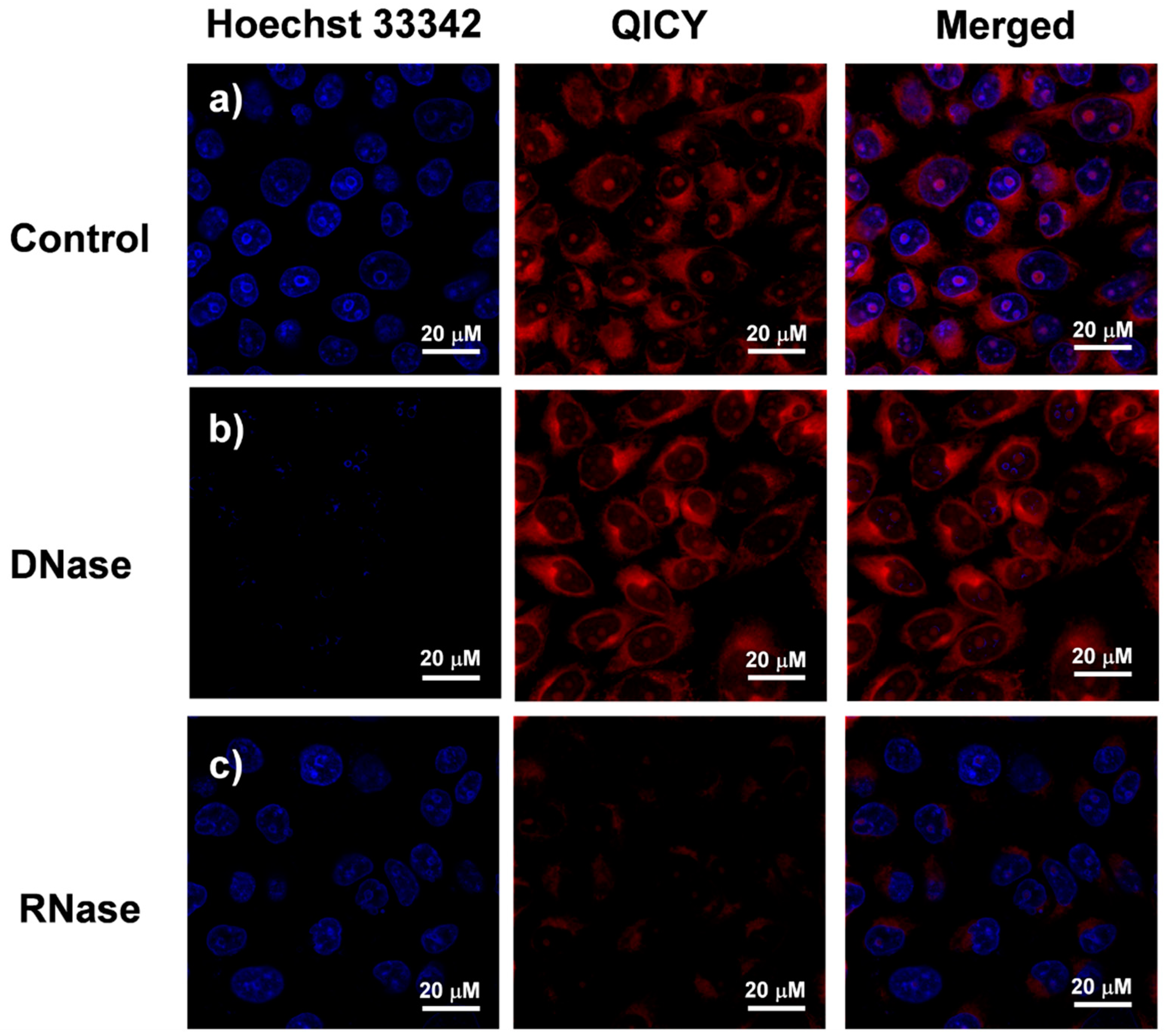
2.2. Spectral Characterization and RNA Interaction of QICY
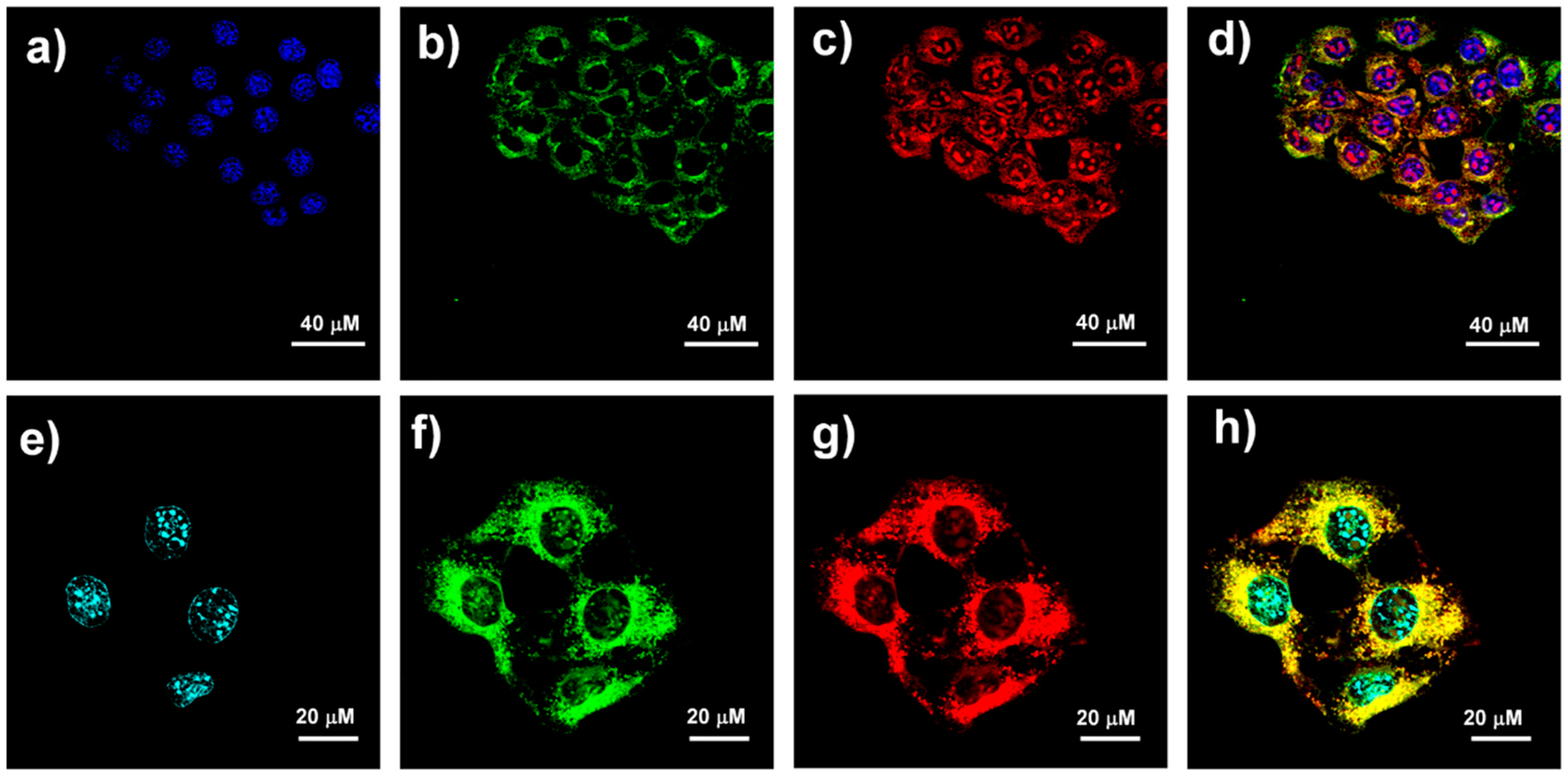
2.3. Subcellular Localization of QICY
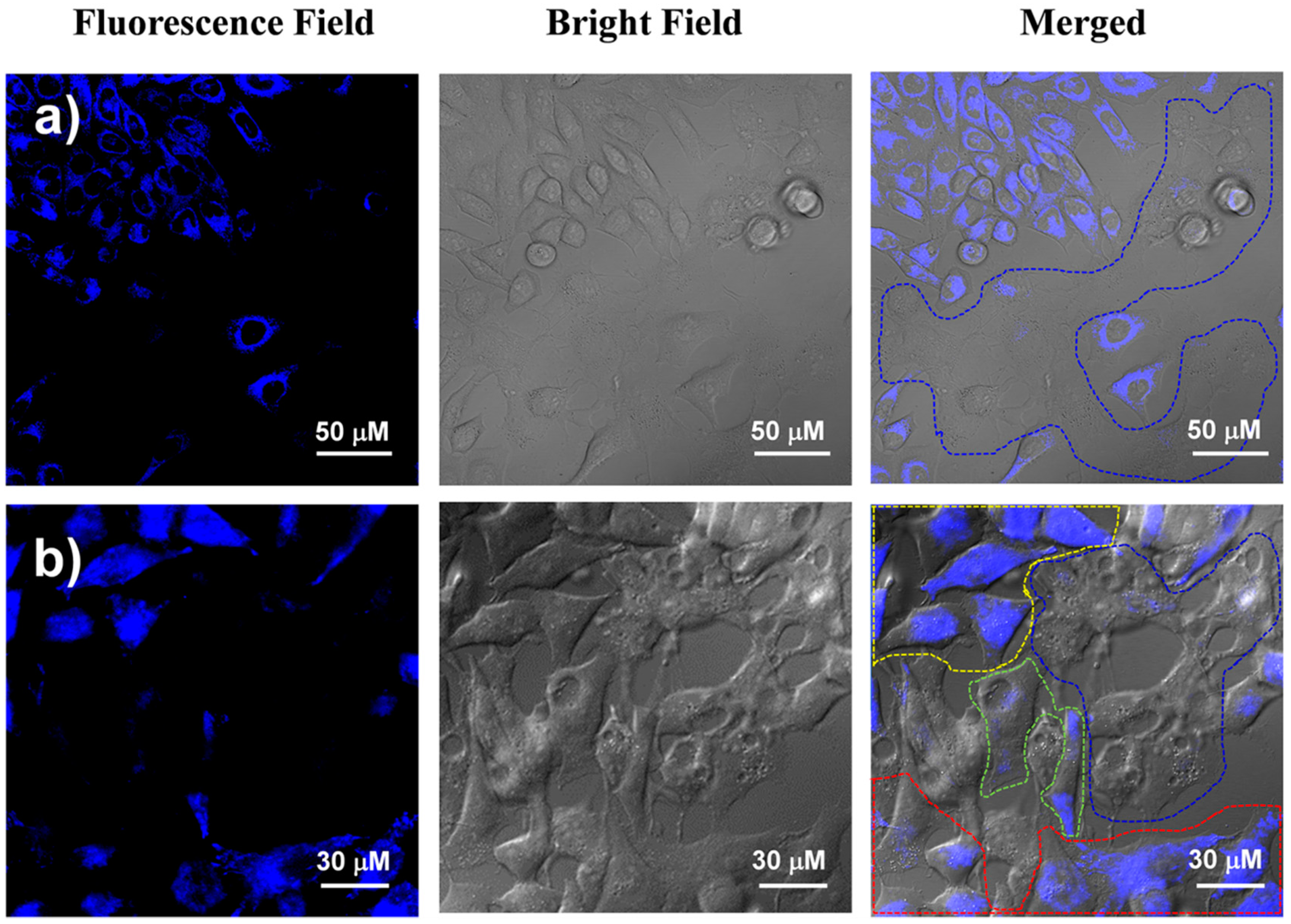
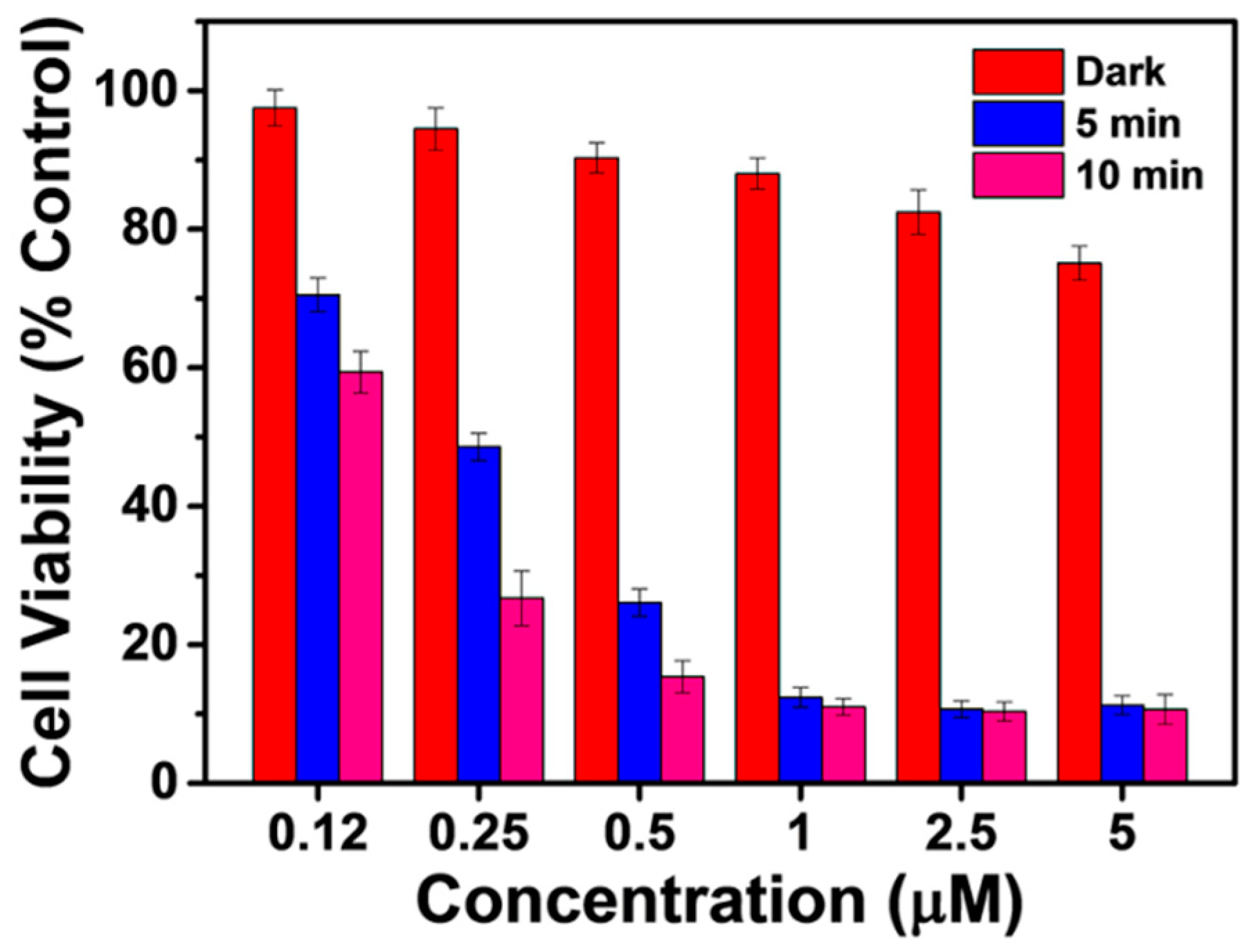
2.4. Cancer Cell Discrimination Assay
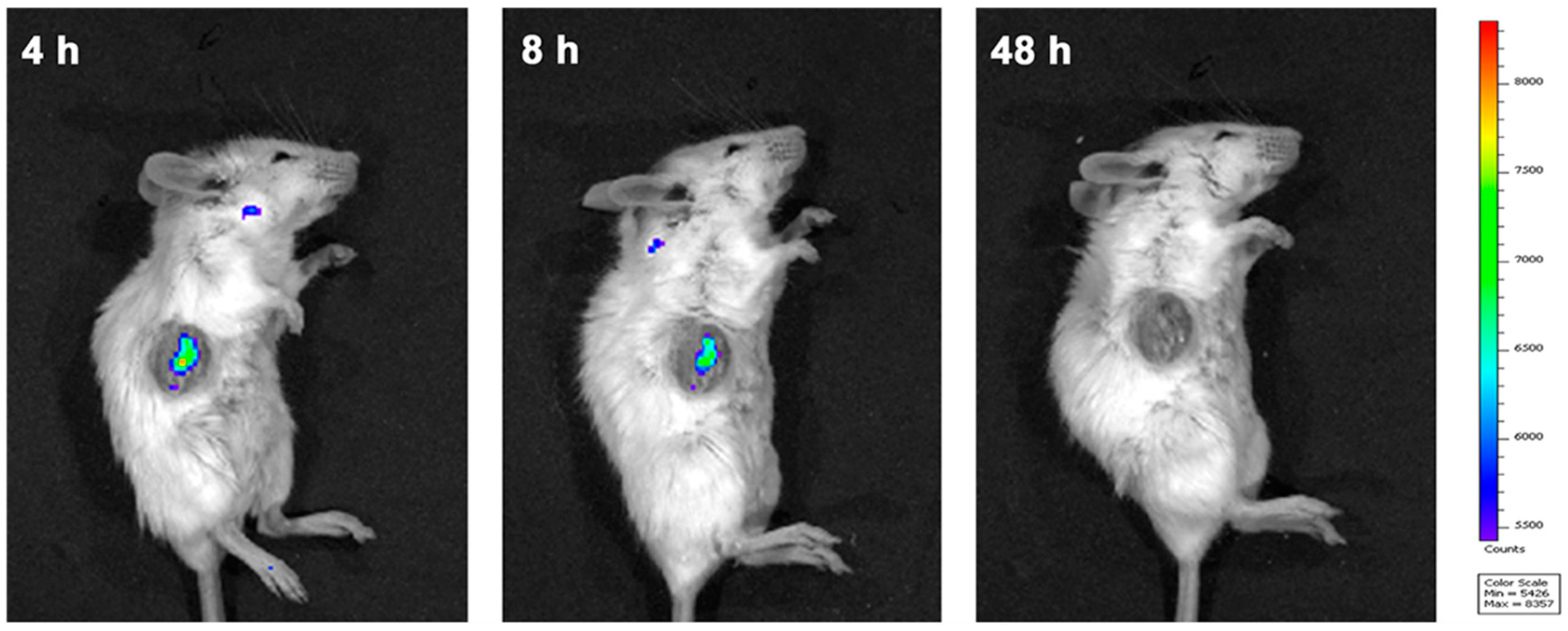
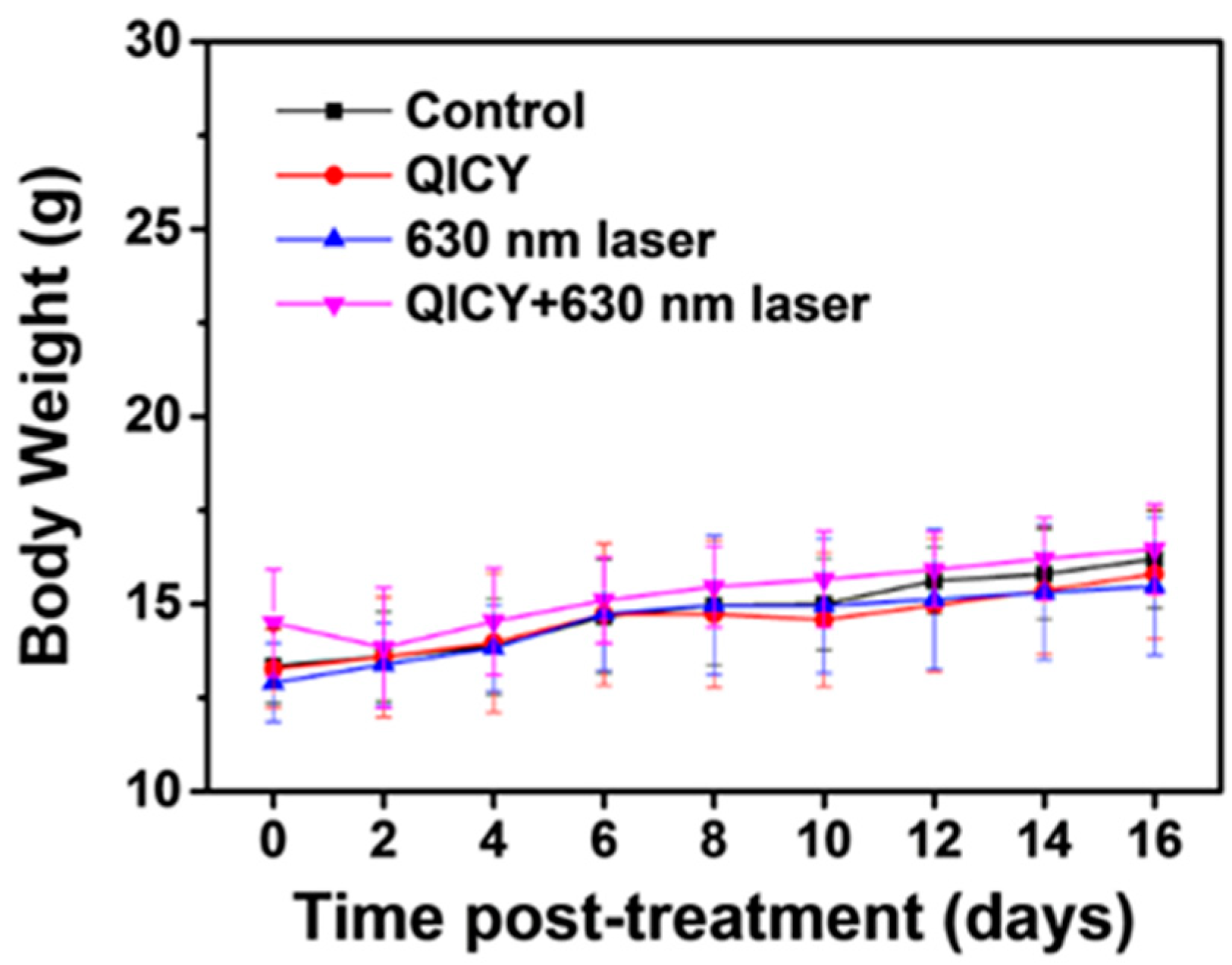
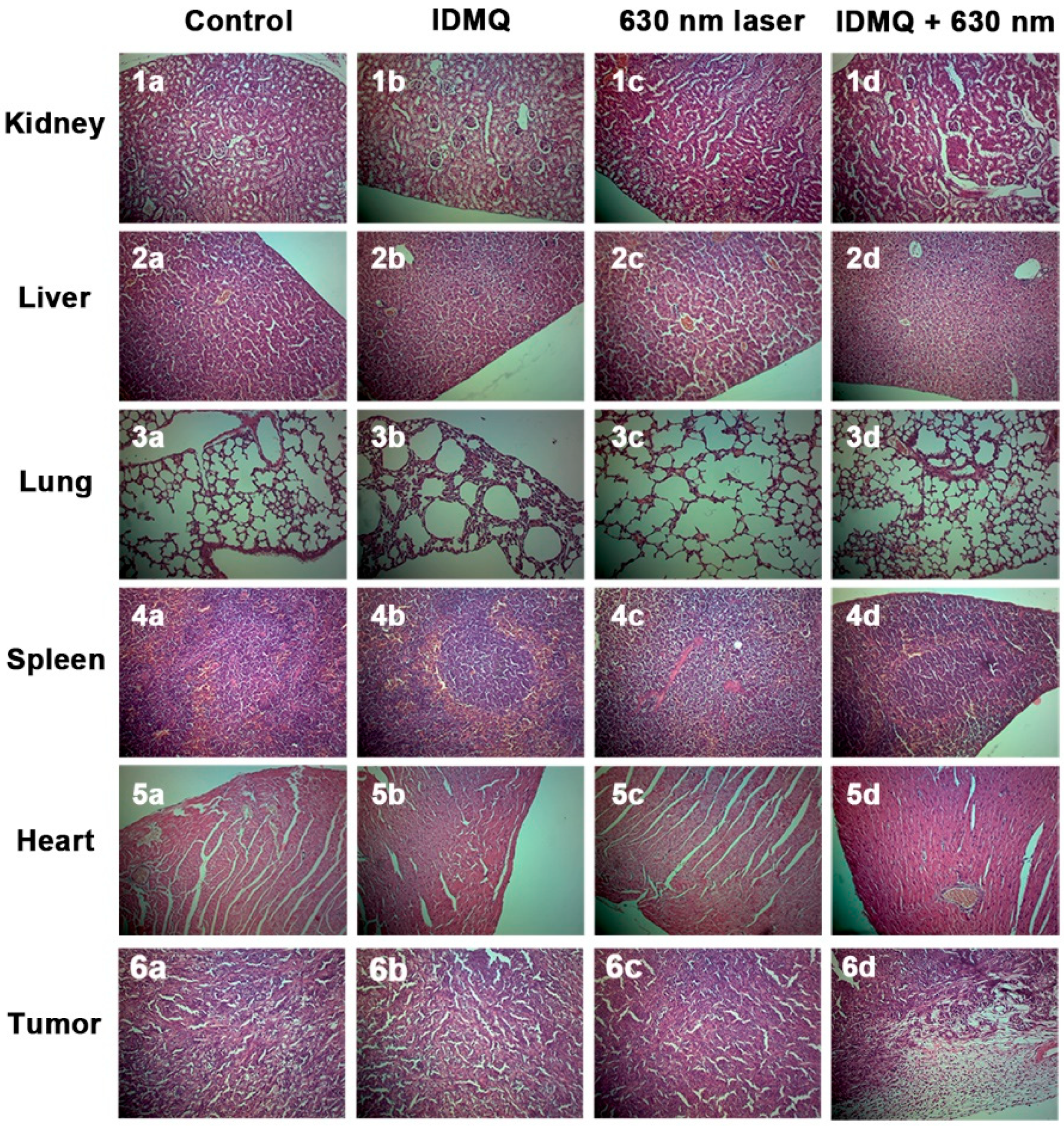
2.5. In Vivo Tumor Imaging and Therapy
3. Materials and Methods
3.1. Materials
3.2. Detection of Spectroscopic Properties
3.3. Cell Culture and Simulation of Tumor Environment
3.4. Establishment of the Animal Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, Z.J.; Song, J.B.; Nie, L.M.; Chen, X.Y. Reactive oxygen species generating systems meeting challenges of photodynamic cancer therapy. Chem. Soc. Rev. 2016, 45, 6597–6626. [Google Scholar] [CrossRef] [Green Version]
- Fan, W.P.; Huang, P.; Chen, X.Y. Overcoming the Achilles’ heel of photodynamic therapy. Chem. Soc. Rev. 2016, 45, 6488–6519. [Google Scholar] [CrossRef]
- Hu, F.; Xu, S.D.; Liu, B. Photosensitizers with Aggregation-Induced Emission: Materials and Biomedical Applications. Adv. Mater. 2018, 30, 1801350. [Google Scholar] [CrossRef]
- Lucky, S.S.; Soo, K.C.; Zhang, Y. Nanoparticles in Photodynamic Therapy. Chem. Rev. 2015, 115, 1990–2042. [Google Scholar] [CrossRef]
- Zhao, X.Z.; Long, S.R.; Li, M.L.; Cao, J.F.; Li, Y.C.; Guo, L.Y.; Sun, W.; Du, J.J.; Fan, J.L.; Peng, X.J. Oxygen-Dependent Regulation of Excited-State Deactivation Process of Rational Photosensitizer for Smart Phototherapy. J. Am. Chem. Soc. 2020, 142, 1510–1517. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.S.; Han, J.; Shi, H.; Koo, S.; Singh, H.; Kim, H.J.; Sessler, J.L.; Lee, J.Y.; Kim, J.H.; Kim, J.S. Overcoming the Limits of Hypoxia in Photodynamic Therapy: A Carbonic Anhydrase IX-Targeted Approach. J. Am. Chem. Soc. 2017, 139, 7595–7602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, G.B.; Xu, L.G.; Chao, Y.; Xu, J.; Sun, X.Q.; Wu, Y.F.; Peng, R.; Liu, Z. Hollow MnO2 as a tumor-microenvironment-responsive biodegradable nano-platform for combination therapy favoring antitumor immune responses. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Li, S.Y.; Cheng, H.; Xie, B.R.; Qiu, W.X.; Zeng, J.Y.; Li, C.X.; Wan, S.S.; Zhang, L.; Liu, W.L.; Zhang, X.Z. Cancer Cell Membrane Camouflaged Cascade Bioreactor for Cancer Targeted Starvation and Photodynamic Therapy. ACS Nano 2017, 11, 7006–7018. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Li, X.Y.; Li, A.J.; Yang, P.; Zhang, C.Y.; Tang, B. H2S-Activable MOF Nanoparticle Photosensitizer for Effective Photodynamic Therapy against Cancer with Controllable Singlet-Oxygen Release. Angew. Chem. Int. Ed. 2017, 56, 13752–13756. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.H.; Yan, G.B.; Zhao, Z.L.; Hu, X.X.; Zhang, W.H.; Liu, H.; Fu, X.Y.; Fu, T.; Zhang, X.B.; Tan, W.H. A Smart Photosensitizer-Manganese Dioxide Nanosystem for Enhanced Photodynamic Therapy by Reducing Glutathione Levels in Cancer Cells. Angew. Chem. Int. Ed. 2016, 55, 5477–5482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.L.; Long, S.R.; Kang, Y.; Guo, L.Y.; Wang, J.Y.; Fan, J.L.; Du, J.J.; Peng, X.J. De Novo Design of Phototheranostic Sensitizers Based on Structure-Inherent Targeting for Enhanced Cancer Ablation. J. Am. Chem. Soc. 2018, 140, 15820–15826. [Google Scholar] [CrossRef]
- Chiba, M.; Ichikawa, Y.; Kamiya, M.; Komatsu, T.; Ueno, T.; Hanaoka, K.; Nagano, T.; Lange, N.; Urano, Y. An Activatable Photosensitizer Targeted to γ-Glutamyltranspeptidase. Angew. Chem. Int. Ed. 2017, 56, 10418–10422. [Google Scholar] [CrossRef]
- Tian, J.W.; Zhou, J.F.; Shen, Z.; Ding, L.; Yu, J.S.; Ju, H.X. A pH-activatable and aniline-substituted photosensitizer for near-infrared cancer theranostics. Chem. Sci. 2015, 6, 5969–5977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Wu, Y.L.; Zeng, F.; Wu, S.Z. An Activatable Near-Infrared Chromophore for Multispectral Optoacoustic Imaging of Tumor Hypoxia and for Tumor Inhibition. Theranostics 2019, 9, 7313–7324. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.X.; Li, H.D.; Li, Y.Q.; Li, M.; Fan, J.L.; Sun, W.; Li, N.; Li, R.J.; Shao, K.; Peng, X.J. In Vivo Coinstantaneous Identification of Hepatocellular Carcinoma Circulating Tumor Cells by Dual-Targeting Magnetic-Fluorescent Nanobeads. Nano Lett. 2021, 21, 634–641. [Google Scholar] [CrossRef]
- Xia, W.X.; Shangguan, X.Y.; Li, M.; Wang, Y.; Xi, D.M.; Sun, W.; Fan, J.L.; Shao, K.; Peng, X.J. Ex vivo identification of circulating tumor cells in peripheral blood by fluorometric “turn on” aptamer nanoparticles. Chem. Sci. 2021, 12, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Singha, S.; Shao, K.; Ellestad, K.K.; Yang, Y.; Santamaria, P. Nanoparticles for Immune Stimulation Against Infection, Cancer, and Autoimmunity. ACS Nano 2018, 12, 10621–10635. [Google Scholar] [CrossRef]
- He, N.Z.; Liu, P.; Wang, Z.Q.; Guo, Z.W.; Yan, X.X.; Chen, H.B.; Zhang, Z.C. Discovery of selective Mcl-1 inhibitors via structure-based design and structure-activity relationship analysis. Biochem. Biophys. Res. Commun. 2019, 512, 921–926. [Google Scholar] [CrossRef]
- Zhang, X.D.; Wang, Z.Q.; Guo, Z.W.; He, N.Z.; Liu, P.; Xia, D.S.; Yan, X.F.; Zhang, Z.C. A novel turn-on fluorescent probe for selective sensing and imaging of glutathione in live cells and organisms. Analyst 2019, 144, 3260–3266. [Google Scholar] [CrossRef]
- Friberg, A.; Vigil, D.; Zhao, B.; Daniels, R.N.; Burke, J.P.; Garcia-Barrantes, P.M.; Camper, D.; Chauder, B.A.; Lee, T.; Olejniczak, E.T.; et al. Discovery of Potent Myeloid Cell Leukemia 1 (Mcl-1) Inhibitors Using Fragment-Based Methods and Structure-Based Design. J. Med. Chem. 2013, 56, 15–30. [Google Scholar] [CrossRef]
- Peng, X.J.; Wu, T.; Fan, J.L.; Wang, J.Y.; Zhang, S.; Song, F.L.; Sun, S.G. An Effective Minor Groove Binder as a Red Fluorescent Marker for Live-Cell DNA Imaging and Quantification. Angew. Chem. Int. Ed. 2011, 50, 4180–4183. [Google Scholar] [CrossRef]
- Yao, Q.C.; Li, H.D.; Xian, L.M.; Xu, F.; Xia, J.; Fan, J.L.; Du, J.J.; Wang, J.Y.; Peng, X.J. Differentiating RNA from DNA by a molecular fluorescent probe based on the “door-bolt” mechanism biomaterials. Biomaterials 2018, 177, 78–87. [Google Scholar] [CrossRef]
- Pan, J. RNA polymerase—An important molecular target of triptolide in cancer cells. Cancer Lett. 2010, 292, 149–152. [Google Scholar] [CrossRef]
- Manzo, S.G.; Zhou, Z.-L.; Wang, Y.-Q.; Marinello, J.; He, J.-X.; Li, Y.-C.; Ding, J.; Capranico, G.; Miao, Z.-H. Natural Product Triptolide Mediates Cancer Cell Death by Triggering CDK7-Dependent Degradation of RNA Polymerase II. Cancer Res. 2012, 72, 5363–5373. [Google Scholar] [CrossRef] [Green Version]
- Yi, J.-M.; Huan, X.-J.; Song, S.-S.; Zhou, H.; Wang, Y.-Q.; Miao, Z.-H. Triptolide Induces Cell Killing in Multidrug-Resistant Tumor Cells via CDK7/RPB1 Rather than XPB or p44. Mol. Cancer Ther. 2016, 15, 1495–1503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, A.; Behera, R.K.; Behera, P.K.; Mishra, B.K.; Behera, G.B. Cyanines during the 1990s: A review. Chem. Rev. 2000, 100, 1973–2011. [Google Scholar] [CrossRef]
- Sun, W.; Guo, S.G.; Hu, C.; Fan, J.L.; Peng, X.J. Recent Development of Chemosensors Based on Cyanine Platforms. Chem. Rev. 2016, 116, 7768–7817. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.P.; Palanikumar, L.; Jeena, M.T.; Kim, K.; Ryu, J.H. Cancer-mitochondria-targeted photodynamic therapy with supramolecular assembly of HA and a water soluble NIR cyanine dye. Chem. Sci. 2017, 8, 8351–8356. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.Z.; Wu, W.H.; Sun, J.F.; Guo, S. Triplet photosensitizers: From molecular design to applications. Chem. Soc. Rev. 2013, 42, 5323–5351. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.Z.; Xu, K.J.; Yang, W.B.; Wang, Z.J.; Zhong, F.F. The triplet excited state of Bodipy: Formation, modulation and application. Chem. Soc. Rev. 2015, 44, 8904–8939. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Fan, J.L.; Wang, S.Z.; Kang, Y.; Du, J.J.; Peng, X.J. Biodegradable Drug-Loaded Hydroxyapatite Nanotherapeutic Agent for Targeted Drug Release in Tumors. ACS Appl. Mater. Interfaces 2018, 10, 7832–7840. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Tan, Y.; Ma, X.; Jin, X.; Tian, Y.; Li, M. Photodynamic Therapy with Tumor Cell Discrimination through RNA-Targeting Ability of Photosensitizer. Molecules 2021, 26, 5990. https://doi.org/10.3390/molecules26195990
Xu Y, Tan Y, Ma X, Jin X, Tian Y, Li M. Photodynamic Therapy with Tumor Cell Discrimination through RNA-Targeting Ability of Photosensitizer. Molecules. 2021; 26(19):5990. https://doi.org/10.3390/molecules26195990
Chicago/Turabian StyleXu, Yuan, Yang Tan, Xiuqin Ma, Xiaoyi Jin, Ye Tian, and Miao Li. 2021. "Photodynamic Therapy with Tumor Cell Discrimination through RNA-Targeting Ability of Photosensitizer" Molecules 26, no. 19: 5990. https://doi.org/10.3390/molecules26195990
APA StyleXu, Y., Tan, Y., Ma, X., Jin, X., Tian, Y., & Li, M. (2021). Photodynamic Therapy with Tumor Cell Discrimination through RNA-Targeting Ability of Photosensitizer. Molecules, 26(19), 5990. https://doi.org/10.3390/molecules26195990





