Workflow to Investigate Subtle Differences in Wine Volatile Metabolome Induced by Different Root Systems and Irrigation Regimes
Abstract
:1. Introduction
2. Results and Discussion
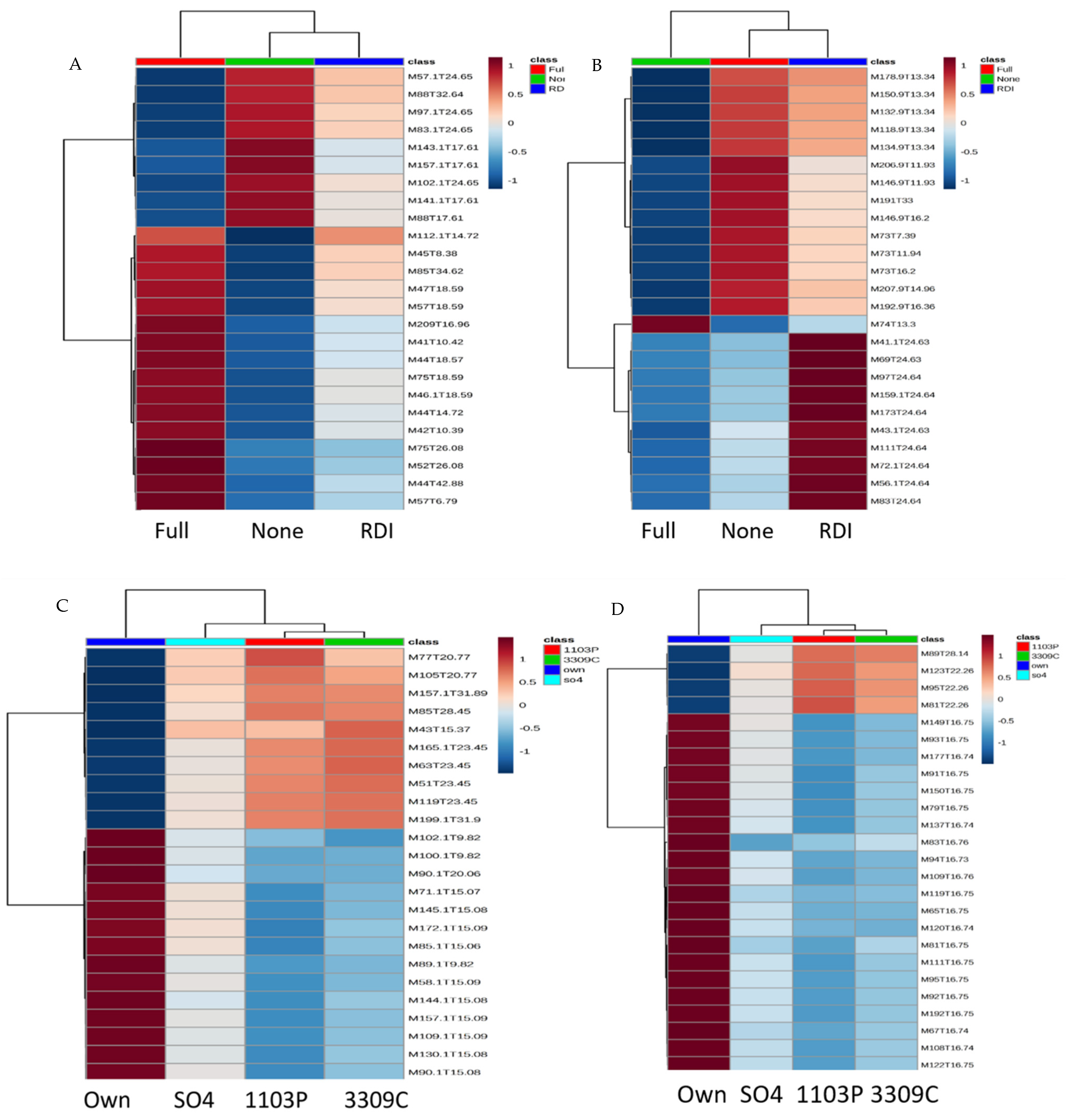
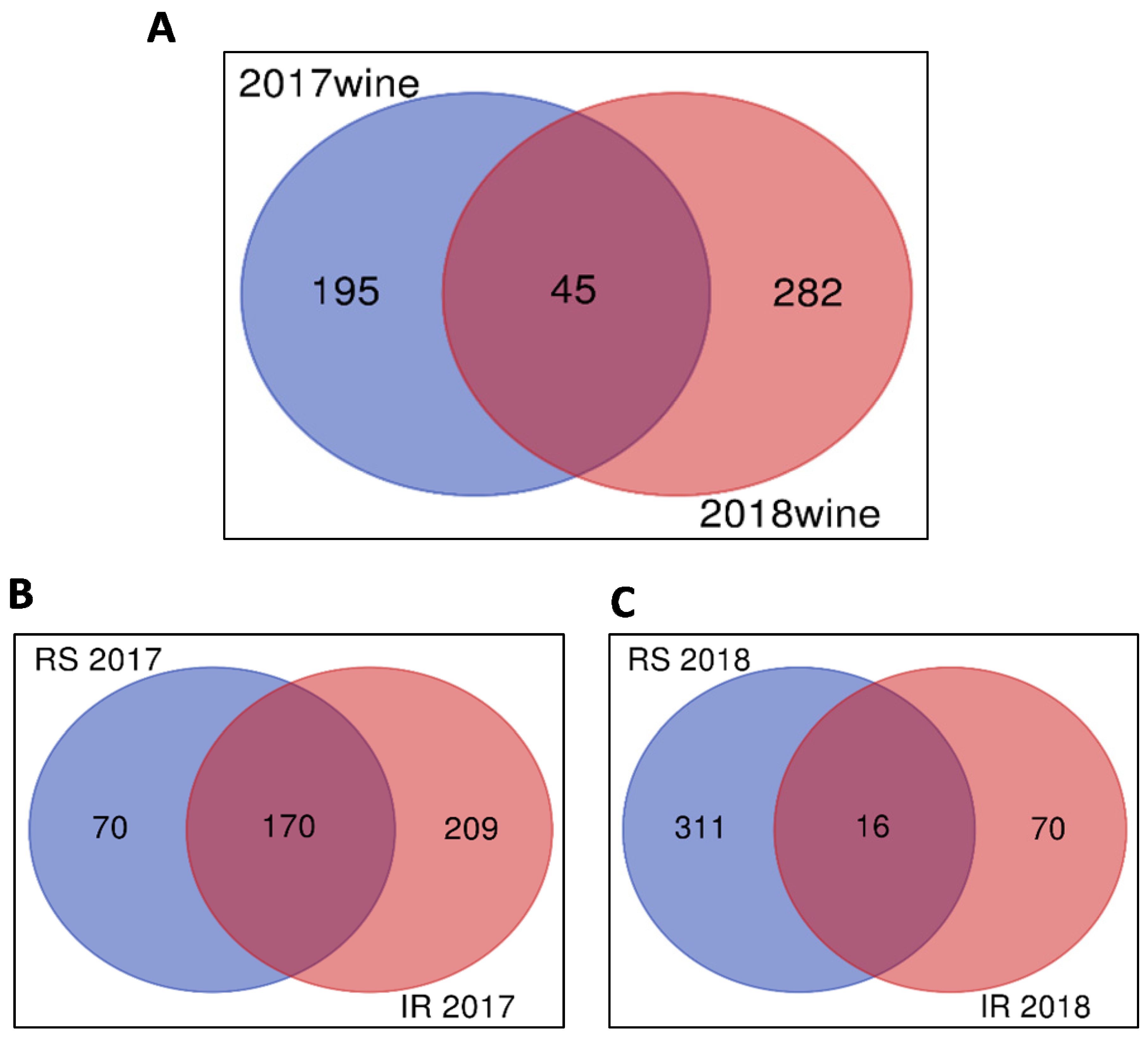
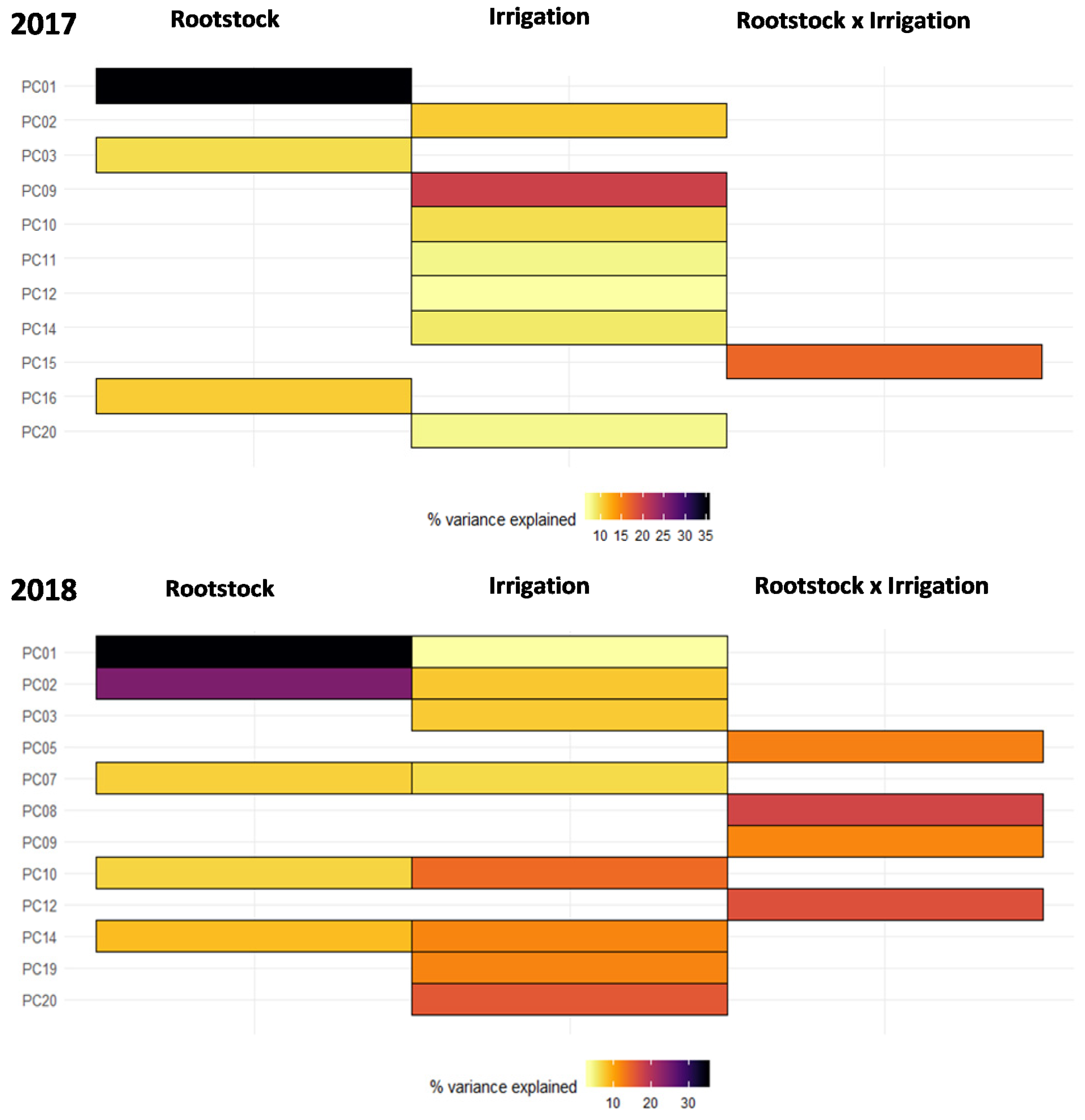
2.1. Untargeted Metabolomics Results
2.2. Identification of Significant Features
2.3. Compound Identification and Confirmation
2.4. Quantitation of the Compounds
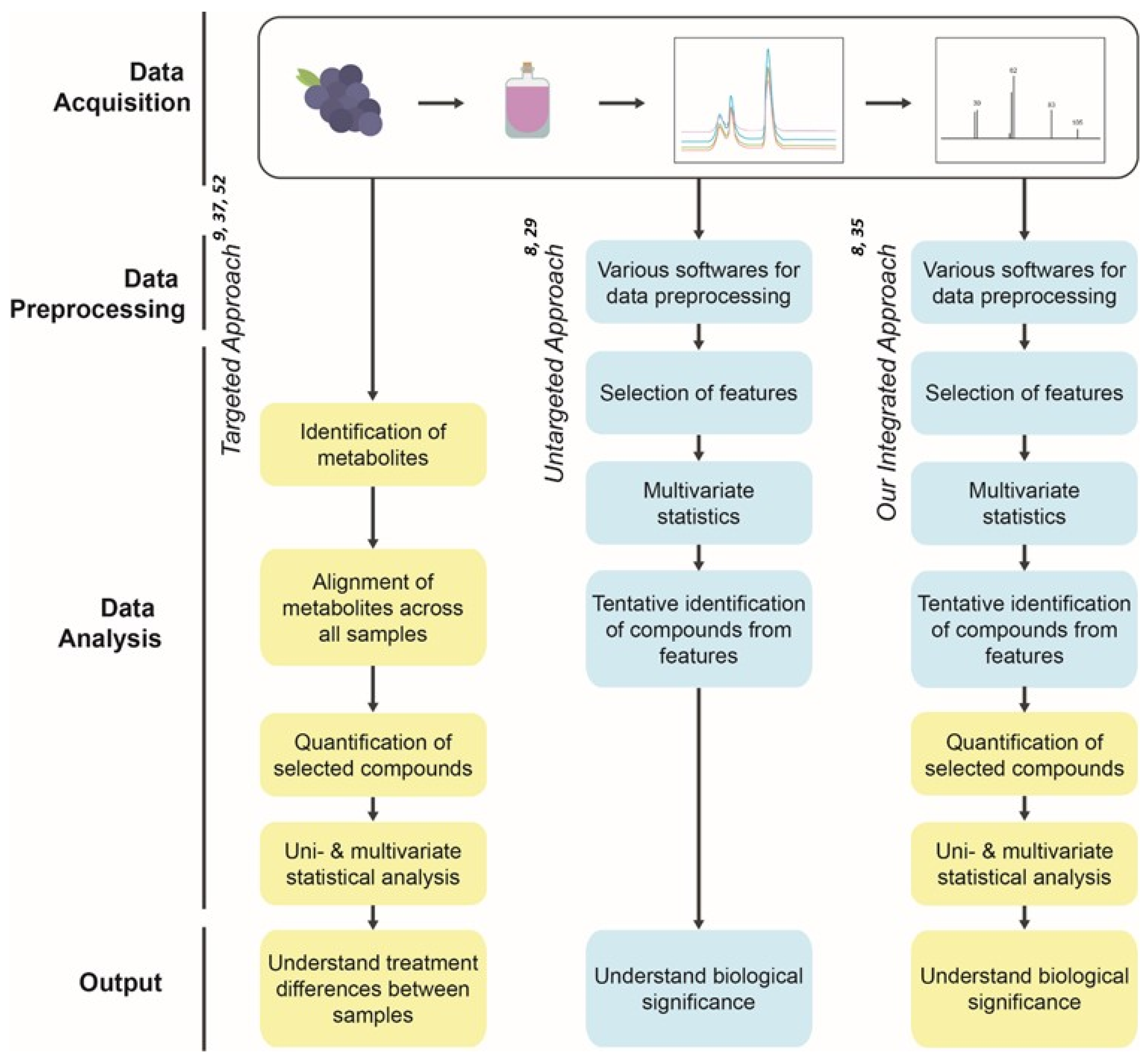
3. Materials and Methods
3.1. Study Design and Sampling
3.2. Winemaking
3.3. Reagents and Chemicals
3.4. Extraction of Wine Volatiles
3.5. HS-SPME-GC-MS/MS
3.6. Data Processing Using Untargeted Metabolomics Analysis
3.7. Identification and Confirmation of the Compounds
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Asproudi, A.; Petrozziello, M.; Cavalletto, S.; Guidoni, S. Grape aroma precursors in cv. Nebbiolo as affected by vine microclimate. Food Chem. 2016, 211, 947–956. [Google Scholar] [CrossRef]
- Ferreira, V.; Lopez, R. The Actual and Potential Aroma of Winemaking Grapes. Biomolecules 2019, 9, 818. [Google Scholar] [CrossRef] [Green Version]
- Sáenz-Navajas, M.-P.; Fernández-Zurbano, P.; Ferreira, V. Contribution of nonvolatile composition to wine flavor. Food Rev. Int. 2012, 28, 389–411. [Google Scholar] [CrossRef] [Green Version]
- Narduzzi, L.; Stanstrup, J.; Mattivi, F. Comparing Wild American Grapes with Vitis vinifera: A Metabolomics Study of Grape Composition. J. Agric. Food Chem. 2015, 63, 6823–6834. [Google Scholar] [CrossRef]
- Schueuermann, C.; Steel, C.C.; Blackman, J.W.; Clark, A.C.; Schwarz, L.J.; Moraga, J.; Collado, I.G.; Schmidtke, L.M. A GC–MS untargeted metabolomics approach for the classification of chemical differences in grape juices based on fungal pathogen. Food Chem. 2019, 270, 375–384. [Google Scholar] [CrossRef]
- Parker, M.; Capone, D.L.; Francis, I.L.; Herderich, M.J. Aroma Precursors in Grapes and Wine: Flavor Release during Wine Production and Consumption. J. Agric. Food Chem. 2017, 66, 2281–2286. [Google Scholar] [CrossRef]
- Francis, I.; Newton, J. Determining wine aroma from compositional data. Aust. J. Grape Wine Res. 2005, 11, 114–126. [Google Scholar] [CrossRef]
- Tikunov, Y.; Lommen, A.; de Vos, C.H.R.; Verhoeven, H.A.; Bino, R.J.; Hall, R.D.; Bovy, A.G. A Novel Approach for Nontargeted Data Analysis for Metabolomics. Large-Scale Profiling of Tomato Fruit Volatiles. Plant Physiol. 2005, 139, 1125–1137. [Google Scholar] [CrossRef] [Green Version]
- Fan, W.; Xu, Y.; Jiang, W.; Li, J. Identification and quantification of impact aroma compounds in 4 nonfloral Vitis vinifera varieties grapes. J. Food Sci. 2010, 75, S81–S88. [Google Scholar] [CrossRef]
- Ollat, N.; Bordenave, L.; Tandonnet, J.-P.; Boursiquot, J.-M.; Marguerit, E. Grapevine Rootstocks: Origins and Perspectives. In Proceedings of the International Symposium on Grapevine Roots, Rauscedo, Italy, 16–17 October 2014; pp. 11–22. [Google Scholar]
- Main, G.; Morris, J.; Striegler, K. Rootstock effects on Chardonel productivity, fruit, and wine composition. Am. J. Enol. Vitic. 2002, 53, 37–40. [Google Scholar]
- Ferris, H.; Zheng, L.; Walker, M. Resistance of grape rootstocks to plant-parasitic nematodes. J. Nematol. 2012, 44, 377. [Google Scholar]
- Serra, I.; Strever, A.; Myburgh, P.; Deloire, A. The interaction between rootstocks and cultivars (Vitis vinifera L.) to enhance drought tolerance in grapevine. Aust. J. Grape Wine Res. 2014, 20, 1–14. [Google Scholar] [CrossRef]
- Zhang, L.; Marguerit, E.; Rossdeutsch, L.; Ollat, N.; Gambetta, G.A. The influence of grapevine rootstocks on scion growth and drought resistance. Theor. Exp. Plant Physiol. 2016, 28, 143–157. [Google Scholar] [CrossRef]
- Ough, C.; Cook, J.; Lider, L. Rootstock-scion interactions concerning wine making. II. Wine compositional and sensory changes attributed to rootstock and fertilizer level differences. Am. J. Enol. Vitic. 1968, 19, 254–265. [Google Scholar]
- Olarte Mantilla, S.M.; Collins, C.; Iland, P.G.; Kidman, C.M.; Ristic, R.; Boss, P.K.; Jordans, C.; Bastian, S.E.P. Shiraz (Vitis vinifera L.) Berry and Wine Sensory Profiles and Composition Are Modulated by Rootstocks. Am. J. Enol. Vitic. 2018, 69, 32–44. [Google Scholar] [CrossRef]
- Romero, P.; Botía, P.; del Amor, F.M.; Gil-Muñoz, R.; Flores, P.; Navarro, J.M. Interactive effects of the rootstock and the deficit irrigation technique on wine composition, nutraceutical potential, aromatic profile, and sensory attributes under semiarid and water limiting conditions. Agric. Water Manag. 2019, 225, 105733. [Google Scholar] [CrossRef]
- Nelson, C.C.; Kennedy, J.A.; Zhang, Y.; Kurtural, S.K. Applied Water and Rootstock Affect Productivity and Anthocyanin Composition of Zinfandel in Central California. Am. J. Enol. Vitic. 2015, 67, 18–28. [Google Scholar] [CrossRef]
- Vivian, Z.; Thibaut, V.; Agnès, D.; Sandrine, B.; Fabrice, L.; Carole, K.; Katia, G.; Jorge, E.S.; Olivier, V.; Jean-Laurent, S. The impact of plant water status on the gas exchange, berry composition and wine quality of Chasselas grapes in Switzerland. OENO One 2018, 52, 4. [Google Scholar]
- Berdeja, M.; Hilbert, G.; Dai, Z.W.; Lafontaine, M.; Stoll, M.; Schultz, H.R.; Delrot, S. Effect of water stress and rootstock genotype on Pinot Noir berry composition. Aust. J. Grape Wine Res. 2014, 20, 409–421. [Google Scholar] [CrossRef]
- Deluc, L.G.; Quilici, D.R.; Decendit, A.; Grimplet, J.; Wheatley, M.D.; Schlauch, K.A.; Mérillon, J.-M.; Cushman, J.C.; Cramer, G.R. Water deficit alters differentially metabolic pathways affecting important flavor and quality traits in grape berries of Cabernet Sauvignon and Chardonnay. BMC Genom. 2009, 10, 212. [Google Scholar] [CrossRef] [Green Version]
- Bindon, K.A.; Dry, P.R.; Loveys, B.R. Influence of Plant Water Status on the Production of C13-Norisoprenoid Precursors in Vitis vinifera L. Cv. Cabernet Sauvignon Grape Berries. J. Agric. Food Chem. 2007, 55, 4493–4500. [Google Scholar] [CrossRef]
- Qian, M.C.; Fang, Y.; Shellie, K. Volatile Composition of Merlot Wine from Different Vine Water Status. J. Agric. Food Chem. 2009, 57, 7459–7463. [Google Scholar] [CrossRef]
- Berdeja, M.; Nicolas, P.; Kappel, C.; Dai, Z.W.; Hilbert, G.; Peccoux, A.; Lafontaine, M.; Ollat, N.; Gomès, E.; Delrot, S. Water limitation and rootstock genotype interact to alter grape berry metabolism through transcriptome reprogramming. Hortic. Res. 2015, 2, 15012. [Google Scholar] [CrossRef] [Green Version]
- Koundouras, S.; Hatzidimitriou, E.; Karamolegkou, M.; Dimopoulou, E.; Kallithraka, S.; Tsialtas, J.T.; Zioziou, E.; Nikolaou, N.; Kotseridis, Y. Irrigation and rootstock effects on the phenolic concentration and aroma potential of Vitis vinifera L. cv. cabernet sauvignon grapes. J. Agric. Food Chem. 2009, 57, 7805–7813. [Google Scholar] [CrossRef]
- Cuadros-Inostroza, A.; Ruíz-Lara, S.; González, E.; Eckardt, A.; Willmitzer, L.; Peña-Cortés, H. GC–MS metabolic profiling of Cabernet Sauvignon and Merlot cultivars during grapevine berry development and network analysis reveals a stage-and cultivar-dependent connectivity of primary metabolites. Metabolomics 2016, 12, 39. [Google Scholar] [CrossRef] [Green Version]
- Flamini, R.; De Rosso, M.; Panighel, A.; Dalla Vedova, A.; De Marchi, F.; Bavaresco, L. Profiling of grape monoterpene glycosides (aroma precursors) by ultra-high performance-liquid chromatography-high resolution mass spectrometry (UHPLC/QTOF). J. Mass Spectrom. 2014, 49, 1214–1222. [Google Scholar] [CrossRef]
- Roullier-Gall, C.; Witting, M.; Gougeon, R.D.; Schmitt-Kopplin, P. High precision mass measurements for wine metabolomics. Front. Chem. 2014, 2, 102. [Google Scholar] [CrossRef] [Green Version]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies—Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [Green Version]
- Toffali, K.; Zamboni, A.; Anesi, A.; Stocchero, M.; Pezzotti, M.; Levi, M.; Guzzo, F. Novel aspects of grape berry ripening and post-harvest withering revealed by untargeted LC-ESI-MS metabolomics analysis. Metabolomics 2011, 7, 424–436. [Google Scholar] [CrossRef]
- Shure, K.B.; Acree, T.E. Changes in the odor-active compounds in Vitis labruscana cv. Concord during growth and development. J. Agric. Food Chem. 1994, 42, 350–353. [Google Scholar] [CrossRef]
- Sun, Q.; Gates, M.J.; Lavin, E.H.; Acree, T.E.; Sacks, G.L. Comparison of odor-active compounds in grapes and wines from Vitis vinifera and non-foxy American grape species. J. Agric. Food Chem. 2011, 59, 10657–10664. [Google Scholar] [CrossRef]
- Poivet, E.; Tahirova, N.; Peterlin, Z.; Xu, L.; Zou, D.-J.; Acree, T.; Firestein, S. Functional odor classification through a medicinal chemistry approach. Sci. Adv. 2018, 4, eaao6086. [Google Scholar] [CrossRef] [Green Version]
- De Vos, R.C.; Moco, S.; Lommen, A.; Keurentjes, J.J.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778. [Google Scholar] [CrossRef]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: A web-based platform to process untargeted metabolomic data. Anal. Chem. 2012, 84, 5035–5039. [Google Scholar] [CrossRef] [Green Version]
- Callejon, R.M.; Margulies, B.; Hirson, G.D.; Ebeler, S.E. Dynamic Changes in Volatile Compounds during Fermentation of Cabernet Sauvignon Grapes with and without Skins. Am. J. Enol. Vitic. 2012, 63, 301–312. [Google Scholar] [CrossRef]
- Canuti, V.; Conversano, M.; Calzi, M.L.; Heymann, H.; Matthews, M.A.; Ebeler, S.E. Headspace solid-phase microextraction-gas chromatography-mass spectrometry for profiling free volatile compounds in Cabernet Sauvignon grapes and wines. J. Chromatogr. A 2009, 1216, 3012–3022. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, W.-K.; Gao, X.-T.; He, L.; Yang, X.-H.; He, F.; Duan, C.-Q.; Wang, J. Rootstock-Mediated Effects on Cabernet Sauvignon Performance: Vine Growth, Berry Ripening, Flavonoids, and Aromatic Profiles. Int. J. Mol. Sci. 2019, 20, 401. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Qian, M.C. Quantification of selected aroma-active compounds in Pinot noir wines from different grape maturities. J. Agric. Food Chem. 2006, 54, 8567–8573. [Google Scholar] [CrossRef]
- Wooldridge, J.; Louw, P.; Conradie, W. Effects of rootstock on grapevine performance, petiole and must composition, and overall wine score of Vitis vinifera cv. Chardonnay and Pinot Noir. S. Afr. J. Enol. Vitic. 2010, 31, 45–48. [Google Scholar] [CrossRef] [Green Version]
- Pinu, F.R. Grape and wine metabolomics to develop new insights using untargeted and targeted approaches. Fermentation 2018, 4, 92. [Google Scholar] [CrossRef] [Green Version]
- Commisso, M.; Strazzer, P.; Toffali, K.; Stocchero, M.; Guzzo, F. Untargeted metabolomics: An emerging approach to determine the composition of herbal products. Comput. Struct. Biotechnol. J. 2013, 4, e201301007. [Google Scholar] [CrossRef] [Green Version]
- Teh, S.L.; Rostandy, B.; Awale, M.; Luby, J.J.; Fennell, A.; Hegeman, A.D. Genetic analysis of stilbenoid profiles in grapevine stems reveals a major mQTL hotspot on chromosome 18 associated with disease-resistance motifs. Hortic. Res. 2019, 6, 121. [Google Scholar] [CrossRef] [Green Version]
- Harbertson, J.F.; Keller, M. Rootstock Effects on Deficit-Irrigated Winegrapes in a Dry Climate: Grape and Wine Composition. Am. J. Enol. Vitic. 2011, 63, 40–48. [Google Scholar] [CrossRef] [Green Version]
- Arapitsas, P.; Ugliano, M.; Marangon, M.; Piombino, P.; Rolle, L.; Gerbi, V.; Versari, A.; Mattivi, F. Use of Untargeted Liquid Chromatography—Mass Spectrometry Metabolome To Discriminate Italian Monovarietal Red Wines, Produced in Their Different Terroirs. J. Agric. Food Chem. 2020, 68, 13353–13366. [Google Scholar] [CrossRef]
- Lee, J.-E.; Hwang, G.-S.; Van Den Berg, F.; Lee, C.-H.; Hong, Y.-S. Evidence of vintage effects on grape wines using 1H NMR-based metabolomic study. Anal. Chim. Acta 2009, 648, 71–76. [Google Scholar] [CrossRef]
- Schueuermann, C.; Khakimov, B.; Engelsen, S.B.; Bremer, P.; Silcock, P. GC-MS Metabolite Profiling of Extreme Southern Pinot noir Wines: Effects of Vintage, Barrel Maturation, and Fermentation Dominate over Vineyard Site and Clone Selection. J. Agric. Food Chem. 2016, 64, 2342–2351. [Google Scholar] [CrossRef]
- Bowen, A.J.; Reynolds, A.G. Aroma compounds in Ontario Vidal and Riesling icewines. I. Effects of harvest date. Food Res. Int. 2015, 76, 540–549. [Google Scholar] [CrossRef]
- Bowen, A.J.; Reynolds, A.G. Aroma compounds in Ontario Vidal and Riesling icewines. II. Effects of crop level. Food Res. Int. 2015, 76, 550–560. [Google Scholar] [CrossRef]
- Bisson, L.F.; Waterhouse, A.L.; Ebeler, S.E.; Walker, M.A.; Lapsley, J.T. The present and future of the international wine industry. Nature 2002, 418, 696. [Google Scholar] [CrossRef]
- Weingart, G.; Kluger, B.; Forneck, A.; Krska, R.; Schuhmacher, R. Establishment and Application of a Metabolomics Workflow for Identification and Profiling of Volatiles from Leaves of Vitis vinifera by HS-SPME-GC-MS. Phytochem. Anal. 2012, 23, 345–358. [Google Scholar] [CrossRef]
- Ferreira, V.; López, R.; Cacho, J.F. Quantitative determination of the odorants of young red wines from different grape varieties. J. Sci. Food Agric. 2000, 80, 1659–1667. [Google Scholar] [CrossRef]
- El-Ansary, D.O.; Okamoto, G. Vine Water Relations and Quality of Muscat of Alexandria Table Grapes Subjected to Partial Root-zone Drying and Regulated Deficit Irrigation. J. Jpn. Soc. Hortic. Sci. 2007, 76, 13–19. [Google Scholar] [CrossRef] [Green Version]
- Bianco, R.L.; Farina, V.; Avellone, G.; Filizzola, F.; Agozzino, P. Fruit quality and volatile fraction of ‘Pink Lady’apple trees in response to rootstock vigor and partial rootzone drying. J. Sci. Food Agric. 2008, 88, 1325–1334. [Google Scholar] [CrossRef]
- des Gachons, C.P.; Leeuwen, C.V.; Tominaga, T.; Soyer, J.P.; Gaudillère, J.P.; Dubourdieu, D. Influence of water and nitrogen deficit on fruit ripening and aroma potential of Vitis vinifera L. cv Sauvignon blanc in field conditions. J. Sci. Food Agric. 2005, 85, 73–85. [Google Scholar] [CrossRef]
- Van Wyk, C.; Augustyn, O.; De Wet, P.; Joubert, W. Isoamyl acetate—A key fermentation volatile of wines of Vitis vinifera cv Pinotage. Am. J. Enol. Vitic. 1979, 30, 167–173. [Google Scholar]
- Plata, C.; Millan, C.; Mauricio, J.; Ortega, J. Formation of ethyl acetate and isoamyl acetate by various species of wine yeasts. Food Microbiol. 2003, 20, 217–224. [Google Scholar] [CrossRef]
- Maimaitiyiming, M.; Ghulam, A.; Bozzolo, A.; Wilkins, J.L.; Kwasniewski, M.T. Early detection of plant physiological responses to different levels of water stress using reflectance spectroscopy. Remote Sens. 2017, 9, 745. [Google Scholar] [CrossRef] [Green Version]
- Migicovsky, Z.; Harris, Z.N.; Klein, L.L.; Li, M.; McDermaid, A.; Chitwood, D.H.; Fennell, A.; Kovacs, L.G.; Kwasniewski, M.; Londo, J.P.; et al. Rootstock effects on scion phenotypes in a ‘Chambourcin’ experimental vineyard. Hortic. Res. 2019, 6, 64. [Google Scholar] [CrossRef] [Green Version]
- Kwasniewski, M.T.; Vanden Heuvel, J.E.; Pan, B.S.; Sacks, G.L. Timing of cluster light environment manipulation during grape development affects C13 norisoprenoid and carotenoid concentrations in Riesling. J. Agric. Food Chem. 2010, 58, 6841–6849. [Google Scholar] [CrossRef]
- Hampel, D.; Robinson, A.L.; Johnson, A.; Ebeler, S.E. Direct hydrolysis and analysis of glycosidically bound aroma compounds in grapes and wines: Comparison of hydrolysis conditions and sample preparation methods. Aust. J. Grape Wine Res. 2014, 20, 361–377. [Google Scholar] [CrossRef]
- Gowda, H.; Ivanisevic, J.; Johnson, C.H.; Kurczy, M.E.; Benton, H.P.; Rinehart, D.; Nguyen, T.; Ray, J.; Kuehl, J.; Arevalo, B.; et al. Interactive XCMS Online: Simplifying Advanced Metabolomic Data Processing and Subsequent Statistical Analyses. Anal. Chem. 2014, 86, 6931–6939. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2016, 55, 14.10.1–14.10.91. [Google Scholar] [CrossRef]

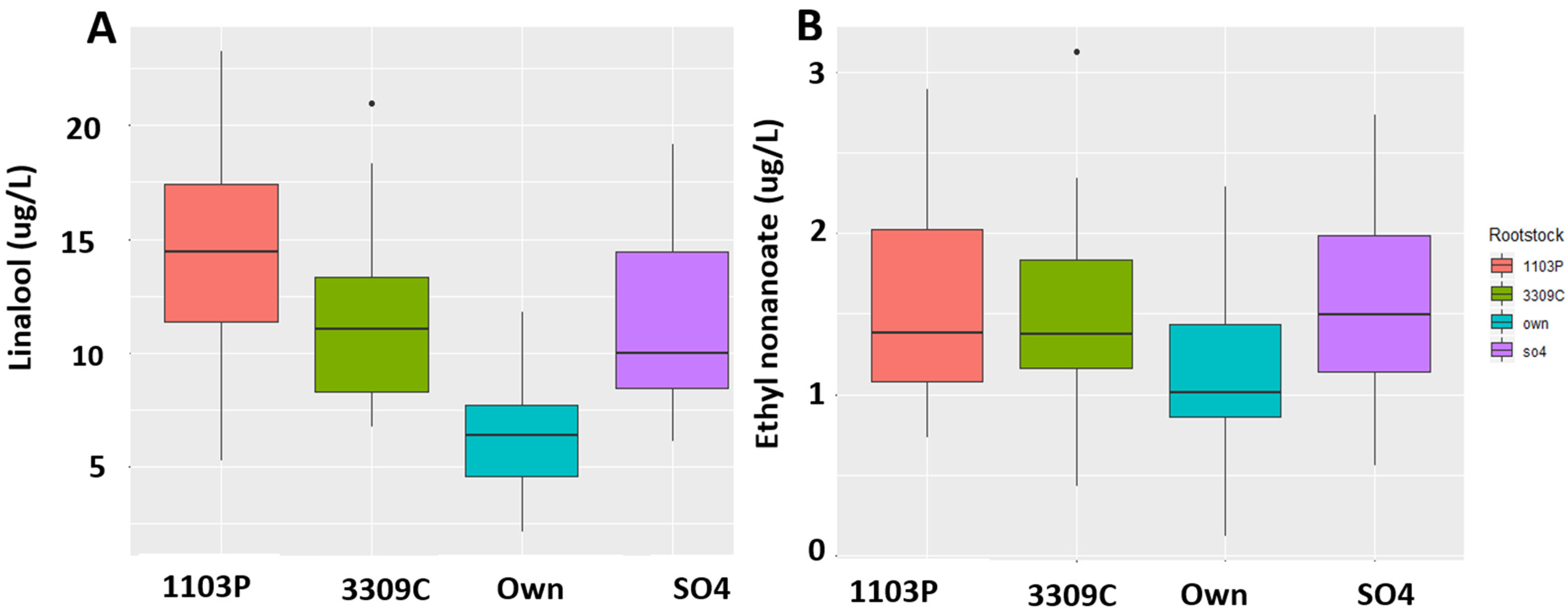
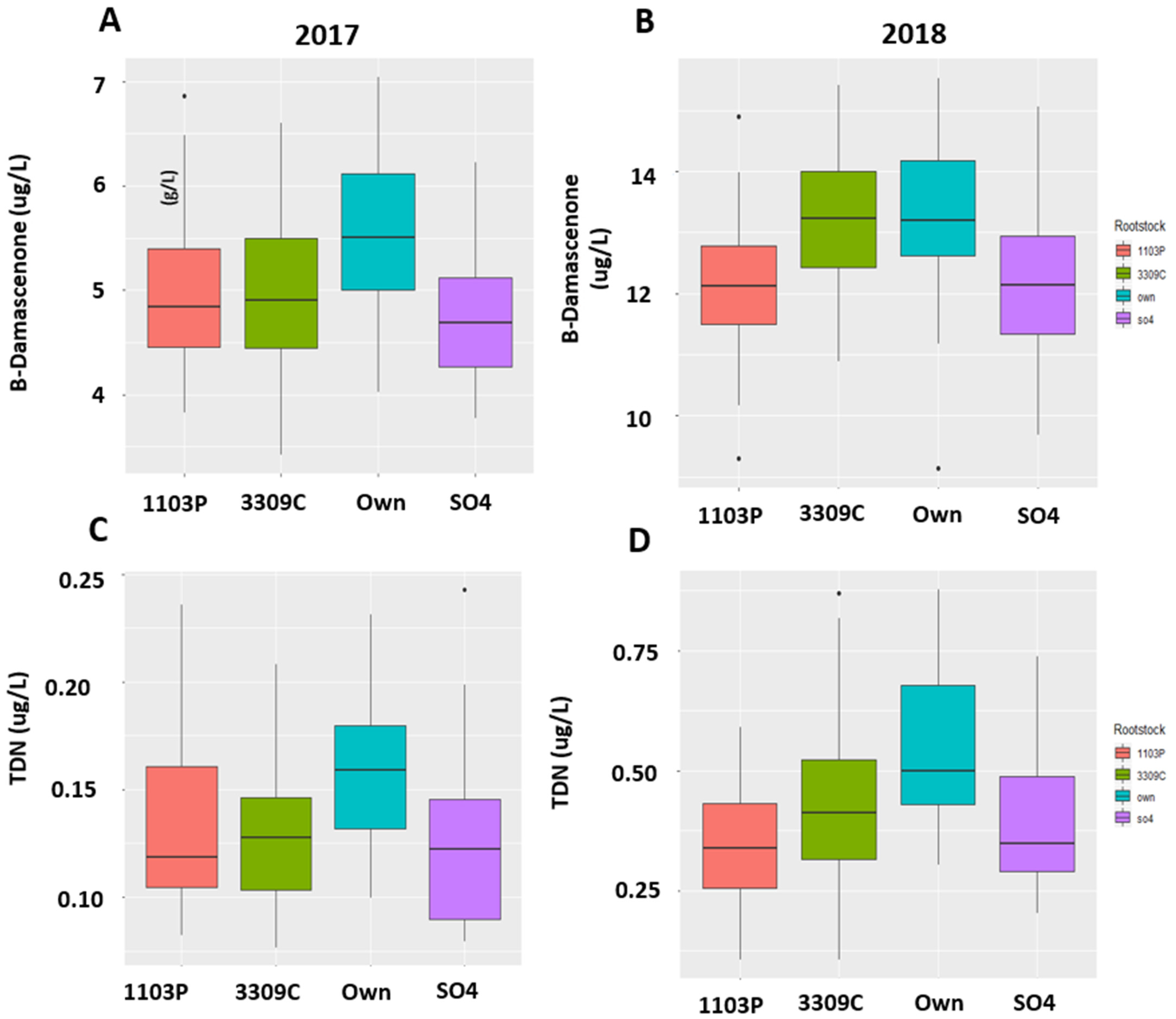
| 2017 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compounds | Full | Full | Full | Full | RDI | RDI | RDI | RDI | None | None | None | None | p-Value |
| 1103P | 3309C | SO4 | Own | 1103P | 3309C | SO4 | Own | 1103P | 3309C | SO4 | Own | ||
| Isoamyl acetate | 407.1 b 1,2 | 1080.8 a | 391.2 b | 381.8 b | 559.5 b | 553.2 b | 431.5 b | 244.9 b | 449.2 b | 573.5 b | 483.4 b | 386.0 b | 0.0203 |
| Ethyl heptanoate | 397.7 c | 295.1 cd | 422.7 c | 589.5 ab | 203.1 d | 443.0 bc | 367.9 c | 640.7 a | 363.8 c | 352.7 c | 366.3 c | 583.2 ab | 0.030 |
| Ethyl octanoate | 38.3 c | 26.7 cd | 44.7 bc | 67.1 a | 15.6 d | 44.3 bc | 38.0 c | 76.6 a | 37.9 c | 35.1 cd | 37.3 c | 61.1 ab | 0.040 |
| 1-Octanol | 15.8 cd | 15.7 cd | 14.5 cd | 14.3 cd | 19.4 a | 15.9 cd | 16.3 bcd | 13.6 d | 14.9 cd | 16.7 bc | 18.7 ab | 14.0 cd | 0.000 |
| 1-Nonanol | 12.3 ab | 13.2 ab | 12.6 ab | 12.3 ab | 14.5 a | 12.3 ab | 14.7 a | 11.0 b | 12.7 ab | 15.2 a | 12.9 ab | 12.5 ab | 0.100 |
| Ethyl hydrocinnamate | 3.0 bc | 2.5 bcde | 1.9 cdef | 1.6 def | 4.7 a | 2.6 bcde | 2.7 bcd | 1.4 f | 2.7 bcd | 2.2 bcdef | 3.2 b | 1.5 ef | 0.000 |
| 1-Dodecanol | 0.4 abcd | 0.3 bcd | 0.4 abc | 0.5 ab | 0.3 d | 0.4 abcd | 0.3 bcd | 0.4 abc | 0.5 a | 0.3 d | 0.4 abcd | 0.5 ab | 0.030 |
| Ethyl-tetradecenoate | 42.6 bc | 46.6 bc | 46.6 bc | 52.2 bc | 56.8 ab | 69.3 a | 48.3 bc | 36.9 c | 55.2 ab | 44.7 bc | 55.9 ab | 40.8 bc | 0.000 |
| Ethyl hexadecanoate | 192.4 bc | 168.6 bc | 186.5 bc | 234.1 abc | 249.0 ab | 317.9 a | 222.5 bc | 153.1 bc | 249.5 ab | 145.2 c | 238.8 abc | 185.2 bc | 0.000 |
| Isoamyl hexanoate | 1.6 bc | 1.5 bc | 1.4 cd | 1.3 cd | 2.3 a | 1.8 b | 1.6 bc | 1.0 d | 1.5 bc | 1.9 b | 1.8 b | 1.24cd | 0.000 |
| 2-Phenylethyl acetate | 1.6 bc | 35.2 bcd | 28.6 cde | 28.2 cde | 60.4 a | 41.8 bc | 39.8 bcd | 15.7 e | 35.2 bcd | 46.70 b | 39.1 bcd | 24.9 de | 0.000 |
| 2018 | |||||||||||||
| Linalool | 15.6 a | 11.5 bcd | 9.7 cd | 8.4 de | 16.1 a | 10.6 bcd | 11.4 bcd | 5.6 e | 11.5 bcd | 12.1 bc | 13.4 ab | 6.1 e | 0.005 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Awale, M.; Liu, C.; Kwasniewski, M.T. Workflow to Investigate Subtle Differences in Wine Volatile Metabolome Induced by Different Root Systems and Irrigation Regimes. Molecules 2021, 26, 6010. https://doi.org/10.3390/molecules26196010
Awale M, Liu C, Kwasniewski MT. Workflow to Investigate Subtle Differences in Wine Volatile Metabolome Induced by Different Root Systems and Irrigation Regimes. Molecules. 2021; 26(19):6010. https://doi.org/10.3390/molecules26196010
Chicago/Turabian StyleAwale, Mani, Connie Liu, and Misha T. Kwasniewski. 2021. "Workflow to Investigate Subtle Differences in Wine Volatile Metabolome Induced by Different Root Systems and Irrigation Regimes" Molecules 26, no. 19: 6010. https://doi.org/10.3390/molecules26196010
APA StyleAwale, M., Liu, C., & Kwasniewski, M. T. (2021). Workflow to Investigate Subtle Differences in Wine Volatile Metabolome Induced by Different Root Systems and Irrigation Regimes. Molecules, 26(19), 6010. https://doi.org/10.3390/molecules26196010







