Rapid Screening Alpha-Glucosidase Inhibitors from Polygoni Vivipari Rhizoma by Multi-Step Matrix Solid-Phase Dispersion, Ultrafiltration and HPLC
Abstract
:1. Introduction
2. Results and Discussion
2.1. Optimization of Multi-Step MSPD Extraction Conditions
2.2. The Optimization of HPLC Conditions
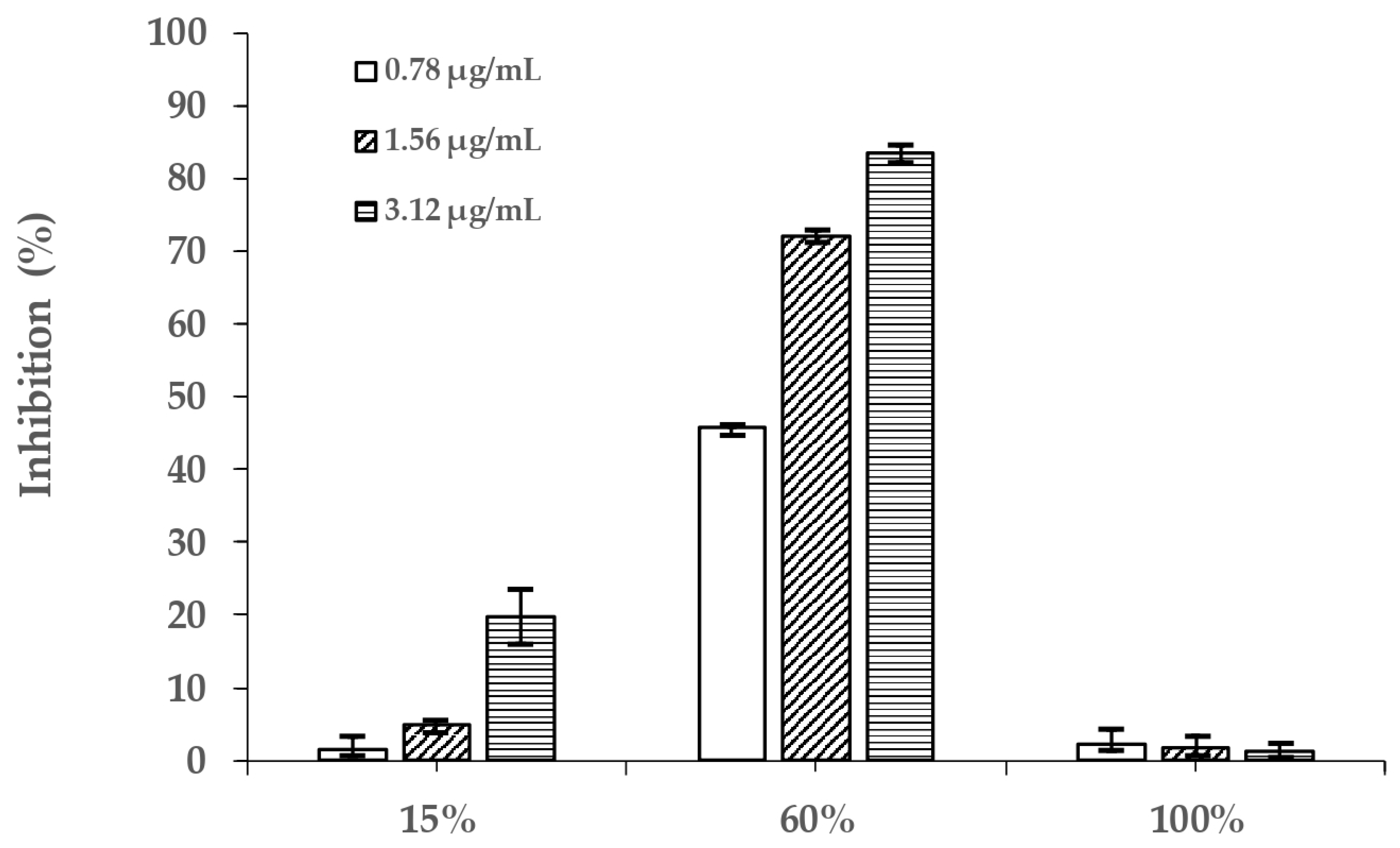
2.3. Alpha-Glucosidase Inhibitory Activity of Different PVR Fractions
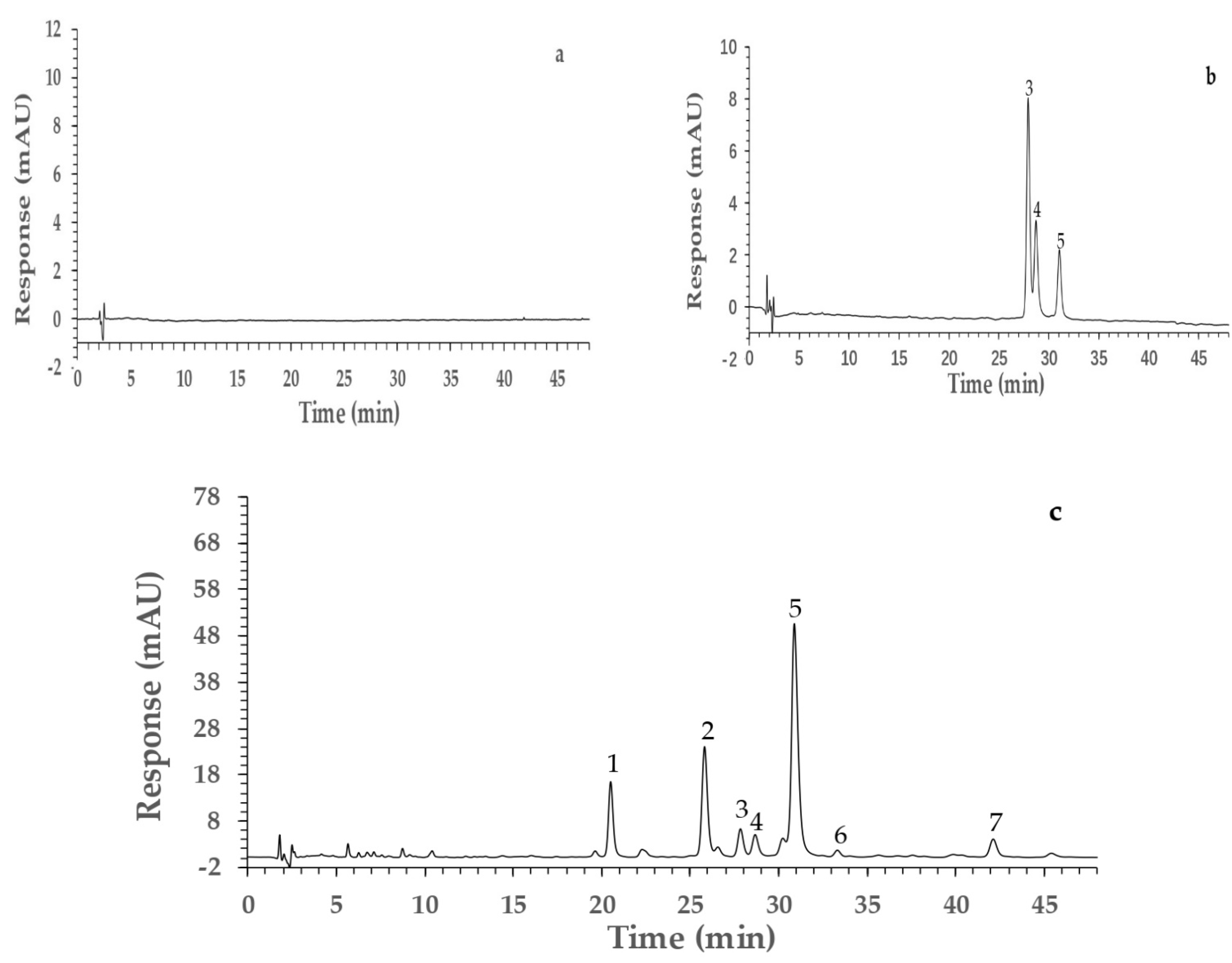
2.4. Identification of Components from PVR
2.5. Screening of Potential AGI by Ultrafiltration and HPLC Analysis
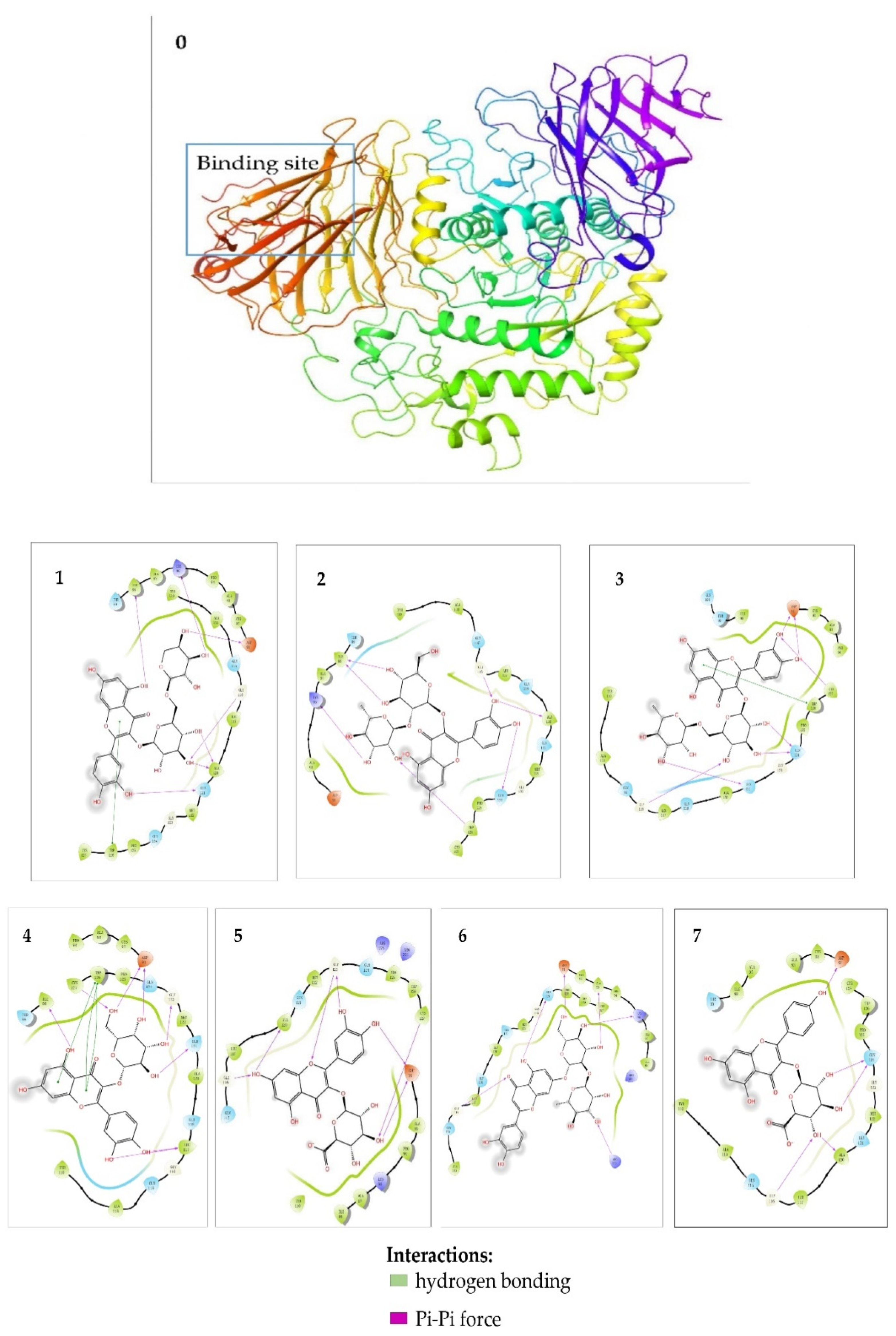
2.6. Molecular Docking Studies
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Sample Preparation
3.2.1. Preparation of Reference Compound Solutions and Blank Solution
3.2.2. Preparation of the Sample by Multi-Step MSPD
3.3. Alpha-Glucosidase Inhibitory Assay
3.4. HPLC-MS Analysis
3.5. Screening Potential AGIs by Ultrafiltration and HPLC Analysis
3.6. Molecular Docking Studies
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Artasensi, A.; Pedretti, A.; Vistoli, G.; Fumagalli, L. Type 2 Diabetes Mellitus: A Review of Multi-Target Drugs. Molecules 2020, 25, 1987. [Google Scholar] [CrossRef]
- Sanchez-Brito, M.; Luna-Rosas, F.J.; Mendoza-Gonzalez, R.; Vazquez-Zapien, G.J.; Martinez-Romo, J.C.; Mata-Miranda, M.M. Type 2 diabetes diagnosis assisted by machine learning techniques through the analysis of FTIR spectra of saliva. Biomed. Signal Process. Control. 2021, 69, 102855. [Google Scholar] [CrossRef]
- Ezzat, S.M.; Salama, M.M. A new α-glucosidase inhibitor from Achillea fragrantissima (Forssk.) Sch. Bip. growing in Egypt. Nat. Prod. Res. 2014, 28, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Z.; Dai, H.; Pan, S.; Wang, H.; Hu, Y.; Ma, W. Isolation and Characterization of an α-Glucosidase Inhibitor from Musa spp. (Baxijiao) Flowers. Molecules 2014, 19, 10563–10573. [Google Scholar] [CrossRef] [Green Version]
- Dhameja, M.; Gupta, P. Synthetic heterocyclic candidates as promising α-glucosidase inhibitors: An overview. Eur. J. Med. Chem. 2019, 176, 343–377. [Google Scholar] [CrossRef]
- Xie, L.; Fu, Q.; Shi, S.; Li, J.; Zhou, X. Rapid and comprehensive profiling of α-glucosidase inhibitors in Buddleja Flos by ultrafiltration HPLC–QTOF-MS/MS with diagnostic ions filtering strategy. Food Chem. 2020, 344, 128651. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, Y.; Sun, L.; Wang, Y.; Gao, X.; Cheng, Y. An ultrafiltration high-performance liquid chromatography coupled with diode array detector and mass spectrometry approach for screening and characterising tyrosinase inhibitors from mulberry leaves. Anal. Chim. Acta 2012, 719, 87–95. [Google Scholar] [CrossRef]
- Liu, L.; Xiao, A.; Ma, L.; Li, D. Analysis of Xanthine Oxidase Inhibitors from Puerariae flos Using Centrifugal Ultrafiltration Coupled with HPLC-MS. J. Braz. Chem. Soc. 2016, 28, 360–366. [Google Scholar] [CrossRef]
- Tu, X.; Chen, W. A Review on the Recent Progress in Matrix Solid Phase Dispersion. Molecules 2018, 23, 2767. [Google Scholar] [CrossRef] [Green Version]
- Qian, Z.; Wu, Z.; Li, C.; Yao, C.; Tan, G.; Li, W.; Guo, D.-A. Rapid Determination of 3 Components with Different Polarities in Medicinal Mushrooms by Multistep Matrix Solid-Phase Dispersion and High-Performance Liquid Chromatography Analysis. Nat. Prod. Commun. 2021, 16, 1–7. [Google Scholar] [CrossRef]
- Qian, Z.; Wu, Z.; Li, C.; Tan, G.; Hu, H.; Li, W. A green liquid chromatography method for rapid determination of ergosterol in edible fungi based on matrix solid-phase dispersion extraction and a core–shell column. Anal. Methods 2020, 12, 3337–3343. [Google Scholar] [CrossRef]
- Pharmacopoeia Commission of the People’s Republic of China. Pharmaceutical Standards of the Minstry of Health of the People’s Republic of China-Volume 1 of Tibetan Medicine; People’s Health Press: Beijing, China, 1995; p. 75.
- Gong, Z.F.; Yang, G.L.; Yan, Z.T.; Xie, J.S. Survey of chemical constituents and bioactivity of Polygonum L. plants. Chin. Trad. Herb Drug. 2002, 33, 82–84. [Google Scholar]
- Zhang, C.X.; Li, Y.L.; Hu, F.Z. Chemical Constituents of fruits of Polygonum viviparum. Acta Bot Boreal-Occident. Sin. 2005, 25, 386–387. [Google Scholar]
- Qian, Z.-M.; Chen, L.; Wu, M.-Q.; Li, D.-Q. Rapid screening and characterization of natural antioxidants in Polygonum viviparum by an on-line system integrating the pressurised liquid micro-extraction, HPLC-DAD-QTOF-MS/MS analysis and antioxidant assay. J. Chromatogr. B 2020, 1137, 121926. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, X.; Qi, X.; Gao, W.; Sun, S.; Li, X.; Jiang, C.; Yu, A.; Zhang, H.; Yu, Y. Determination of sulfonamides in livers using matrix solid-phase dispersion extraction high-performance liquid chromatography. J. Sep. Sci. 2011, 35, 45–52. [Google Scholar] [CrossRef]
- Xu, Y.L.; Dong, Q.; Hu, F.Z. Simultaneous quantitative determination of viterxin, quercetin and quercitrin in Polygonum viviparum in Tibet Plateau by RP-HPLC. Nat. Prod. Res. Dev. 2011, 23, 894–897. [Google Scholar] [CrossRef]
- Lage, G.A.; Medeiros, F.D.S.; Furtado, W.D.L.; Takahashi, J.; Filho, J.D.D.S.; Pimenta, L. The first report on flavonoid isolation from Annona crassiflora Mart. Nat. Prod. Res. 2014, 28, 808–811. [Google Scholar] [CrossRef] [PubMed]
- Su, S.L.; Xue, P.; Ouyang, Z.; Zhou, W.; Duan, J.A. Study on antiplatelet and antithrombin activitives and effective components variation of Puhuang-Wulingzhi before and after compatibility. China J. Chin. Mater. Med. 2015, 40, 3187–3193. [Google Scholar] [CrossRef]
- Li, L.; Zhao, Y.; Liu, W.; Feng, F.; Xie, N. HPLC with quadrupole TOF-MS and chemometrics analysis for the characterization of Folium Turpiniae from different regions. J. Sep. Sci. 2013, 36, 2552–2561. [Google Scholar] [CrossRef]
- Li, W.L.; Ding, J.X.; Zang, B.S.; Gao, S.; Yang, B.; Ji, Y.B. Analytisis of Purified Active Ingredients of Estrogen Mimic Effect in Cuscuta chinensis by HPLC-Q TOF-MS/MS. Chin. Pharm. J. 2014, 49, 1791–1795. [Google Scholar] [CrossRef]
- Wang, Z.; Hwang, S.; Huang, B.; Lim, S. Identification of tyrosinase specific inhibitors from Xanthium strumarium fruit extract using ultrafiltration-high performance liquid chromatography. J. Chromatogr. B. 2015, 1002, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Song, H.-P.; Li, P.; Zhou, P.; Dong, X.; Chen, J. Screening of direct thrombin inhibitors from Radix Salviae Miltiorrhizae by a peak fractionation approach. J. Pharm. Biomed. Anal. 2015, 109, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Shi, S.; Chen, X.; Peng, M. Analysis of tyrosinase binders from Glycyrrhiza uralensis root: Evaluation and comparison of tyrosinase immobilized magnetic fishing-HPLC and reverse ultrafiltration-HPLC. J. Chromatogr. B. 2013, 932, 19–25. [Google Scholar] [CrossRef] [PubMed]
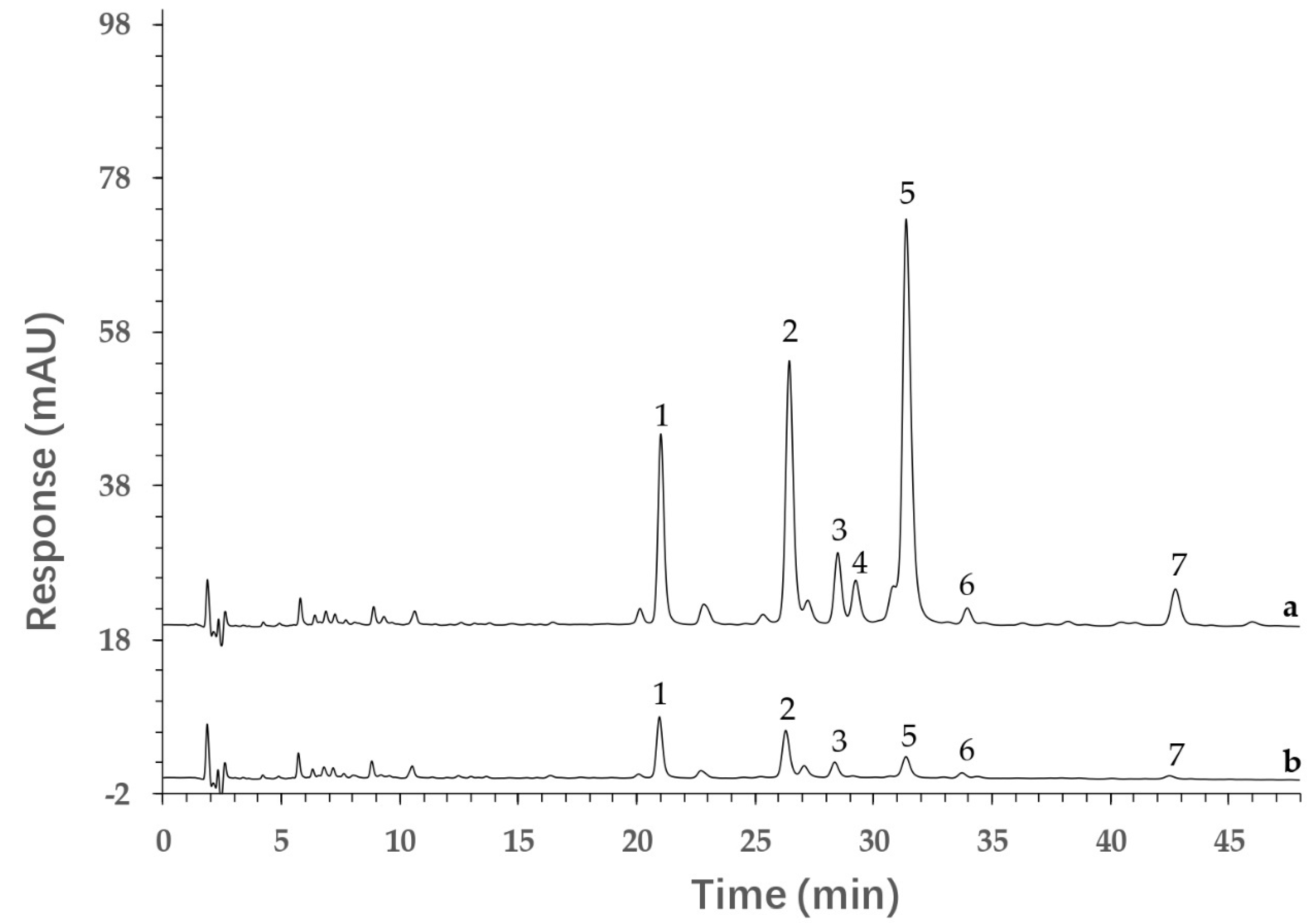

| Peak No. | tR (min) | [M-H]− (m/z) | Fragmentation | Molecular Formula | Identification |
|---|---|---|---|---|---|
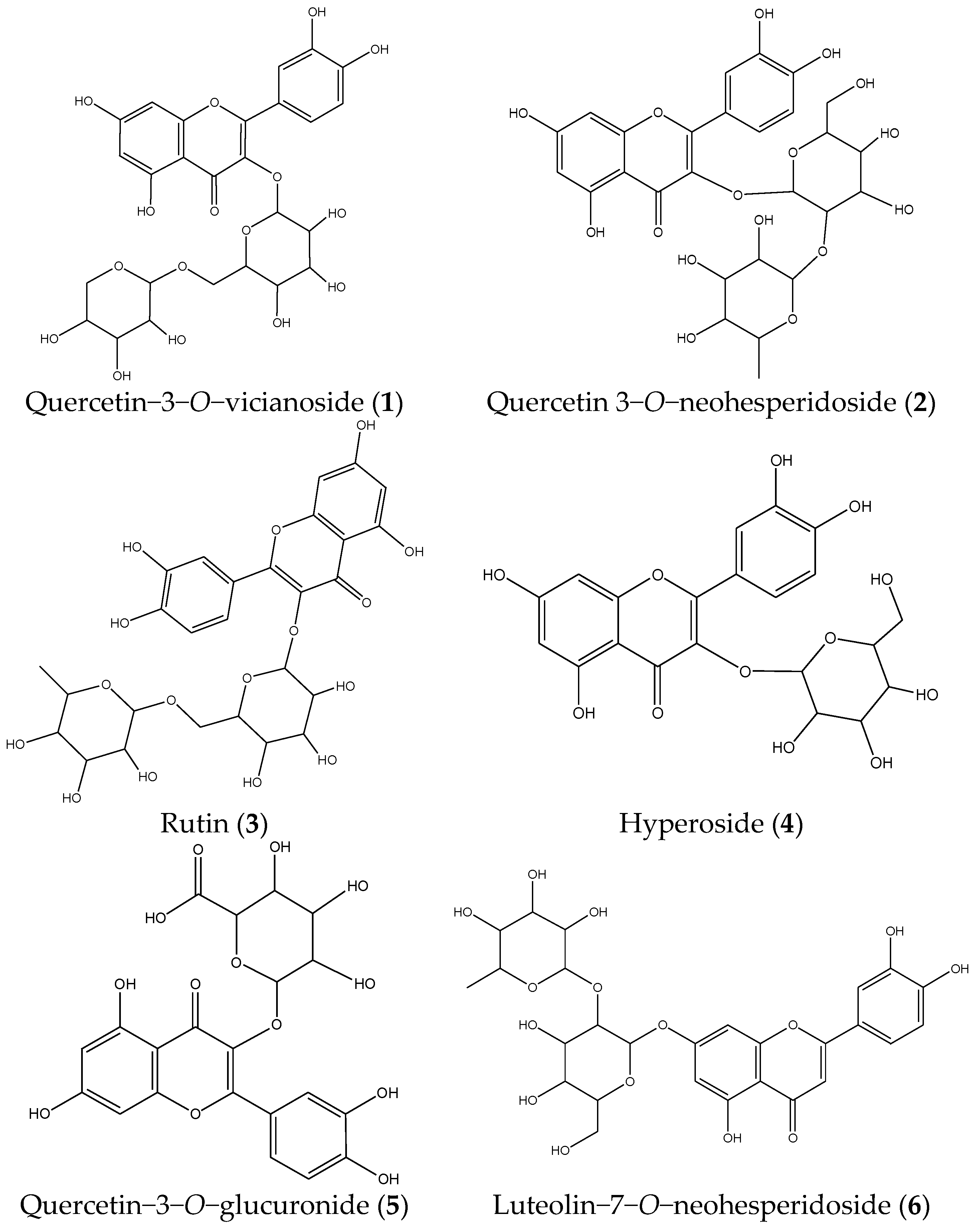
| 1 | 21.008 | 595.1353 | 300.0291, 271.0263, 255.0315, 151.0047 | C26H28O16 | Quercetin−3−O−vicianoside |
| 2 | 26.434 | 609.1494 | 300.0291, 271.0255 | C27H30O16 | Quercetin 3−O−neohesperidoside |
| 3 | 28.487 | 609.1439 | 300.0291, 271.0255 | C27H30O16 | Rutin |
| 4 | 29.233 | 463.0860 | 300.0245, 271.0221, 151.0007 | C21H20O12 | Hyperoside |
| 5 | 31.379 | 477.0654 | 384.9314, 301.0330, 255.0279, 151.0014 | C21H18O13 | Quercetin−3−O−glucuronide |
| 6 | 33.952 | 593.1520 | 284.0324, 255.0301, 227.0344 | C27H30O15 | Luteolin−7−O−neohesperidoside |
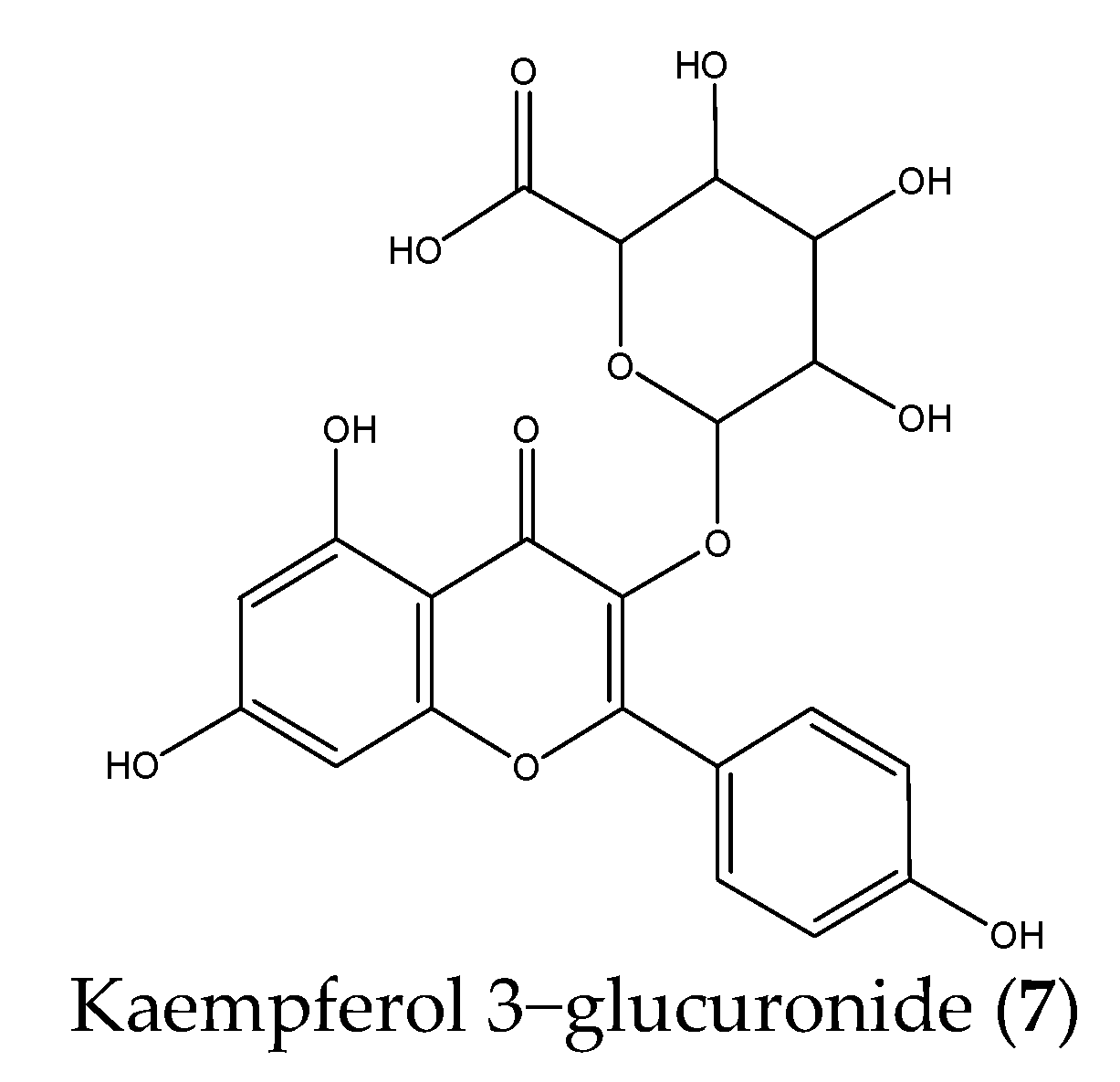
| 7 | 42.750 | 461.0755 | 285.0414, 257.0462, 229.0521 | C21H18O12 | Kaempferol 3−glucuronide |
| Peak No. | Compounds | Binding Degree (%) | Peak No. | Compounds | Binding Degree (%) |
|---|---|---|---|---|---|
| 1 | Quercetin−3−O−vicianoside | 68.70 | 5 | Quercetin−3−O−glucuronide | 96.29 |
| 2 | Quercetin 3−O−neohesperidoside | 83.15 | 6 | Luteolin−7−O−neohesperidoside | 71.35 |
| 3 | Rutin | 78.97 | 7 | Kaempferol 3−glucuronide | 91.63 |
| 4 | Hyperoside | 100.00 |
| Compounds | Binding Energy (Kcal/mol) | Amino Acid Residues | Hydrogen Bonds |
|---|---|---|---|
| Quercetin−3−O−vicianoside | −7.204 | Asp91, Lys96, Ile98, Gly116, Ala120, Gln121, Trp126 | Asp91, Lys96, Ile98, Gly116, Ala120, Gln121 |
| Quercetin 3−O−neohesperidoside | −7.032 | Lys96, Ile98, Gly116, Ala120, Gln124, Trp126 | Lys96, Ile98, Gly116, Ala120, Gln124, Trp126 |
| Rutin | −7.842 | Asp91, Gly116, Gln121, Gln124, Trp126, Cys27 | Asp91, Gly116, Gln121, Gln124, Cys127 |
| Hyperoside | −7.922 | Asp91, Ile98, Ieu117, Gln121, Gly123, Trp126, Cys127 | Asp91, Ile98, Ieu117, Gln121, Gly123, Cys127 |
| Quercetin 3−O−glucuronide | −7.718 | Asp91, Gly116, Ala120, Gly123, Cys127 | Asp91, Gly116, Ala120, Gly123, Cys127 |
| Luteolin−7−O−neohesperidoside | −7.948 | Asp91, Ala93, Lys96, Gly116, Gln124, Arg275 | Asp91, Ala93, Lys96, Gly116, Gln124, Arg275 |
| Kaempferol 3−glucuronide | −7.250 | Asp91, Gly116, Ala120, Gln124 | Asp91, Gly116, Ala120, Gln124 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; He, Z.; Shen, Q.; Fan, W.; Tan, G.; Zou, Y.; Mei, Q.; Qian, Z. Rapid Screening Alpha-Glucosidase Inhibitors from Polygoni Vivipari Rhizoma by Multi-Step Matrix Solid-Phase Dispersion, Ultrafiltration and HPLC. Molecules 2021, 26, 6111. https://doi.org/10.3390/molecules26206111
Li H, He Z, Shen Q, Fan W, Tan G, Zou Y, Mei Q, Qian Z. Rapid Screening Alpha-Glucosidase Inhibitors from Polygoni Vivipari Rhizoma by Multi-Step Matrix Solid-Phase Dispersion, Ultrafiltration and HPLC. Molecules. 2021; 26(20):6111. https://doi.org/10.3390/molecules26206111
Chicago/Turabian StyleLi, Haoxiang, Zhuobin He, Qianhui Shen, Weifeng Fan, Guoying Tan, Yuansheng Zou, Quanxi Mei, and Zhengming Qian. 2021. "Rapid Screening Alpha-Glucosidase Inhibitors from Polygoni Vivipari Rhizoma by Multi-Step Matrix Solid-Phase Dispersion, Ultrafiltration and HPLC" Molecules 26, no. 20: 6111. https://doi.org/10.3390/molecules26206111
APA StyleLi, H., He, Z., Shen, Q., Fan, W., Tan, G., Zou, Y., Mei, Q., & Qian, Z. (2021). Rapid Screening Alpha-Glucosidase Inhibitors from Polygoni Vivipari Rhizoma by Multi-Step Matrix Solid-Phase Dispersion, Ultrafiltration and HPLC. Molecules, 26(20), 6111. https://doi.org/10.3390/molecules26206111






