Green Oxidation of Amines by a Novel Cold-Adapted Monoamine Oxidase MAO P3 from Psychrophilic Fungi Pseudogymnoascus sp. P3
Abstract
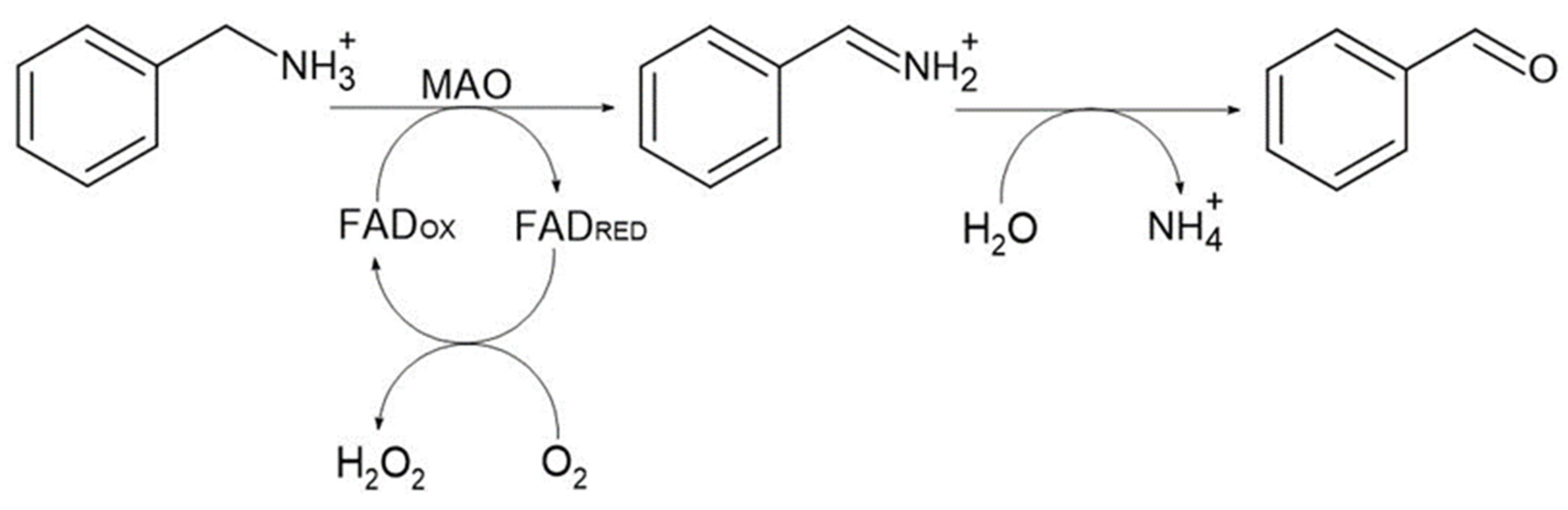
:1. Introduction
2. Results and Discussion
2.1. Identification of a Potential MAO Producer from Antarctic Fungi Strains
2.2. Characteristics of the P3 Strain
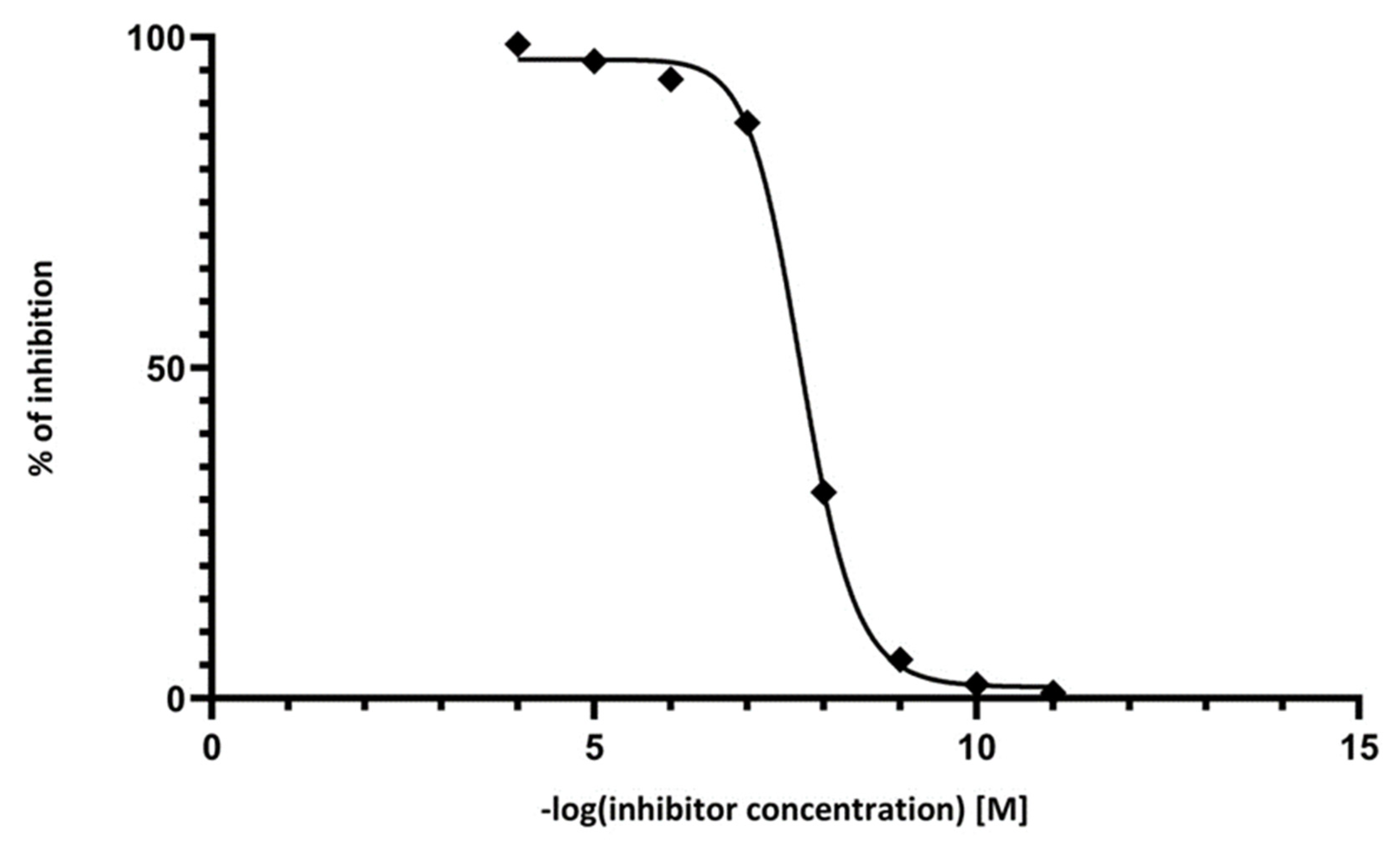
2.3. Inhibition Analysis
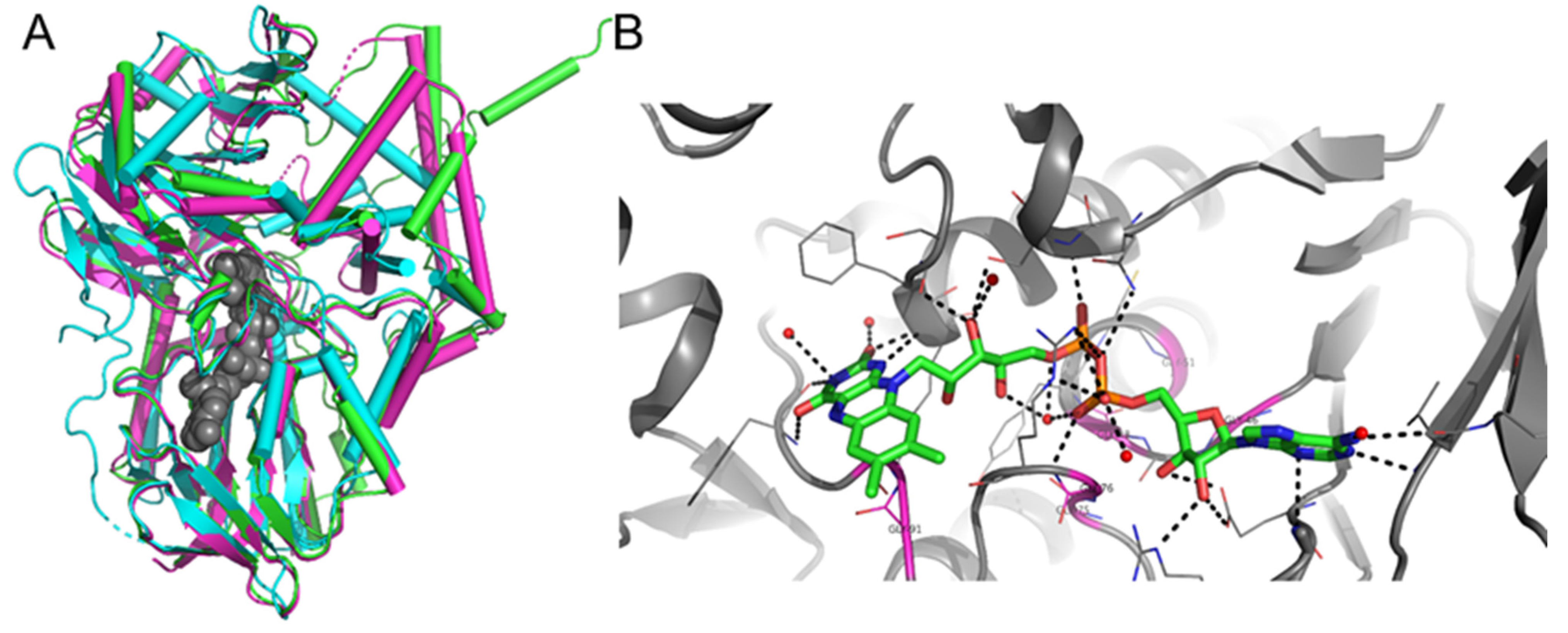
2.4. Purification of MAO P3
2.5. Temperature Profile of MAO P3
2.6. pH Profile of MAO P3
2.7. Bioconversion of Amines and MAO P3 Substrate Specificity
3. Materials and Methods
3.1. Microorganisms and Growth Conditions
3.2. Genetic Identification of Antarctic Fungal Strain P3
3.3. Protein Extraction
3.4. Assay of MAO P3 Activity
3.5. Inhibition Analysis of MAO P3
3.6. Protein Purification
3.6.1. FAD Removal
3.6.2. Affinity Chromatography
3.6.3. Reconstitution of Flavoprotein
3.7. Effect of Temperature
3.8. Effect of pH
3.9. Biotransformations of Amine Substrates by MAO P3
3.10. Analytical Method—GC-MS
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Sample Availability
References
- Zhou, B.P.; Wu, B.; Kwan, S.-W.; Abell, C.W. Characterization of a Highly Conserved FAD-binding Site in Human Monoamine Oxidase B. J. Biol. Chem. 1998, 273, 14862–14868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsugeno, Y.; Ito, A. A Key Amino Acid Responsible for Substrate Selectivity of Monoamine Oxidase A and B. J. Biol. Chem. 1997, 272, 14033–14036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, H.; Adachi, O.; Ogata, K. Amine Oxidases of Microorganisms. Agric. Biol. Chem. 1965, 29, 117–123. [Google Scholar] [CrossRef] [Green Version]
- Edmondson, D.E.; Binda, C.; Mattevi, A. Structural insights into the mechanism of amine oxidation by monoamine oxidases A and B. Arch. Biochem. Biophys. 2007, 464, 269–276. [Google Scholar] [CrossRef] [Green Version]
- Gaweska, H.; Fitzpatrick, P.F. Structures and mechanism of the monoamine oxidase family. Biomol. Concepts 2011, 2, 365–377. [Google Scholar] [CrossRef] [Green Version]
- Geha, R.M.; Rebrin, I.; Chen, K.; Shih, J.C. Substrate and Inhibitor Specificities for Human Monoamine Oxidase A and B Are Influenced by a Single Amino Acid. J. Biol. Chem. 2001, 276, 9877–9882. [Google Scholar] [CrossRef] [Green Version]
- Shih, J.C.; Chen, K.; Ridd, M.J. Monoamine Oxidase: From Genes to Behavior. Annu. Rev. Neurosci. 1999, 22, 197–217. [Google Scholar] [CrossRef] [Green Version]
- Schilling, B.; Lerch, K. Amine oxidases from Aspergillus niger: Identification of a novel flavin-dependent enzyme. Biochim. Biophys. Acta Gen. Subj. 1995, 1243, 529–537. [Google Scholar] [CrossRef]
- Schilling, B.; Lerch, K. Cloning, sequencing and heterologous expression of the monoamine oxidase gene from Aspergillus niger. Mol. Genet. Genom. 1995, 247, 430–438. [Google Scholar] [CrossRef]
- Atkin, K.E.; Reiss, R.; Koehler, V.; Bailey, K.R.; Hart, S.; Turkenburg, J.P.; Turner, N.J.; Brzozowski, A.M.; Grogan, G. The Structure of Monoamine Oxidase from Aspergillus niger Provides a Molecular Context for Improvements in Activity Obtained by Directed Evolution. J. Mol. Biol. 2008, 384, 1218–1231. [Google Scholar] [CrossRef]
- Edmondson, D. The FAD Binding Sites of Human Monoamine Oxidases A and B. NeuroToxicology 2004, 25, 63–72. [Google Scholar] [CrossRef]
- Ghislieri, D.; Green, A.P.; Pontini, M.; Willies, S.C.; Rowles, I.; Frank, A.; Grogan, G.; Turner, N.J. Engineering an Enantioselective Amine Oxidase for the Synthesis of Pharmaceutical Building Blocks and Alkaloid Natural Products. J. Am. Chem. Soc. 2013, 135, 10863–10869. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, E.; Iglesias, C.; Ghislieri, D.; Hopwood, J.; Galman, J.L.; Lloyd, R.C.; Turner, N.J. A Regio- and Stereoselective ω-Transaminase/Monoamine Oxidase Cascade for the Synthesis of Chiral 2,5-Disubstituted Pyrrolidines. Angew. Chem. Int. Ed. 2014, 53, 2447–2450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carr, R.; Alexeeva, M.; Dawson, M.; Gotor-Fernández, V.; Humphrey, C.E.; Turner, N.J. Directed Evolution of an Amine Oxidase for the Preparative Deracemisation of Cyclic Secondary Amines. ChemBioChem 2005, 6, 637–639. [Google Scholar] [CrossRef]
- Toscani, A.; Risi, C.; Black, G.W.; Brown, N.L.; Shaaban, A.; Turner, N.J.; Castagnolo, D. Monoamine Oxidase (MAO-N) Whole Cell Biocatalyzed Aromatization of 1,2,5,6-Tetrahydropyridines into Pyridines. ACS Catal. 2018, 8, 8781–8787. [Google Scholar] [CrossRef]
- Siddiqui, K.S. Some like it hot, some like it cold: Temperature dependent biotechnological applications and improvements in extremophilic enzymes. Biotechnol. Adv. 2015, 33, 1912–1922. [Google Scholar] [CrossRef]
- D’Amico, S.; Collins, T.; Marx, J.; Feller, G.; Gerday, C. Psychrophilic microorganisms: Challenges for life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef]
- Alexeeva, M.; Enright, A.; Dawson, M.J.; Mahmoudian, M.; Turner, N.J. Deracemization of α-Methylbenzylamine Using an Enzyme Obtained by In Vitro Evolution†. Angew. Chem. Int. Ed. 2002, 41, 3177–3180. [Google Scholar] [CrossRef]
- Leushkin, E.V.; Logacheva, M.D.; Penin, A.A.; Sutormin, R.A.; Gerasimov, E.S.; Kochkina, G.A.; Ivanushkina, N.E.; Vasilenko, O.V.; Kondrashov, A.S.; Ozerskaya, S.M. Comparative genome analysis of Pseudogymnoascus spp. reveals primarily clonal evolution with small genome fragments exchanged between lineages. BMC Genom. 2015, 16, 400. [Google Scholar] [CrossRef] [Green Version]
- Hefti, M.H.; Vervoort, J.; Van Berkel, W.J.H. Deflavination and reconstitution of flavoproteins. JBIC J. Biol. Inorg. Chem. 2003, 270, 4227–4242. [Google Scholar] [CrossRef]
- Zlateva, T.; Boteva, R.; Filippi, B.; Veenhuis, M.; van der Klei, I.J. Deflavination of flavo-oxidases by nucleophilic reagents. Biochim. Biophys. Acta Protein Struct. Mol. Enzym. 2001, 1548, 213–219. [Google Scholar] [CrossRef] [Green Version]
- Osborne, T.H.; Heath, M.D.; Martin, A.C.R.; Pankowski, J.A.; Hudson-Edwards, K.A.; Santini, J.M. Cold-adapted arsenite oxidase from a psychrotolerant Polaromonas species. Metallomics 2013, 5, 318–324. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, S.; Furukawara, M.; Omae, K.; Tadokoro, N.; Saito, Y.; Abe, K.; Kera, Y. A Highly Stable d-Amino Acid Oxidase of the Thermophilic Bacterium Rubrobacter xylanophilus. Appl. Environ. Microbiol. 2014, 80, 7219–7229. [Google Scholar] [CrossRef] [Green Version]
- Dym, O.; Eisenberg, D. Sequence-structure analysis of FAD-containing proteins. Protein Sci. 2001, 10, 1712–1728. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Jiang, X.; Liu, W.; Wang, Y.; Huang, H.; Bai, Y.; Su, X.; Yao, B.; Luo, H. Characterization, stability improvement, and bread baking applications of a novel cold-adapted glucose oxidase from Cladosporium neopsychrotolerans SL16. Food Chem. 2020, 310, 125970. [Google Scholar] [CrossRef] [PubMed]
- Yuivar, Y.; Barahona, S.; Alcaíno, J.; Cifuentes, V.; Baeza, M. Biochemical and Thermodynamical Characterization of Glucose Oxidase, Invertase, and Alkaline Phosphatase Secreted by Antarctic Yeasts. Front. Mol. Biosci. 2017, 4, 86. [Google Scholar] [CrossRef] [PubMed]
- Rios-Solis, L.; Mothia, B.; Yi, S.; Zhou, Y.; Micheletti, M.; Lye, G. High throughput screening of monoamine oxidase (MAO-N-D5) substrate selectivity and rapid kinetic model generation. J. Mol. Catal. B Enzym. 2015, 120, 100–110. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, S.; Osugi, K.; Shimekake, Y.; Shinbo, A.; Abe, K.; Kera, Y. Characterization and improvement of substrate-binding affinity of D-aspartate oxidase of the thermophilic fungus Thermomyces dupontii. Appl. Microbiol. Biotechnol. 2019, 103, 4053–4064. [Google Scholar] [CrossRef] [PubMed]
- Shimekake, Y.; Furuichi, T.; Abe, K.; Kera, Y.; Takahashi, S. A novel thermostable d-amino acid oxidase of the thermophilic fungus Rasamsonia emersonii strain YA. Sci. Rep. 2019, 9, 11948. [Google Scholar] [CrossRef] [PubMed]
- Binda, C.; Mattevi, A.; Edmondson, D.E. Structure-Function Relationships in Flavoenzyme-dependent Amine Oxidations. J. Biol. Chem. 2002, 277, 23973–23976. [Google Scholar] [CrossRef] [Green Version]
- Köhler, V.; Bailey, K.R.; Znabet, A.; Raftery, J.; Helliwell, M.; Turner, N.J. Enantioselective Biocatalytic Oxidative Desymmetrization of Substituted Pyrrolidines. Angew. Chem. Int. Ed. 2010, 49, 2182–2184. [Google Scholar] [CrossRef]
- Dunsmore, C.J.; Carr, R.; Fleming, T.; Turner, N. A Chemo-Enzymatic Route to Enantiomerically Pure Cyclic Tertiary Amines. J. Am. Chem. Soc. 2006, 128, 2224–2225. [Google Scholar] [CrossRef]
- Li, T.; Liang, J.; Ambrogelly, A.; Brennan, T.; Gloor, G.; Huisman, G.; LaLonde, J.; Lekhal, A.; Mijts, B.; Muley, S.; et al. Efficient, Chemoenzymatic Process for Manufacture of the Boceprevir Bicyclic [3.1.0]Proline Intermediate Based on Amine Oxidase-Catalyzed Desymmetrization. J. Am. Chem. Soc. 2012, 134, 6467–6472. [Google Scholar] [CrossRef]
- Znabet, A.; Polak, M.M.; Janssen, E.; de Kanter, F.J.J.; Turner, N.J.; Orru, R.V.A.; Ruijter, E. A highly efficient synthesis of telaprevir by strategic use of biocatalysis and multicomponent reactions. Chem. Commun. 2010, 46, 7918–7920. [Google Scholar] [CrossRef]
- O’Reilly, E.; Turner, N.J. Enzymatic cascades for the regio- and stereoselective synthesis of chiral amines. Perspect. Sci. 2015, 4, 55–61. [Google Scholar] [CrossRef] [Green Version]
- Pelckmans, M.; Renders, T.; Van de Vyver, S.; Sels, B.F. Bio-based amines through sustainable heterogeneous catalysis. Green Chem. 2017, 19, 5303–5331. [Google Scholar] [CrossRef]
- Trytek, M.; Jędrzejewski, K.; Fiedurek, J. Bioconversion of α-pinene by a novel cold-adapted fungus Chrysosporium pannorum. J. Ind. Microbiol. Biotechnol. 2015, 42, 181–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, T.; Hallagan, J.; Putnam, J.; Gierke, T.; Doull, J.; Munro, I.; Newberne, P.; Portoghese, P.; Smith, R.; Wagner, B.; et al. The FEMA GRAS assessment of alicyclic substances used as flavour ingredients. Food Chem. Toxicol. 1996, 34, 763–828. [Google Scholar] [CrossRef]
- Store, M.R. Cyclopentanone Market Set for Explosive Growth, to Reach around USD 130.0 Globally Million by 2020, Growing at 4.5% CAGR-Market Research Store. Available online: https://www.globenewswire.com/news-release/2016/01/25/804190/0/en/Cyclopentanone-Market-Set-for-Explosive-Growth-To-Reach-Around-USD-130-0-Globally-Million-by-2020-Growing-at-4-5-CAGR-Market-Research-Store.html (accessed on 13 August 2021).
- Marx, J.-C.; Collins, T.; D’Amico, S.; Feller, G.; Gerday, C. Cold-Adapted Enzymes from Marine Antarctic Microorganisms. Mar. Biotechnol. 2006, 9, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]



| Concentration of n-Butylamine [%] | Relative Activity of MAO P3 [%] |
|---|---|
| 0.01 | 6.4 ± 0.31 |
| 0.03 | 19.9 ± 0.31 |
| 0.05 | 30.7 ± 0.20 |
| 0.10 | 100.0 |
| 0.15 | 45.9 ± 0.34 |
| 0.20 | 9.6 ± 0.20 |
| 0.50 | 0.0 |
| Substrate Number | Name | Structure | Rate of Oxidation [%] |
|---|---|---|---|
| 1 | sec-butylamine | 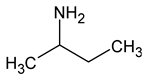 | 29 |
| 2 | cyclopentylamine |  | 31 |
| 3 | pyrrolidine | 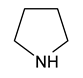 | 36 |
| 4 | 1-methylpyrrolidine | 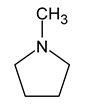 | 0 |
| 5 | α-methylbenzylamine |  | 41 |
| 6 | hexamethyleneimine | 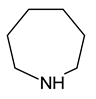 | 30 |
| 7 | 2-azabicyclo[2.2.1]hept-5-en-3-one | 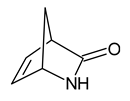 | 0 |
| 8 | pyridine | 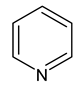 | 0 |
| 9 | 6,6-dimethyl-3-azabicyclo[3.1.0]hexane | 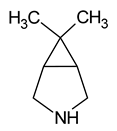 | 100 |
| 10 | 3-azabicyclo[3.3.0]octane | 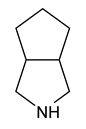 | 67 |
| 11 | cis-8-azabicyclo[4.3.0]nonane | 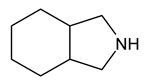 | 0 |
| 12 | indoline | 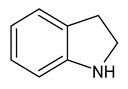 | 9 |
| 13 | 1-(2-aminoethyl)pyrrolidine | 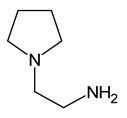 | 0 |
| 14 | benzylhydrylamine | 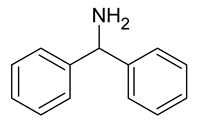 | 0 |
| 15 | putrescine | 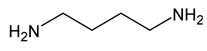 | 0 |
| 16 | spermidine | 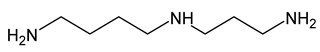 | 0 |
| Substrate Number | Structure | Vmax [mM h−1] | Km [mM] |
|---|---|---|---|
| 9 |  | 1.1 | 2.40 |
| 6 |  | 0.72 | 2.56 |
| 5 |  | 0.27 | 4.13 |
| 2 |  | 0.47 | 8.22 |
| 10 |  | 0.24 | 10.4 |
| 3 |  | 0.16 | 14.3 |
| 1 |  | 0.17 | 18.0 |
| 12 |  | 0.14 | 20.4 |
| Name | Sequence (5′→3′) |
|---|---|
| RLR3R | GGTCCGTGTTTCAAGAC |
| V9 | TGCGTTGATTACGTCCCTGC |
| ITS5 | GGAAGTAAAGTCGTAACAAGG |
| LR6 | CGCCAGTTCTGCTTACC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jodłowska, I.; Twarda-Clapa, A.; Szymczak, K.; Białkowska, A.M. Green Oxidation of Amines by a Novel Cold-Adapted Monoamine Oxidase MAO P3 from Psychrophilic Fungi Pseudogymnoascus sp. P3. Molecules 2021, 26, 6237. https://doi.org/10.3390/molecules26206237
Jodłowska I, Twarda-Clapa A, Szymczak K, Białkowska AM. Green Oxidation of Amines by a Novel Cold-Adapted Monoamine Oxidase MAO P3 from Psychrophilic Fungi Pseudogymnoascus sp. P3. Molecules. 2021; 26(20):6237. https://doi.org/10.3390/molecules26206237
Chicago/Turabian StyleJodłowska, Iga, Aleksandra Twarda-Clapa, Kamil Szymczak, and Aneta M. Białkowska. 2021. "Green Oxidation of Amines by a Novel Cold-Adapted Monoamine Oxidase MAO P3 from Psychrophilic Fungi Pseudogymnoascus sp. P3" Molecules 26, no. 20: 6237. https://doi.org/10.3390/molecules26206237
APA StyleJodłowska, I., Twarda-Clapa, A., Szymczak, K., & Białkowska, A. M. (2021). Green Oxidation of Amines by a Novel Cold-Adapted Monoamine Oxidase MAO P3 from Psychrophilic Fungi Pseudogymnoascus sp. P3. Molecules, 26(20), 6237. https://doi.org/10.3390/molecules26206237






