Characterization of Conjugates between α-Lactalbumin and Benzyl Isothiocyanate—Effects on Molecular Structure and Proteolytic Stability
Abstract
1. Introduction
2. Results
2.1. Determination of Free Amino Groups Using O-Phthaldialdehyde
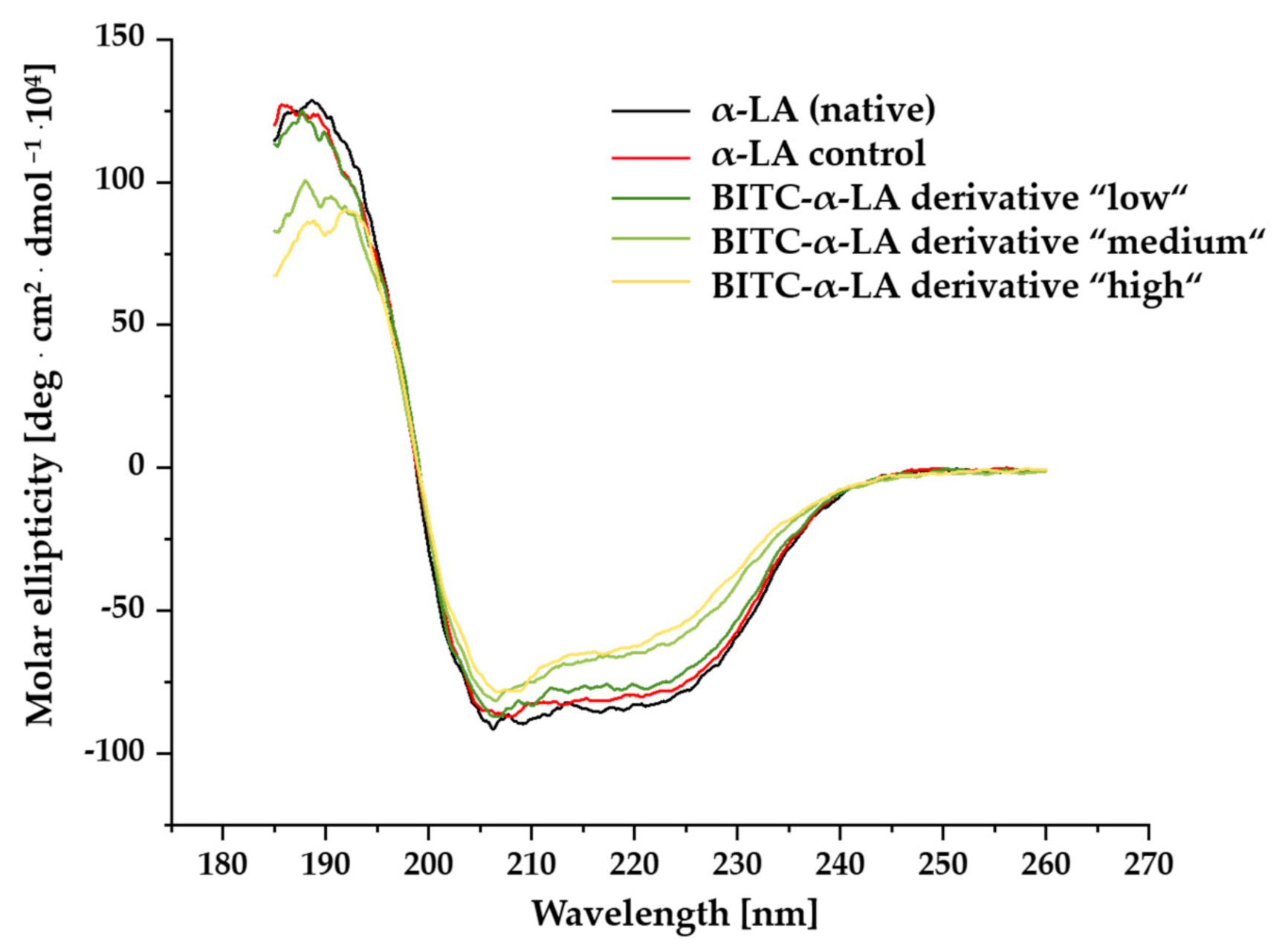
2.2. Investigation of Secondary Structure by Far-UV Circular Dichroism Spectroscopy
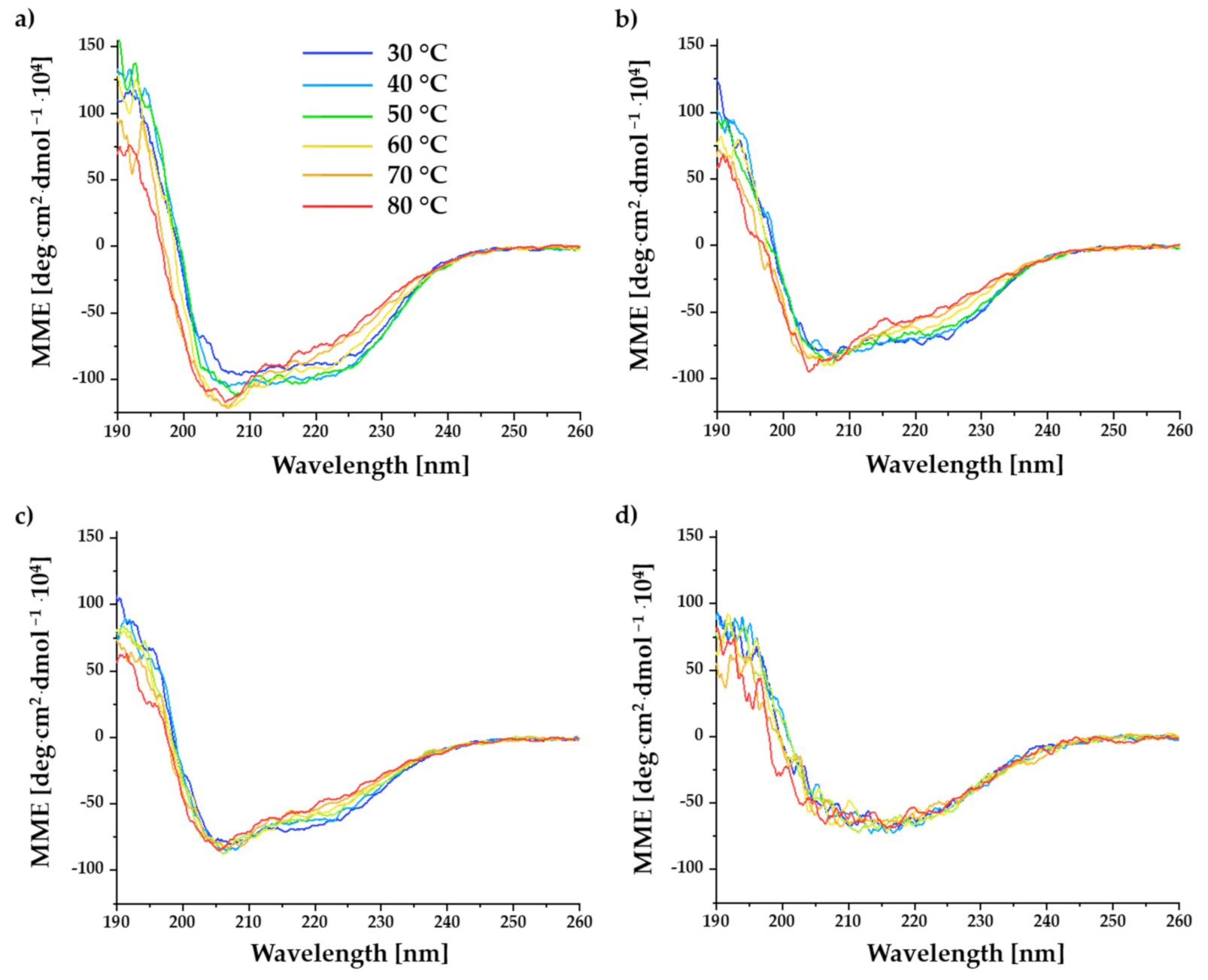
2.3. Investigation of Temperature-Induced Conformational Changes Using Far-UV CD Spectroscopy
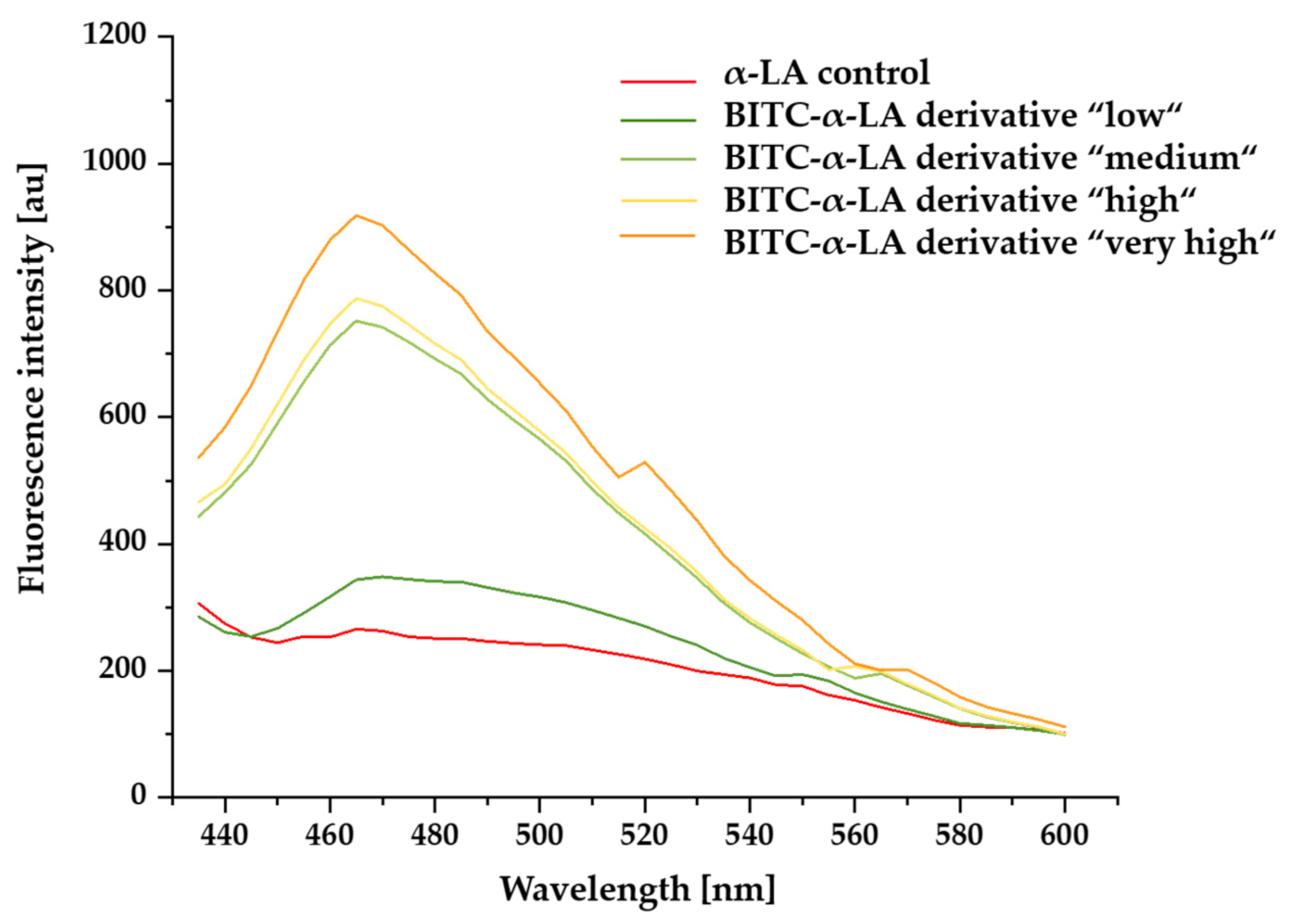
2.4. Investigation of Surface Hydrophobicity Using Anilino-1-Naphthalenesulfonic Acid (ANS) Fluorescence
2.5. Determination of the Hydrodynamic Radius Using Dynamic Light Scattering
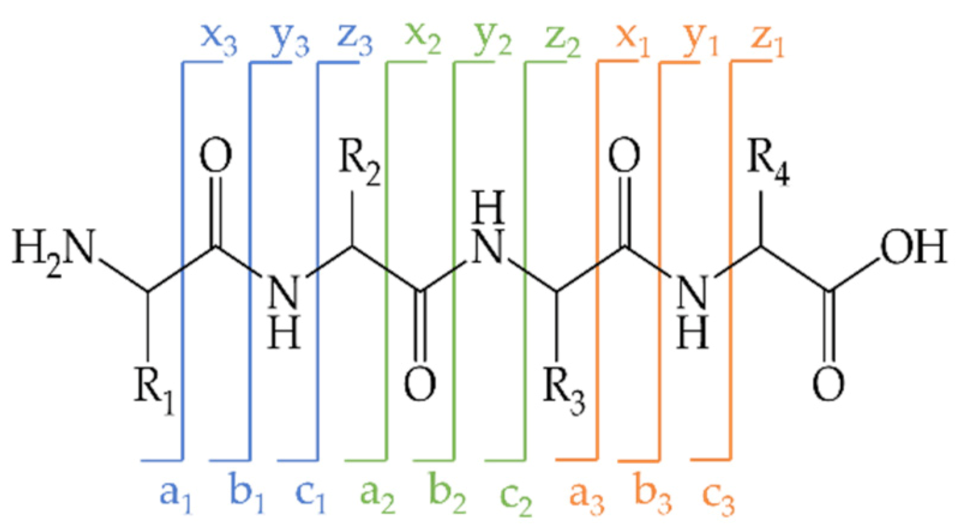
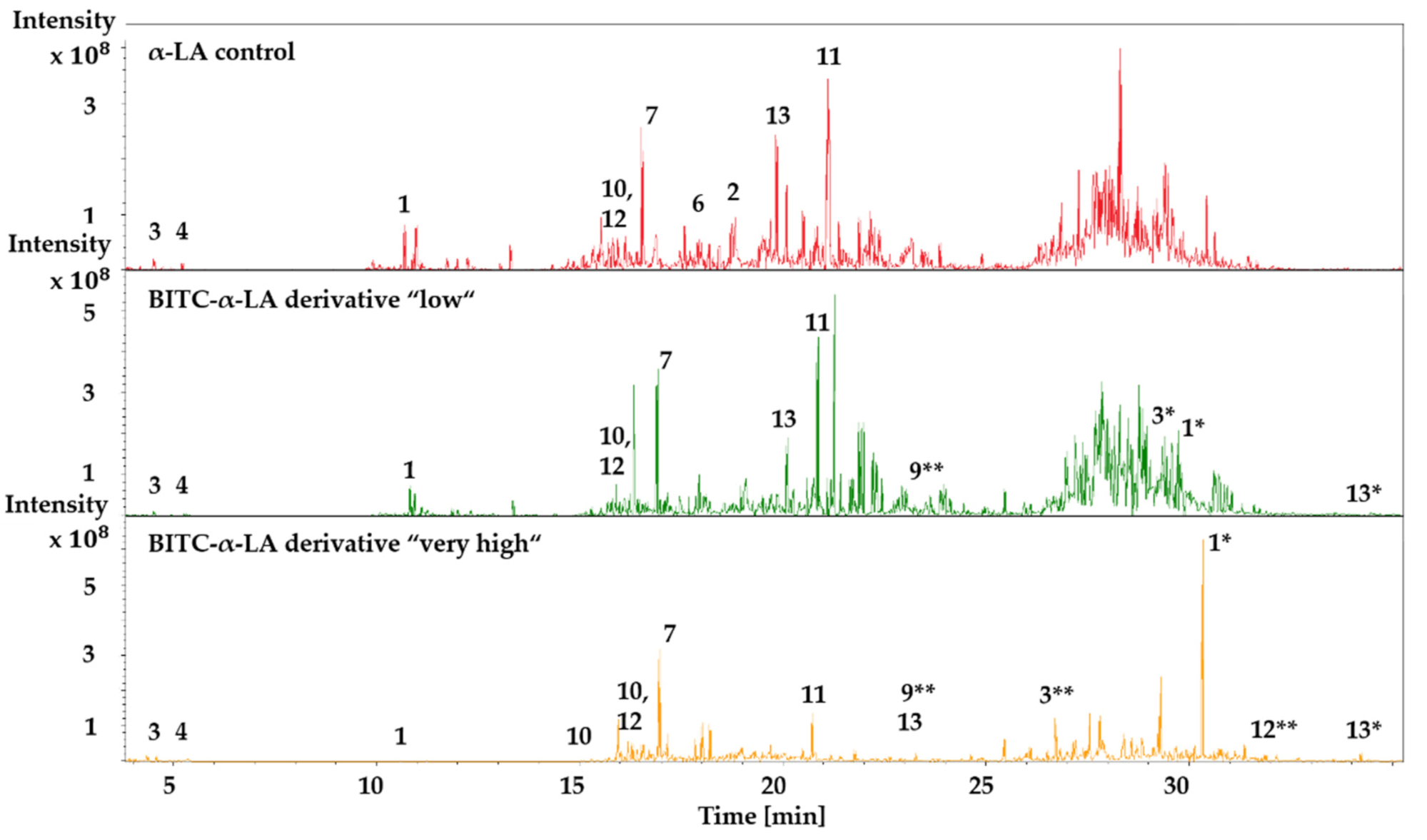
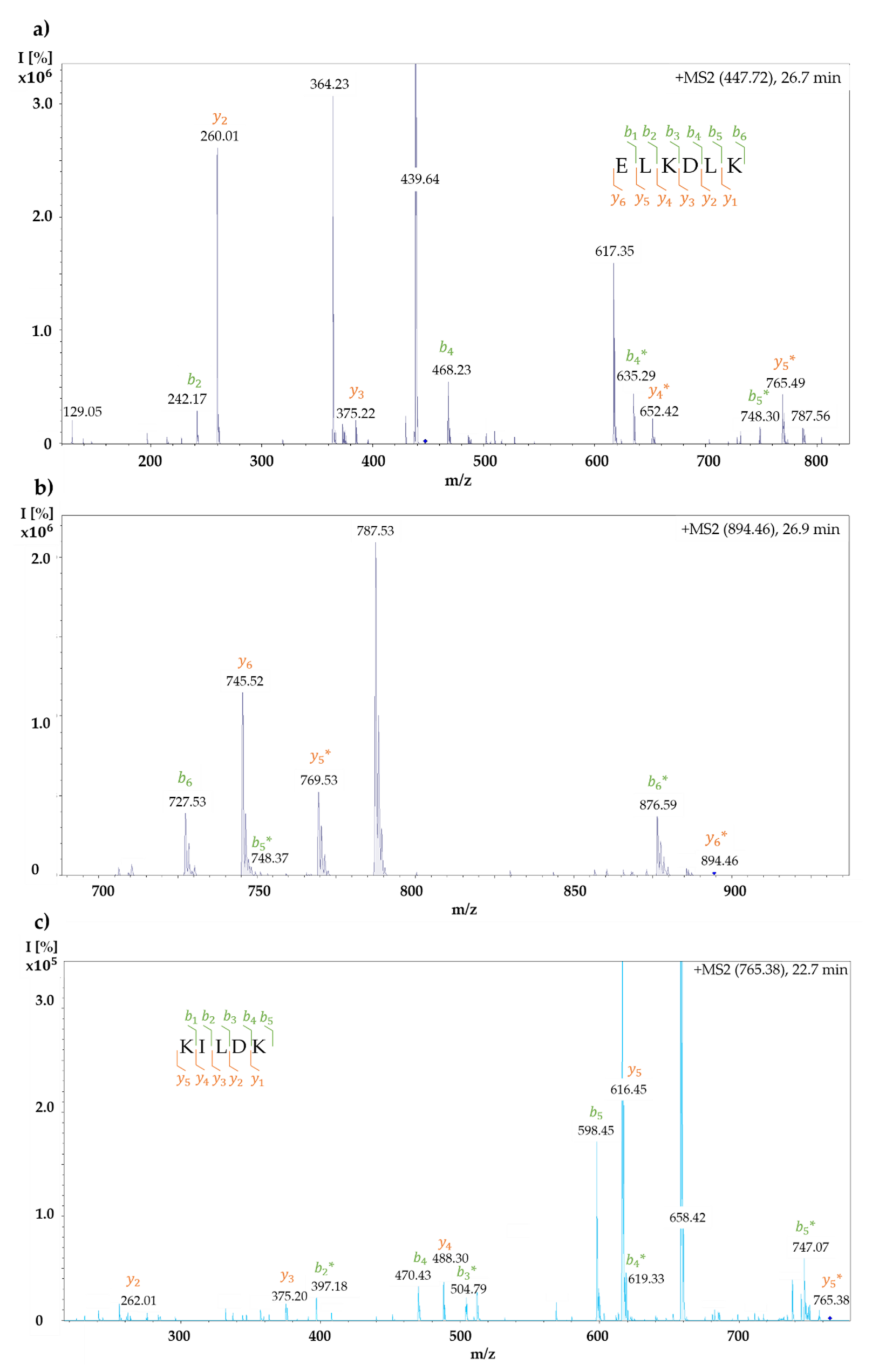
2.6. LC-ESI-MS/MS Analysis of Unmodified and BITC-Modified α-LA Hydrolysates
Identification of BITC-Modified Amino Acids
3. Discussion
3.1. Determination of Free Amino Groups Using O-Phthaldialdehyde
3.2. Influence of BITC Conjugation on Secondary Structure of α-LA
3.3. Influence of BITC Conjugation on Temperature-Induced Conformational Changes of α-LA
3.4. Influence of BITC Conjugation on Surface Hydrophobicity of α-LA
3.5. Influence of BITC Conjugation on the Hydrodynamic Radius of α-LA
3.6. Influence of BITC Conjugation on Tryptic Digestion of α-LA
4. Materials and Methods
4.1. Materials
4.2. Methods
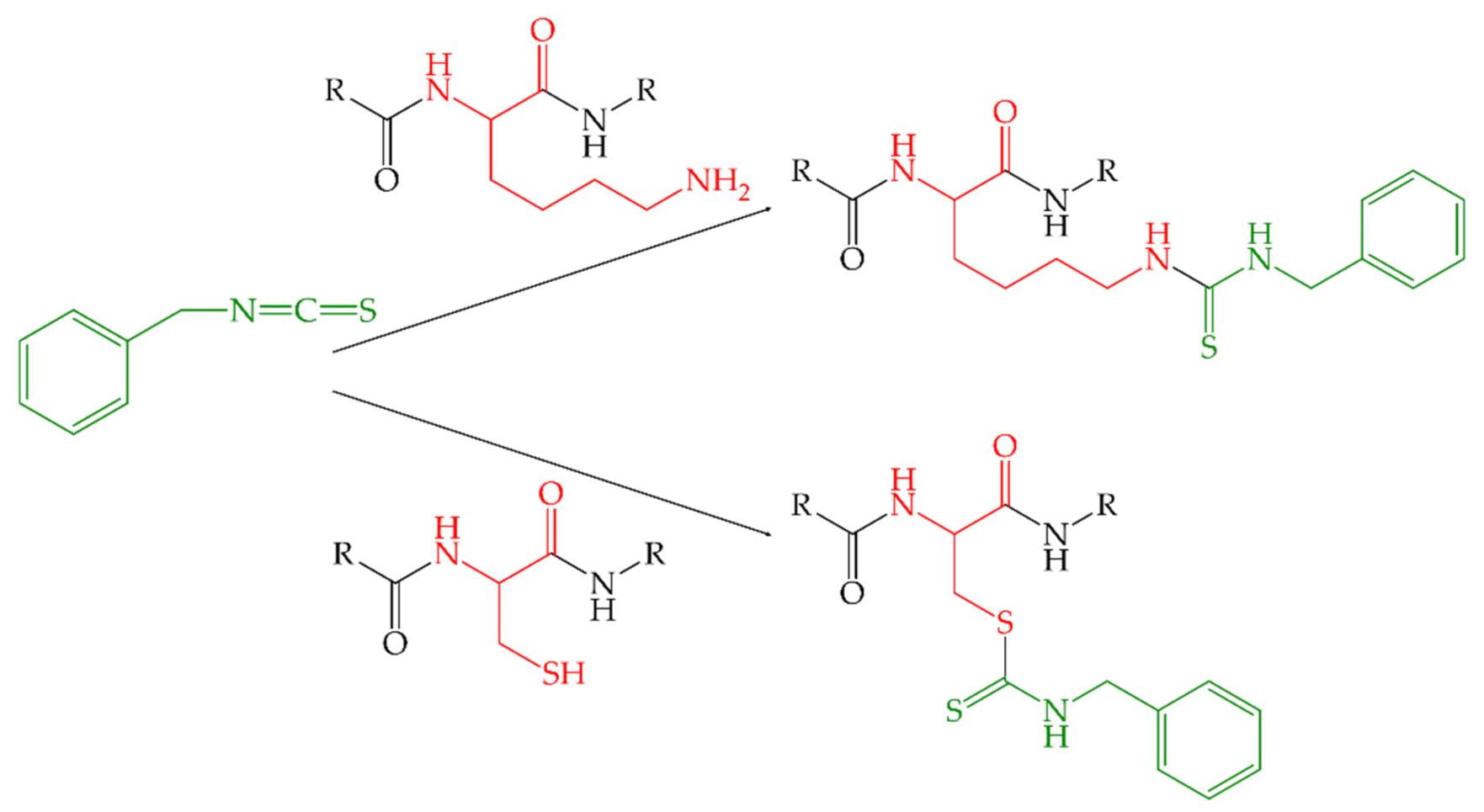
4.2.1. Preparation of ITC-α-LA Conjugates
4.2.2. Determination of Free Amino Groups Using O-Phthaldialdehyde
4.2.3. Investigation of Secondary Structure by Far-UV CD Spectroscopy
4.2.4. Investigation of Temperature-Induced Conformational Changes Using Far UV CD Spectroscopy
4.2.5. Investigation of Surface Hydrophobicity Using ANS Fluorescence
4.2.6. Determination of the Hydrodynamic Radius Using Dynamic Light Scattering
4.2.7. Tryptic Hydrolysis of Unmodified and BITC-Modified α-LA
4.2.8. LC-ESI-MS/MS of Unmodified and BITC-Modified α-LA
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Rade-Kukic, K.; Schmitt, C.; Rawel, H.M. Formation of conjugates between β-lactoglobulin and allyl isothiocyanate: Effect on protein heat aggregation, foaming and emulsifying properties. Food Hydrocoll. 2011, 25, 694–706. [Google Scholar] [CrossRef]
- Sønderby, I.E.; Geu-Flores, F.; Halkier, B.A. Biosynthesis of glucosinolates—Gene discovery and beyond. Trends Plant Sci. 2010, 15, 283–290. [Google Scholar] [CrossRef]
- Fernando, R.C.; Weiß-Schmidt, P. Sekundäre Pflanzenstoffe: Bioaktive substanzen aus obst und gemüse in der krebsprävention. EHK 2007, 56, 192–197. [Google Scholar] [CrossRef]
- Herr, I.; Büchler, M. Glucosinolate der kreuzblütlerfamilie in prävention und therapie maligner tumore. Dtsch. Z. Onkol. 2009, 41, 109–114. [Google Scholar] [CrossRef]
- Haller, D.; Grune, T.; Rimbach, G. Biofunktionalität der Lebensmittelinhaltsstoffe; Springer Spektrum: Berlin/Heidelberg, Germany, 2013; ISBN 9783642293733. [Google Scholar]
- Kühn, C.; von Oesen, T.; Hanschen, F.S.; Rohn, S. Determination of isothiocyanate-protein conjugates in milk and curd after adding garden cress (Lepidium sativum L.). Food Res. Int. 2018, 108, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Keppler, J.K.; Koudelka, T.; Palani, K.; Tholey, A.; Schwarz, K. Interaction of β-lactoglobulin with small hydrophobic ligands—Influence of covalent AITC modification on β-LG tryptic cleavage. Food Biophys. 2014, 9, 349–358. [Google Scholar] [CrossRef]
- Juge, N.; Mithen, R.F.; Traka, M. Molecular basis for chemoprevention by sulforaphane: A comprehensive review. Cell. Mol. Life Sci. 2007, 64, 1105–1127. [Google Scholar] [CrossRef] [PubMed]
- Keppler, J.K.; Martin, D.; Garamus, V.M.; Berton-Carabin, C.; Nipoti, E.; Coenye, T.; Schwarz, K. Functionality of whey proteins covalently modified by allyl isothiocyanate. Part 1 physicochemical and antibacterial properties of native and modified whey proteins at pH 2 to 7. Food Hydrocoll. 2017, 65, 130–143. [Google Scholar] [CrossRef]
- Kühn, C.; Von Oesen, T.; Herz, C.; Schreiner, M.; Hanschen, F.S.; Lamy, E.; Rohn, S. In vitro determination of protein conjugates in human cells by LC-ESI-MS/MS after benzyl isothiocyanate exposure. J. Agric. Food Chem. 2018, 66, 6727–6733. [Google Scholar] [CrossRef] [PubMed]
- Beevi, S.S.; Mangamoori, L.N.; Dhand, V.; Ramakrishna, D.S. Isothiocyanate profile and selective antibacterial activity of root, stem, and leaf extracts derived from Raphanus sativus L. Foodborne Pathog. Dis. 2009, 6, 129–136. [Google Scholar] [CrossRef]
- Lee, Y.M.; Seon, M.R.; Cho, H.J.; Kim, J.S.; Park, J.H.Y. Benzyl isothiocyanate exhibits anti-inflammatory effects in murine macrophages and in mouse skin. J. Mol. Med. 2009, 87, 1251–1261. [Google Scholar] [CrossRef] [PubMed]
- Sofrata, A.; Santangelo, E.M.; Azeem, M.; Borg-Karlson, A.K.; Gustafsson, A.; Pütsep, K. Benzyl isothiocyanate, a major component from the roots of Salvadora persica is highly active against gram-negative bacteria. PLoS ONE 2011, 6, e23045. [Google Scholar] [CrossRef] [PubMed]
- Guzmán-Pérez, V.; Bumke-Vogt, C.; Schreiner, M.; Mewis, I.; Borchert, A.; Pfeiffer, A.F.H. Benzylglucosinolate derived isothiocyanate from Tropaeolum majus reduces gluconeogenic gene and protein expression in human cells. PLoS ONE 2016, 11, e162397. [Google Scholar] [CrossRef] [PubMed]
- Kreis, W. Sekundäre Pflanzenstoffe und Krebs. Dtsch. Z. Onkol. 2009, 41, 100–108. [Google Scholar] [CrossRef]
- Zhang, Y. Allyl isothiocyanate as a cancer chemopreventive phytochemical. Mol. Nutr. Food Res. 2010, 54, 127–135. [Google Scholar] [CrossRef]
- Hanschen, F.S.; Lamy, E.; Schreiner, M.; Rohn, S. Reactivity and stability of glucosinolates and their breakdown products in foods. Angew. Chemie-Int. Ed. 2014, 53, 11430–11450. [Google Scholar] [CrossRef]
- Cejpek, K.; Valusek, J.; Velisek, J. Reactions of allyl isothiocyanate with alanine, glycine, and several peptides in model systems. J. Agric. Food Chem. 2000, 48, 3560–3565. [Google Scholar] [CrossRef]
- Hanschen, F.S.; Brüggemann, N.; Brodehl, A.; Mewis, I.; Schreiner, M.; Rohn, S.; Kroh, L.W. Characterization of products from the reaction of glucosinolate-derived isothiocyanates with cysteine and lysine derivatives formed in either model systems or broccoli sprouts. J. Agric. Food Chem. 2012, 60, 7735–7745. [Google Scholar] [CrossRef]
- Zhang, Y.; Talalay, P. Anticarcinogenic activities of organic isothiocyanates: Chemistry and mechanisms. Cancer Res. 1994, 54, 19761981. [Google Scholar]
- Keppler, J.K.; Martin, D.; Garamus, V.M.; Schwarz, K. Differences in binding behavior of (-)-epigallocatechin gallate to β-lactoglobulin heterodimers (AB) compared to homodimers (A) and (B). J. Mol. Recognit. 2015, 28, 656–666. [Google Scholar] [CrossRef]
- Keppler, J.K.; Steffen-Heins, A.; Berton-Carabin, C.C.; Ropers, M.H.; Schwarz, K. Functionality of whey proteins covalently modified by allyl isothiocyanate. Part 2: Influence of the protein modification on the surface activity in an O/W system. Food Hydrocoll. 2018, 81, 286–299. [Google Scholar] [CrossRef]
- Bock, A.; Steinhäuser, U.; Drusch, S. Partitioning behavior and interfacial activity of phenolic acid derivatives and their impact on β-lactoglobulin at the oil-water interface. Food Biophys. 2021, 16, 191–202. [Google Scholar] [CrossRef]
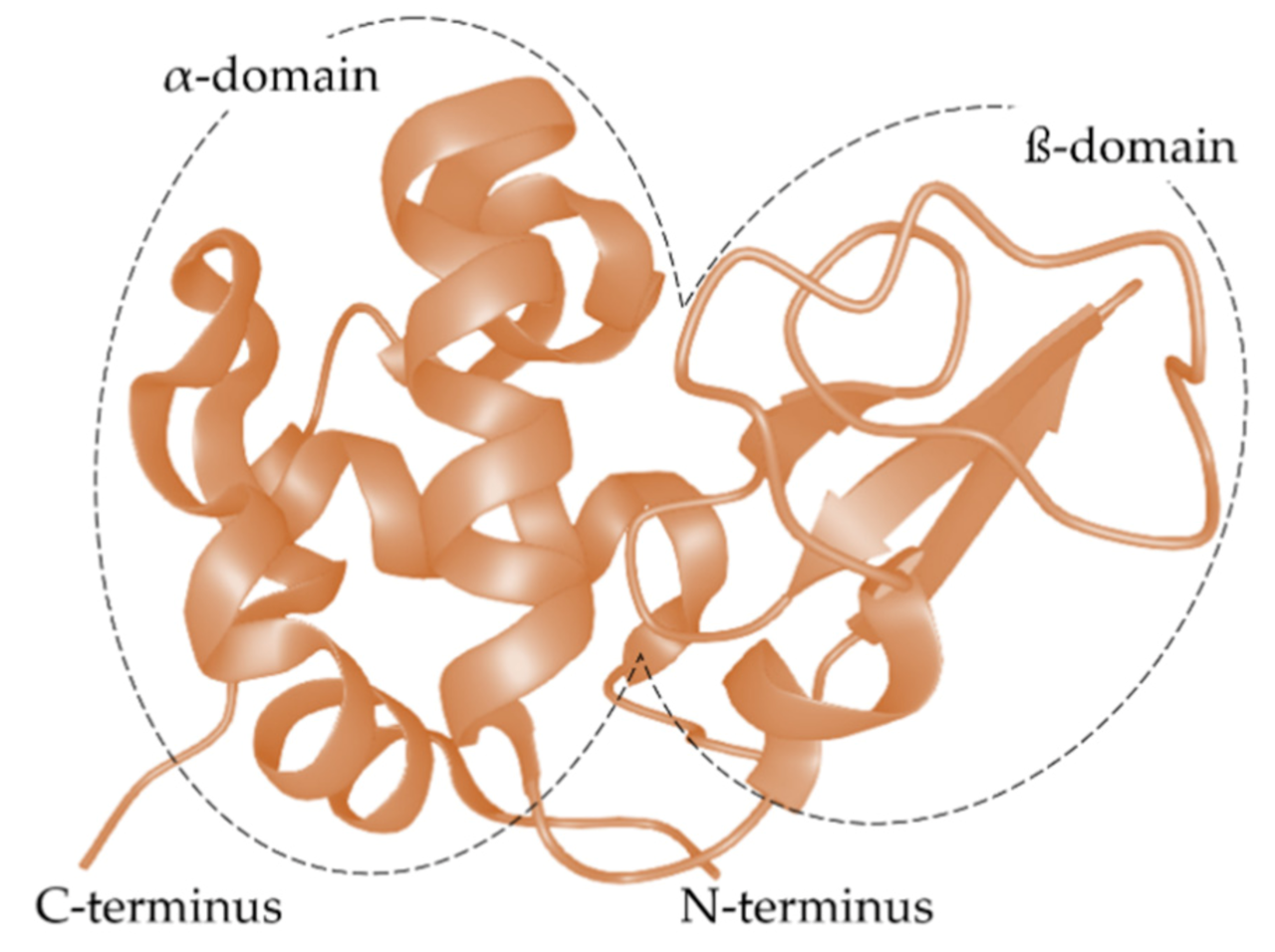
- Stanciuc, N.; Râpeanu, G. An overview of bovine a-lactalbumin structure and functionality. Ann. Univ. Dunarea Jos Galati-Fascicle VI–Food Technol. 2010, 34, 82–93. [Google Scholar]
- Smithers, G.W. Whey and whey proteins-From “gutter-to-gold”. Int. Dairy J. 2008, 18, 695–704. [Google Scholar] [CrossRef]
- Smithers, G.W.; Ballard, F.J.; Copeland, A.D.; De Silva, K.J.; Dionysius, D.A.; Francis, G.L.; Goddard, C.; Grieve, P.A.; Mcintosh, G.H.; Mitchell, I.R.; et al. New Opportunities from the isolation and utilization of whey proteins. J. Dairy Sci. 1996, 79, 1454–1459. [Google Scholar] [CrossRef]
- Kinsella, J.E. Milk proteins: Physicochemical and functional properties. CRC Crit. Rev. Food Sci. Nutr. 1984, 21, 197–262. [Google Scholar] [CrossRef]
- Lam, R.S.H.; Nickerson, M.T. The effect of pH and temperature pre-treatments on the structure, surface characteristics and emulsifying properties of alpha-lactalbumin. Food Chem. 2015, 173, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Farrell, H.M.; Jimenez-Flores, R.; Bleck, G.T.; Brown, E.M.; Butler, J.E.; Creamer, L.K.; Hicks, C.L.; Hollar, C.M.; Ng-Kwai-Hang, K.F.; Swaisgood, H.E. Nomenclature of the proteins of cows’ milk—Sixth revision. J. Dairy Sci. 2004, 87, 1641–1674. [Google Scholar] [CrossRef]
- Kamau, S.M.; Cheison, S.C.; Chen, W.; Liu, X.M.; Lu, R.R. Alpha-lactalbumin: Its production technologies and bioactive peptides. Compr. Rev. Food Sci. Food Saf. 2010, 9, 197–212. [Google Scholar] [CrossRef]
- Brew, K.; Castellino, F.J.; Vanaman, T.C.; Hill, R.L. The complete amino acid sequence of bovine alpha-lactalbumin. J. Biol. Chem. 1970, 245, 4570–4582. [Google Scholar] [CrossRef]
- Permyakov, E.A.; Berliner, L.J. α-lactalbumin: Structure and function. FEBS Lett. 2000, 473, 269–274. [Google Scholar] [CrossRef]
- Bramaud, C.; Aimar, P.; Daufin, G. Thermal isoelectric precipitation of α-lactalbumin from a whey protein concentrate: Influence of protein–calcium complexation. Biotechnol. Bioeng. 1995, 47, 121–130. [Google Scholar] [CrossRef]
- Jackson, J.G.; Janszen, D.B.; Lonnerdal, B.; Lien, E.L.; Pramuk, K.P.; Kuhlman, C.F. A multinational study of α-lactalbumin concentrations in human milk. J. Nutr. Biochem. 2004, 15, 517–521. [Google Scholar] [CrossRef]
- Permyakov, E.A. α-Lactalbumin, amazing calcium-binding protein. Biomolecules 2020, 10, 1210. [Google Scholar] [CrossRef]
- Deeth, H.; Bansal, N. Whey Proteins; Elsevier Inc.: Amsterdam, The Netherlands, 2019; ISBN 9780128121245. [Google Scholar]
- Jakopović, K.L.; Barukčić, I.; Božanić, R. Physiological significance, structure and isolation of α-lactalbumin. Mljekarstvo 2016, 66, 3–11. [Google Scholar] [CrossRef]
- Chang, J.Y.; Li, L. Pathway of oxidative folding of α-lactalbumin: A model for illustrating the diversity of disulfide folding pathways. Biochemistry 2002, 41, 8405–8413. [Google Scholar] [CrossRef]
- Rao, K.R.; Brew, K. Calcium regulates folding and disulfide-bond formation in α-lactalbumin. Biochem. Biophys. Res. Commun. 1989, 163, 1390–1396. [Google Scholar] [CrossRef]
- Pripp, A.H.; Vreeker, R.; Van Duynhoven, J. Binding of olive oil phenolics to food proteins. J. Sci. Food Agric. 2005, 85, 354–362. [Google Scholar] [CrossRef]
- Katsuragi, Y.; Kashiwayanagi, M.; Kurihara, K. Specific inhibitor for bitter taste: Inhibition of frog taste nerve responses and human taste sensation to bitter stimuli. Brain Res. Protoc. 1997, 1, 292–298. [Google Scholar] [CrossRef]
- Rawel, H.M.; Kroll, J.; Schröder, I. Reactions of isothiocyanates with food proteins: Influence on enzyme activity and tryptical degradation. Nahrung-Food 1998, 42, 197–199. [Google Scholar] [CrossRef]
- Kroll, J.; Noack, J.; Rawel, H.; Kroeck, R.; Proll, J. Chemical reactions of benzyl isothiocyanate with egg-white protein fractions. J. Sci. Food Agric. 1994, 65, 337–345. [Google Scholar] [CrossRef]
- Keppler, J.K.; Schwarz, K. Increasing the emulsifying capacity of whey proteins at acidic pH values through covalent modification with allyl isothiocyanate. Colloids Surf. A Physicochem. Eng. Asp. 2017, 522, 514–524. [Google Scholar] [CrossRef]
- Kroll, J.; Rawel, H.; Kröck, R.; Proll, J.; Schnaak, W. Interactions of isothiocyanates with egg white proteins. Food Nahrung 1994, 38, 53–60. [Google Scholar] [CrossRef]
- Rawel, H.; Kroll, J.; Haebel, S.; Peter, M. Reactions of a glucosinolate breakdown product (benzyl isothiocyanate) with myoglobin. Phytochemistry 1998, 48, 1305–1311. [Google Scholar] [CrossRef]
- Spöttel, J.; Brockelt, J.; Badekow, S.; Rohn, S. Immunological analysis of isothiocyanate-modified α-lactalbumin using high-performance thin layer chromatography. Molecules 2021, 26, 1842. [Google Scholar] [CrossRef]
- Kelly, S.M.; Price, N.C. The application of circular dichroism to studies of protein folding and unfolding. Biochim. Biophys. Acta-Protein Struct. Mol. Enzymol. 1997, 1338, 161–185. [Google Scholar] [CrossRef]
- Venyaminov, S.Y.; Yang, J.T. Determination of protein secondary structure. Circ. Dichroism Conform. Anal. Biomol. 1996, 69–107. [Google Scholar] [CrossRef]
- Keppler, J.K.; Schwarz, K.; van der Goot, A.J. Covalent modification of food proteins by plant-based ingredients (polyphenols and organosulphur compounds): A commonplace reaction with novel utilization potential. Trends Food Sci. Technol. 2020, 101, 38–49. [Google Scholar] [CrossRef]
- Sun, Q.; Gao, X.; Bi, H.; Xie, Y.; Tang, L. Assessment of binding interaction between bovine lactoferrin and tetracycline hydrochloride: Multi-spectroscopic analyses and molecular modeling. Molecules 2018, 23, 1900. [Google Scholar] [CrossRef]
- Hawe, A.; Sutter, M.; Jiskoot, W. Extrinsic fluorescent dyes as tools for protein characterization. Pharm. Res. 2008, 25, 1487–1499. [Google Scholar] [CrossRef]
- Acharya, P.; Rao, N.M. Stability studies on a lipase from Bacillus subtilis in guanidinium chloride. J. Protein Chem. 2003, 22, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Anraku, M.; Yamasaki, K.; Maruyama, T.; Kragh-Hansen, U.; Otagiri, M. Effect of oxidative stress on the structure and function of human serum albumin. Pharm. Res. 2001, 18, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Fink, A.L. Conformational states of β-lactamase: Molten-globule states at acidic and alkaline pH with high salt. Biochemistry 1989, 28, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Cardamone, M.; Puri, N.K. Spectrofluorimetric assessment of the surface hydrophobicity of proteins. Biochem. J. 1992, 282, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Qadeer, A.; Rabbani, G.; Zaidi, N.; Ahmad, E.; Khan, J.M.; Khan, R.H. 1-Anilino-8-Naphthalene Sulfonate (ANS) Is not a desirable probe for determining the molten globule state of chymopapain. PLoS ONE 2012, 7, e50633. [Google Scholar] [CrossRef]
- Alizadeh-Pasdar, N.; Li-Chan, E.C.Y. Comparison of protein surface hydrophobicity measured at various pH values using three different fluorescent probes. J. Agric. Food Chem. 2000, 48, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, S.; Nakai, S. Relationships of hydrophobicity and net charge to the solubility of milk and soy proteins. J. Food Sci. 1985, 50, 486–491. [Google Scholar] [CrossRef]
- Gerbanowski, A.; Malabat, C.; Rabiller, C.; Guéguen, J. Grafting of aliphatic and aromatic probes on rapeseed 2S and 12S proteins: Influence on their structural and physicochemical properties. J. Agric. Food Chem. 1999, 47, 5218–5226. [Google Scholar] [CrossRef]
- Lottspeich, F.; Engels, J. Bioanalytik; Spektrum Academischer: Heidelberg, Germany, 2012. [Google Scholar]
- Biemann, K. Mass spectrometry of peptides and proteins. Annu. Rev. Biochem. 1992, 61, 977–1010. [Google Scholar] [CrossRef]
- Roepstorff, P.; Fohlman, P. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 1984, 11, 601. [Google Scholar] [CrossRef]
- Keppler, J.K.; Koudelka, T.; Palani, K.; Stuhldreier, M.C.; Temps, F.; Tholey, A.; Schwarz, K. Characterization of the covalent binding of allyl isothiocyanate to β-lactoglobulin by fluorescence quenching, equilibrium measurement, and mass spectrometry. J. Biomol. Struct. Dyn. 2014, 32, 1103–1117. [Google Scholar] [CrossRef] [PubMed]
- Keppler, J.K.; Sönnichsen, F.D.; Lorenzen, P.C.; Schwarz, K. Differences in heat stability and ligand binding among β-lactoglobulin genetic variants A, B and C using 1H NMR and fluorescence quenching. Biochim. Biophys. Acta-Proteins Proteomics 2014, 1844, 1083–1093. [Google Scholar] [CrossRef] [PubMed]
- Kroll, J.; Rawel, H.; Kröck, R.; Schnaak, W. Interaction of benzyl isothiocyanate with egg white proteins. Food/Nahrung 1993, 37, 179–181. [Google Scholar] [CrossRef]
- Kroll, J.; Rawel, H. Chemical reactions of benzyl isothiocyanate with myoglobin. J. Sci. Food Agric. 1996, 72, 376–384. [Google Scholar] [CrossRef]
- Kawakishi, S.; Kaneko, T. Interaction of proteins with allyl isothiocyanate. J. Agric. Food Chem. 1987, 35, 85–88. [Google Scholar] [CrossRef]
- Rawel, H.M.; Kroll, J. Some aspects of reactions of benzyl isothiocyanate with bovine sarcoplasmic proteins. Food Nahrung 1995, 39, 465–474. [Google Scholar] [CrossRef]
- Rutherfurd, S.M. Food Composition and additives: Methodology for determining degree of hydrolysis of proteins in hydrolysates: A review. J. AOAC Int. 2010, 93, 1–8. [Google Scholar] [CrossRef]
- Kishore Kumar Murthy, N.V.; Narasinga Rao, M.S. Interaction of allyl isothiocyanate with mustard 12S Protein. J. Agric. Food Chem. 1986, 34, 448–452. [Google Scholar] [CrossRef]
- Woody, R.W. Circular dichroism. Methods Enzymol. 1995, 246, 34–71. [Google Scholar] [CrossRef]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta-Proteins Proteomics 2005, 1751, 119–139. [Google Scholar] [CrossRef] [PubMed]
- Drake, A.F. Polarisation modulation—The measurement of linear and circular dichroism. J. Phys. Sci. Instrum. 1986, 19, 170–181. [Google Scholar] [CrossRef]
- Bulheller, B.M.; Rodger, A.; Hirst, J.D. Circular and linear dichroism of proteins. Phys. Chem. Chem. Phys. 2007, 9, 2020–2035. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, Y.; Ohta, D.; Hachiya, K.; Mitsui, Y.; Takeda, K. Fluorescence behavior of tryptophan residues of bovine and human serum albumins in ionic surfactant solutions: A comparative study of the two and one tryptophan(s) of bovine and human albumins. J. Protein Chem. 1996, 15, 265–272. [Google Scholar] [CrossRef]
- Moriyama, Y.; Takeda, K. Protective effects of small amounts of bis(2-ethylhexyl)sulfosuccinate on the helical structures of human and bovine serum albumins in their thermal denaturations. Langmuir 2005, 21, 5524–5528. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, Y.; Takeda, K. Re-formation of the helical structure of human serum albumin by the addition of small amounts of sodium dodecyl sulfate after the disruption of the structure by urea. A comparison with bovine serum albumin. Langmuir 1999, 15, 2003–2008. [Google Scholar] [CrossRef]
- Parker, W.; Song, P.S. Protein structures in SDS micelle-protein complexes. Biophys. J. 1992, 61, 1435–1439. [Google Scholar] [CrossRef]
- Alcala, J.R.; Gratton, E.; Prendergast, F.G. Interpretation of fluorescence decays in proteins using continuous lifetime distributions. Biophys. J. 1987, 51, 925–936. [Google Scholar] [CrossRef]
- Santra, M.K.; Banerjee, A.; Rahaman, O.; Panda, D. Unfolding pathways of human serum albumin: Evidence for sequential unfolding and folding of its three domains. Int. J. Biol. Macromol. 2005, 37, 200–204. [Google Scholar] [CrossRef]
- Moriyama, Y.; Kawasaka, Y.; Takeda, K. Protective effect of small amounts of sodium dodecyl sulfate on the helical structure of bovine serum albumin in thermal denaturation. J. Colloid Interface Sci. 2003, 257, 41–46. [Google Scholar] [CrossRef]
- Sreerama, N.; Venyaminov, S.Y.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Inclusion of denatured proteins with native proteins in the analysis. Anal. Biochem. 2000, 287, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Corrêa, D.; Ramos, C. The use of circular dichroism spectroscopy to study protein folding, form and function. Afr. J. Biochem. Res. 2009, 3, 164–173. [Google Scholar]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2007, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Sreerama, N.; Woody, R.W. Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 2004, 383, 318–351. [Google Scholar] [CrossRef]
- Johnson, W.C. Protein secondary structure and circular dichroism: A practical guide. Proteins Struct. Funct. Bioinform. 1990, 7, 205–214. [Google Scholar] [CrossRef]
- Chen, Y.-H.; Yang, J.; Chau, K. Determination of the helix and β form of proteins in aqueous solution by circular dichroism. Biochemistry 1974, 13, 3350–3359. [Google Scholar] [CrossRef]
- Wei, Y.; Thyparambil, A.A.; Latour, R.A. Protein helical structure determination using CD spectroscopy for solutions with strong background absorbance from 190 to 230 nm. Biochim. Biophys. Acta-Proteins Proteom. 2014, 1844, 2331–2337. [Google Scholar] [CrossRef]
- Rawel, H.M.; Rohn, S.; Kruse, H.P.; Kroll, J. Structural changes induced in bovine serum albumin by covalent attachment of chlorogeanic acid. Food Chem. 2002, 78, 443–455. [Google Scholar] [CrossRef]
- Polverino de Laureto, P.; Frare, E.; Gottardo, R.; van Dael, H.; Fontana, A. Partly folded states of members of the lysozyme/lactalbumin superfamily: A comparative study by circular dichroism spectroscopy and limited proteolysis. Protein Sci. 2002, 11, 2932–2946. [Google Scholar] [CrossRef]
- Brew, K. α-Lactalbumin. In Advanced Dairy Chemistry—Proteins; Fox, P.F., McSweeney, P.L.H., Eds.; Springer: Boston, MA, USA, 2003; pp. 387–419. [Google Scholar]
- Acharya, K.R.; Stuart, D.I.; Walker, N.P.C.; Lewis, M.; Phillips, D.C. Refined structure of baboon α-lactalbumin at 1.7 Å resolution. Comparison with C-type lysozyme. J. Mol. Biol. 1989, 208, 99–127. [Google Scholar] [CrossRef]
- Delavari, B.; Saboury, A.A.; Atri, M.S.; Ghasemi, A.; Bigdeli, B.; Khammari, A.; Maghami, P.; Moosavi-Movahedi, A.A.; Haertlé, T.; Goliaei, B. Alpha-lactalbumin: A new carrier for vitamin D3 food enrichment. Food Hydrocoll. 2015, 45, 124–131. [Google Scholar] [CrossRef]
- Pike, A.C.W.; Brew, K.; Acharya, K.R. Crystal structures of guinea-pig, goat and bovine α-lactalbumin highlight the enhanced conformational flexibility of regions that are significant for its action in lactose synthase. Structure 1996, 4, 691–703. [Google Scholar] [CrossRef]
- Chrysina, E.D.; Brew, K.; Acharya, K.R. Crystal structures of Apo- and holo-bovine α-lactalbumin at 2.2-Å resolution reveal an effect of calcium on inter-lobe interactions. J. Biol. Chem. 2000, 275, 37021–37029. [Google Scholar] [CrossRef]
- Boggione Santos, I.J.; Hernandez Hernandez, H.L.; Cardoso Costa, M.H.; de Queiroz Lafetá, J.A.; dos Reis Coimbra, J.S. Conjugates of α-lactalbumin, β-lactoglobulin, and lysozyme with polysaccharides: Characterization and techno-functional properties. Food Res. Int. 2019, 116, 492–498. [Google Scholar] [CrossRef]
- Wilde, S.C.; Treitz, C.; Keppler, J.K.; Koudelka, T.; Palani, K.; Tholey, A.; Rawel, H.M.; Schwarz, K. β-Lactoglobulin as nanotransporter—Part II: Characterization of the covalent protein modification by allicin and diallyl disulfide. Food Chem. 2016, 197, 1022–1029. [Google Scholar] [CrossRef]
- Wu, X.; Dey, R.; Wu, H.; Liu, Z.; He, Q.; Zeng, X. Studies on the interaction of -epigallocatechin-3-gallate from green tea with bovine β-lactoglobulin by spectroscopic methods and docking. Int. J. Dairy Technol. 2013, 66, 7–13. [Google Scholar] [CrossRef]
- Wu, X.; Liu, M.; Xia, L.; Wu, H.; Liu, Z.; Xu, X. Conjugation of functional oligosaccharides reduced in vitro allergenicity of β-lactoglobulin. Food Agric. Immunol. 2013, 24, 379–391. [Google Scholar] [CrossRef]
- Das, A.; Thakur, R.; Dagar, A.; Chakraborty, A. A spectroscopic investigation and molecular docking study on the interaction of hen egg white lysozyme with liposomes of saturated and unsaturated phosphocholines probed by an anticancer drug ellipticine. Phys. Chem. Chem. Phys. 2014, 16, 5368–5381. [Google Scholar] [CrossRef] [PubMed]
- Schwenke, K.D.; Knopfe, C.; Seifert, A.; Görnitz, E.; Zirwer, D. Acetylation of faba bean legumin: Conformational changes and aggregation. J. Sci. Food Agric. 2001, 81, 126–134. [Google Scholar] [CrossRef]
- Rawel, H.M.; Rohn, S.; Kroll, J. Influence of a sugar moiety (rhamnosylglucoside) at 3-O position on the reactivity of quercetin with whey proteins. Int. J. Biol. Macromol. 2003, 32, 109–120. [Google Scholar] [CrossRef]
- Anand, U.; Jash, C.; Mukherjee, S. Protein unfolding and subsequent refolding: A spectroscopic investigation. Phys. Chem. Chem. Phys. 2011, 13, 20418–20426. [Google Scholar] [CrossRef]
- Fang, Y.; Dalgleish, D.G. The conformation of α-lactalbumin as a function of pH, heat treatment and adsorption at hydrophobic surfaces studied by FTIR. Food Hydrocoll. 1998, 12, 121–126. [Google Scholar] [CrossRef]
- Kelly, S.; Price, N. The Use of Circular Dichroism in the Investigation of Protein Structure and Function. Curr. Protein Pept. Sci. 2005, 1, 349–384. [Google Scholar] [CrossRef]
- Walters, J.; Milam, S.L.; Clark, A.C. Chapter 1 Practical Approaches to Protein Folding and Assembly. Spectroscopic Strategies in Thermodynamics and Kinetics; Elsevier Inc.: Amsterdam, The Netherlands, 2009; ISBN 9780123745965. [Google Scholar]
- Semisotnov, G.V.; Rodionova, N.A.; Razgulyaev, O.I.; Uversky, V.N.; Gripas’, A.F.; Gilmanshin, R.I. Study of the “molten globule” intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers 1991, 31, 119–128. [Google Scholar] [CrossRef]
- Stryer, L. The interaction of a naphthalene dye with apomyoglobin and apohemoglobin: A fluorescent probe of non-polar binding sites. J. Mol. Biol. 1965, 13, 482–495. [Google Scholar] [CrossRef]
- Gasymov, O.K.; Glasgow, B.J. ANS fluorescence: Potential to augment the identification of the external binding sites of proteins. Biochim. Biophys. Acta-Proteins Proteom. 2007, 1774, 403–411. [Google Scholar] [CrossRef]
- Slavík, J. Anilinonaphthalene sulfonate as a probe of membrane composition and function. Biochim. Biophys. Acta 1982, 694, 1–25. [Google Scholar] [CrossRef]
- Tanford, C. The Hydrophobic Effect: Formation of micelles and biological membranes. FEBS Lett. 1981, 124, 127. [Google Scholar] [CrossRef]
- Singh, S.K.; Kishore, N. Elucidating the Binding Thermodynamics of 8-Anilino-1-Naphthalene Sulfonic Acid with the A-State of a-Lactalbumin: An Isothermal Titration Calorimetric Investigation. Biopolymers 2006, 83, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Asthana, S.; Bhutia, S.K.; Sahoo, H.; Jha, S. Chaotropes trigger conformational rearrangements differently in Concanavalin A. J. Chem. Sci. 2017, 129, 1267–1276. [Google Scholar] [CrossRef]
- Li, T.; Wang, C.; Li, T.; Ma, L.; Sun, D.; Hou, J.; Jiang, Z. Surface hydrophobicity and functional properties of citric acid cross-linked whey protein isolate: The impact of pH and concentration of citric acid. Molecules 2018, 23, 2383. [Google Scholar] [CrossRef] [PubMed]
- Lakkis, J.; Villota, R. Effect of acylation on substructural properties of proteins: A study using fluorescence and circular dichroism. J. Agric. Food Chem. 1992, 40, 553–560. [Google Scholar] [CrossRef]
- Kato, A.; Nakai, S. Hydrophobicity determined by a fluorescence probe method and its correlation with surface properties of proteins. Biochim. Biophys. Acta 1980, 624, 13–20. [Google Scholar] [CrossRef]
- Kroll, J.; Rawel, H.M.; Seidelmann, N. Physicochemical properties and susceptibility to proteolytic digestion of myoglobin-phenol derivatives. J. Agric. Food Chem. 2000, 48, 1580–1587. [Google Scholar] [CrossRef]
- Rawel, H.M.; Kroll, J.; Hohl, U.C. Model studies on reactions of plant phenols with whey proteins. Nahrung-Food 2001, 45, 72–81. [Google Scholar] [CrossRef]
- Goldburg, W.I. Dynamic light scattering. Am. J. Phys 1999, 67, 1152–1160. [Google Scholar] [CrossRef]
- Lorber, B.; Fischer, F.; Bailly, M.; Roy, H.; Kern, D. Protein analysis by dynamic light scattering: Methods and techniques for students. Biochem. Mol. Biol. Educ. 2012, 40, 372–382. [Google Scholar] [CrossRef]
- Sandhu, R.; Singh, N.; Dhankhar, J.; Kama, G.; Sharma, R. Dynamic light scattering (DLS) technique, principle, theoretical considerations and applications. Nanotechnol. Biochem. Tech. Assess. Qual. Saf. Milk Milk Prod. 2018, pp. 135–137. Available online: https://www.researchgate.net/publication/331022012 (accessed on 14 October 2021).
- Harding, S.; Jumel, K. Analysis of Proteins. Curr. Protoc. Mol. Biol. 1998, 44, 1–14. [Google Scholar] [CrossRef]
- Harding, S.E. Protein Hydrodynamics; Allen, G., Ed.; JAI Press Inc.: Greenwich, CT, USA, 1999; ISBN 9781559386722. [Google Scholar]
- Kehoe, J.J.; Brodkorb, A. Interactions between sodium oleate and α-lactalbumin: The effect of temperature and concentration on complex formation. Food Hydrocoll. 2014, 34, 217–226. [Google Scholar] [CrossRef][Green Version]
- Kataoka, M.; Kuwajima, K.; Tokunaga, F.; Goto, Y. Structural characterization of the molten globule of α-lactalbumin by solution X-ray scattering. Protein Sci. 1997, 6, 422–430. [Google Scholar] [CrossRef]
- Abbasi, A.; Emam-Djomeh, Z.; Mousavi, M.A.E.; Davoodi, D. Stability of vitamin D3 encapsulated in nanoparticles of whey protein isolate. Food Chem. 2014, 143, 379–383. [Google Scholar] [CrossRef]
- Lin, F.Y.; Chen, W.Y.; Ruaan, R.C.; Huang, H.M. Microcalorimetric studies of interactions between protein and hydrophobic ligands in hydrophobic interaction chromatography: Effects of ligand chain length, density, and the amount of bound protein. Prog. Biotechnol. 2000, 16, 59–62. [Google Scholar] [CrossRef]
- Hoffmann, M.A.M.; Sala, G.; Olieman, C.; De Kruif, K.G. Molecular mass distributions of heat-induced β-lactoglobulin aggregates. J. Agric. Food Chem. 1997, 45, 2949–2957. [Google Scholar] [CrossRef]
- Barth, H. Modern Methods of Particle Size Analysi; Wiley: New York, NY, USA, 1984. [Google Scholar]
- Olsen, J.V.; Ong, S.E.; Mann, M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell. Proteomics 2004, 3, 608–614. [Google Scholar] [CrossRef]
- Rawel, H.M.; Kroll, J.; Schröder, I. In vitro enzymatic digestion of benzyl- and phenylisothiocyanate-derivatized food proteins. J. Agric. Food Chem. 1998, 46, 5103–5109. [Google Scholar] [CrossRef]
- Chevalier, F.; Chobert, J.; Molle, D.; Haertle, T. Maillard glycation of ß-lactoglobulin with several sugars: Comparative study of the properties of the obtained polymers and of the substituted sites. Lait 2001, 81, 651–666. [Google Scholar] [CrossRef]
- Chevalier, F.; Chobert, J.M.; Dalgalarrondo, M.; Haertlé, T. Characterization of the Maillard reaction products of β-lactoglobulin glucosylated in mild conditions. J. Food Biochem. 2001, 25, 33–55. [Google Scholar] [CrossRef]
- Syka, J.E.P.; Coon, J.J.; Schroeder, M.J.; Shabanowitz, J.; Hunt, D.F. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. USA 2004, 101, 9528–9533. [Google Scholar] [CrossRef] [PubMed]
- Bertran-Vicente, J.; Schümann, M.; Hackenberger, C.P.R.; Krause, E. Gas-phase rearrangement in lysine phosphorylated peptides during electron-transfer dissociation tandem mass spectrometry. Anal. Chem. 2015, 87, 6990–6994. [Google Scholar] [CrossRef]
- Hernández, M.J.M.; Domingo, E.B.; Camañas, R.M.V.; Alvarez-Coque, M.C.G. Evaluation of the proteolysis degree with the o-phthalaldehyde/N-acetyl-L-cysteine reagent. Fresenius. J. Anal. Chem. 1990, 338, 62–65. [Google Scholar] [CrossRef]
- García Alvarez-Coque, M.C.; Medina Hernández, M.J.; Villanueva Camañas, R.M.; Mongay Fernández, C. Formation and instability of o-phthalaldehyde derivatives of amino acids. Anal. Biochem. 1989, 178, 1–7. [Google Scholar] [CrossRef]
- Hernández, M.J.M.; Camañas, R.M.V.; Cuenca, E.M.; Alvarez-Coque, M.C.G. Determination of the protein and free amino acid content in a sample using o-phthalaldehyde and N-acetyl-L-CYSTEINE. Analyst 1990, 115, 1125–1128. [Google Scholar] [CrossRef]
- Spellman, D.; McEvoy, E.; O’Cuinn, G.; FitzGerald, R.J. Proteinase and exopeptidase hydrolysis of whey protein: Comparison of the TNBS, OPA and pH stat methods for quantification of degree of hydrolysis. Int. Dairy J. 2003, 13, 447–453. [Google Scholar] [CrossRef]
- Hernández, M.J.M.; Domingo, E.B.; Camañas, R.M.V.; Alvarez-Coque, M.C.G. Use of the o-Phthalaldehyde and N-Acetyl-L-Cysteine the Evaluation of Milk Proteins. J. Dairy Sci. 1991, 74, 1779–1785. [Google Scholar] [CrossRef]
- Goodno, C.C.; Swaisgood, H.E.; Catignani, G.L. A fluorimetric assay for available lysine in proteins. Anal. Biochem. 1981, 115, 203–211. [Google Scholar] [CrossRef]
- Roth, M. Fluorescence Reaction for Amino Acids. Anal. Chem. 1971, 43, 880–882. [Google Scholar] [CrossRef]
- Interchim. In OPA, Amine Detection Reagent; Interchim: Montlucon, France, 2016; pp. 10–12. Available online: https://www.interchim.fr/ft/0/02727A.pdf (accessed on 14 October 2021).
- Andrade, M.A.; Chacón, P.; Merelo, J.J.; Morán, F. Evaluation of secondary structure of proteins from uv circular dichroism spectra using an unsupervised learning neural network. Protein Eng. Des. Sel. 1993, 6, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, L.; Wallace, B.A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004, 32, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Takehara, K.; Yuki, K.; Shirasawa, M.; Yamasaki, S.; Yamada, S. Binding Properties of Hydrophobic Molecules to Human Serum Albumin Studied by Fluorescence Titration. Anal. Sci. 2009, 25, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Lam, R.S.H.; Nickerson, M.T. The effect of pH and temperature pre-treatments on the physicochemical and emulsifying properties of whey protein isolate. LWT-Food Sci. Technol. 2015, 60, 427–434. [Google Scholar] [CrossRef]
- Hoffman, R.E.; Shabtai, E.; Rabinovitz, M.; Iyer, S.V.; Müllen, K.; Rai, A.K.; Bayrd, E.; Scott, L.T. Self-diffusion measurements of polycyclic aromatic hydrocarbon alkali metal salts. J. Chem. Soc. Perkin Trans. 1998, 2, 1659–1664. [Google Scholar] [CrossRef]
- Roberts, C.J. Non-Native Protein Aggregation Kinetics Christopher. Biotechnol. Bioeng. 2007, 98, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Rutherfurd, S.M.; Moughan, P.J. Digestible reactive lysine in selected milk-based products. J. Dairy Sci. 2005, 88, 40–48. [Google Scholar] [CrossRef]
- Soto, C.; Sigurdsson, E.M.; Morelli, L.; Asok Kumar, R.; Castaño, E.M.; Frangione, B. Solubility as funtion of proteins structure and solvent components. Nat. Med. 1998, 4, 822–826. [Google Scholar] [CrossRef] [PubMed]


| Sample | BBITC/α-LA | Free-NH2 Content (%) | SD (%) |
|---|---|---|---|
| α-LA control | 0 | 100 | - |
| BITC-α-LA derivative “minimal” | 10 | 87.2 | 3.6 |
| BITC-α-LA derivative “minor” | 25 | 88.0 | 2.0 |
| BITC-α-LA derivative “low” | 50 | 79.3 | 2.1 |
| BITC-α-LA derivative “medium” | 500 | 61.8 | 17.5 |
| BITC-α-LA derivative “high” | 1000 | 59.9 | 15.4 |
| BITC-α-LA derivative “very high” | 1500 | 59.0 | 8.8 |
| Sample | Temperature (°C) | α-Helix (%) | β-Sheet (%) | Random Coil (%) |
|---|---|---|---|---|
| LA (native) | 20 | 30 | 12 | 58 |
| α-LA control | 20 | 29 | 13 | 58 |
| 80 | 15 | 15 | 70 | |
| BITC-α-LA-derivative “low” | 20 | 24 | 19 | 57 |
| 80 | 19 | 12 | 69 | |
| BITC-α-LA-derivative “medium” | 20 | 20 | 25 | 56 |
| 80 | 18 | 19 | 63 | |
| BITC-α-LA-derivative “high” | 20 | 17 | 28 | 55 |
| 80 | 16 | 26 | 58 |
| Sample | No. | RH (± SD) (nm) | No. | RH (± SD) (nm) |
|---|---|---|---|---|
| α-LA control | I | 1.8 (±0.2) | ||
| BITC-α-LA-derivative “low” | I | 2.4 (±0.5) | II | 2.9 (±0.7) |
| III | 13.8 (±3.2) | IV | 15.8 (±3.5) | |
| V | 57.8 (±11.5) | |||
| BITC-α-LA-derivative “medium” | I | 83.8 (±18.3) | II | 89.4 (±14.9) |
| BITC-α-LA-derivative “high” | I | 2.0 (±0.3) | II | 55.5 (±11.3) |
| III | 61.0 (±13.6) | |||
| BITC-α-LA-derivative “very high” | I | 51.6 (±11.7) | II | 62.1 (±11.8) |
| III | 68.6 (±15.6) |
| No. | Peptide Sequence | Mass (m/z) | Control | Low | Medium | High | Very High |
|---|---|---|---|---|---|---|---|
| 1 | EQLTK | 618.35 | 10.7 | 10.9 | 11.2 | 11 | 11.2 |
| 1 * | EQLTK + BITC | 767.56 | - | 29.7 | 29.8 | 30.5 | 30.3 |
| 2 | CEVFR | 653.31 | 19.1 | - | - | - | - |
| 3 | ELK | 389.24 | 4.5 | 4.5 | 4.6 | 4.5 | 4.6 |
| 3 * | ELK + BITC | 538.45 | - | - | - | - | 29.3 |
| 3 ** | ELKDLK + BITC | 894.17 | - | - | 27.0 | 26.9 | 26.8 |
| 4 | DLK | 375.22 | 5.2 | 5.3 | 5.3 | 5.2 | 5.2 |
| 5 | GYGGVSLPEWVCTTFHTSGYD-TQAIVQNNDST-EYGLFQINNK | 4654.15 | - | - | - | - | - |
| 6 | IWCK | 549.29 | 18 | - | - | - | - |
| 7 | DDQNPHSSNICNISCDK | 1889.78 | 16.6 | 17.0 | 16.8 | 17 | 18.1 |
| 8 | FLDDDLTDDIMCVK | 1642.73 | - | - | - | - | - |
| 9 | K | 147.11 | - | - | - | - | - |
| 9 ** | KILDK + BITC | 765.61 | - | 23.4 | 23.3 | 23.2 | 23.3 |
| 10 | ILDK | 488.31 | 16.1 | 16.1 | 16.1 | 16.1 | 16.0 |
| 10 * | ILDK + BITC | 637.52 | - | - | - | 32.5 | 32.7 |
| 11 | VGINYWLAHK | 1220.65 | 20.9 | 20.9 | 20.3 | 21.3 | 20.7 |
| 12 | ALCSEK | 650.32 | 16.4 | 15.9 | 15.8 | 15.7 | 16.2 |
| 12 ** | ALCSEKLDQWLCEK + BITC | 1815.1 | - | - | - | - | 31.8 |
| 13 | LDQWLCEK | 1034.50 | 20 | 21.2 | 23.5 | 24.5 | 23.2 |
| 13 * | LDQWLCEK + BITC | 1183.71 | - | 34.1 | 33.8 | 34.4 | 33.4 |
| 14 | L | 132.10 | - | - | - | - | - |
| Sequence | No. | b-Ions | b*-Ions | y-Ions | y*-Ions | No. |
|---|---|---|---|---|---|---|
| E | 1 | 130.05 | 279.26 | 745.45 | 894.66 | 6 |
| L | 2 | 243.13 | 392.34 | 616.40 | 765.61 | 5 |
| K | 3 | 371.23 | 520.44 | 503.32 | 652.53 | 4 |
| D | 4 | 486.26 | 635.47 | 375.22 | 524.43 | 3 |
| L | 5 | 599.34 | 748.55 | 260.20 | 409.41 | 2 |
| K | 6 | 727.43 | 876.64 | 247.11 | 296.32 | 1 |
| Sequence | No. | b-Ions | b*-Ions | y-Ions | y*-Ions | No. |
|---|---|---|---|---|---|---|
| K | 1 | 129.10 | 278.31 | 616.40 | 765.61 | 5 |
| I | 2 | 242.19 | 391.40 | 488.31 | 637.52 | 4 |
| L | 3 | 355.27 | 504.48 | 375.22 | 524.43 | 3 |
| D | 4 | 470.30 | 619.51 | 262.14 | 411.35 | 2 |
| K | 5 | 598.39 | 747.60 | 147.11 | 296.32 | 1 |
| Sample | c(α-LA) (mM) | c(BITC) (mM) | BBITC/α-LA |
|---|---|---|---|
| α-LA control | 0.07 | - | 0 |
| BITC-α-LA derivate ”minimal” | 0.07 | 0.6 | 10 |
| BITC-α-LA derivate “minor” | 0.07 | 1.9 | 25 |
| BITC-α-LA derivate ”low” | 0.07 | 3.8 | 50 |
| BITC-α-LA derivate “medium” | 0.07 | 38 | 500 |
| BITC-α-LA derivate “high” | 0.07 | 75 | 1000 |
| BITC-α-LA derivate “very high” | 0.07 | 113 | 1500 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spöttel, J.; Brockelt, J.; Falke, S.; Rohn, S. Characterization of Conjugates between α-Lactalbumin and Benzyl Isothiocyanate—Effects on Molecular Structure and Proteolytic Stability. Molecules 2021, 26, 6247. https://doi.org/10.3390/molecules26206247
Spöttel J, Brockelt J, Falke S, Rohn S. Characterization of Conjugates between α-Lactalbumin and Benzyl Isothiocyanate—Effects on Molecular Structure and Proteolytic Stability. Molecules. 2021; 26(20):6247. https://doi.org/10.3390/molecules26206247
Chicago/Turabian StyleSpöttel, Jenny, Johannes Brockelt, Sven Falke, and Sascha Rohn. 2021. "Characterization of Conjugates between α-Lactalbumin and Benzyl Isothiocyanate—Effects on Molecular Structure and Proteolytic Stability" Molecules 26, no. 20: 6247. https://doi.org/10.3390/molecules26206247
APA StyleSpöttel, J., Brockelt, J., Falke, S., & Rohn, S. (2021). Characterization of Conjugates between α-Lactalbumin and Benzyl Isothiocyanate—Effects on Molecular Structure and Proteolytic Stability. Molecules, 26(20), 6247. https://doi.org/10.3390/molecules26206247