AGSE: A Novel Grape Seed Extract Enriched for PP2A Activating Flavonoids That Combats Oxidative Stress and Promotes Skin Health
Abstract
:1. Introduction
2. Results and Discussion
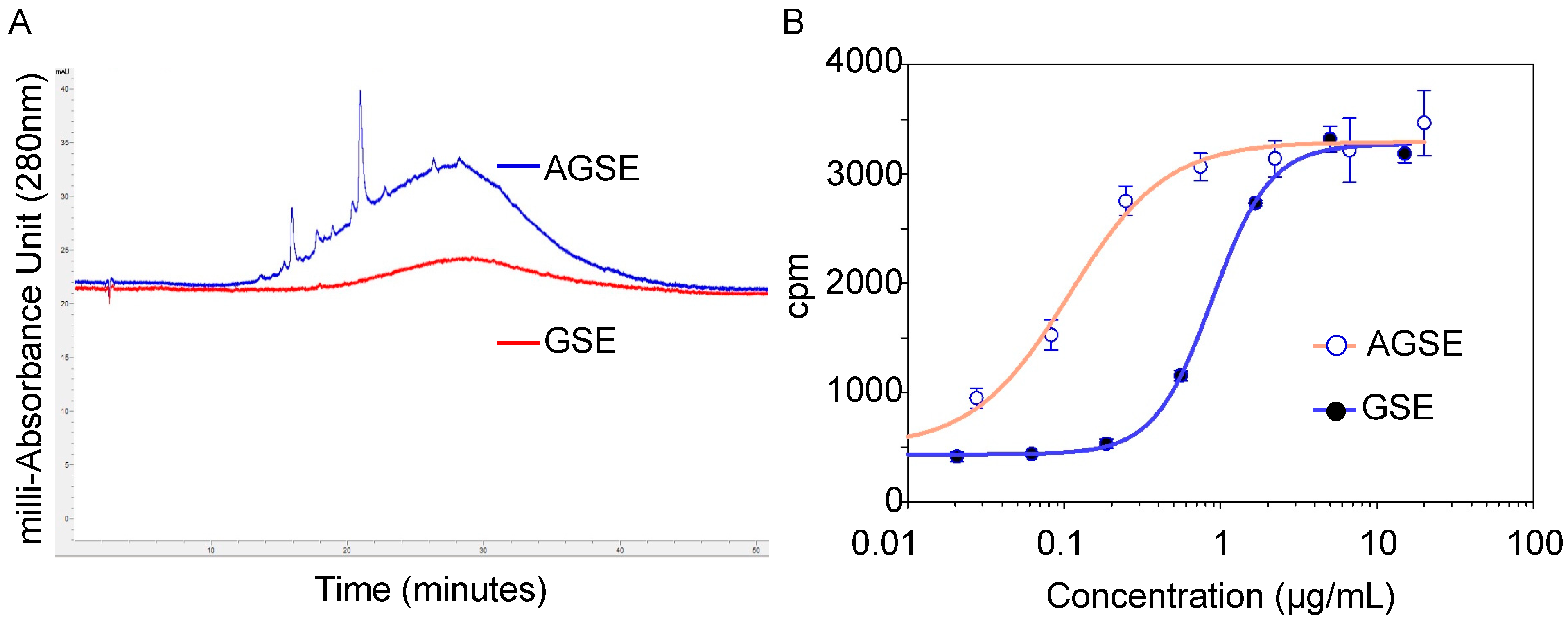
2.1. Botanical Extracts as PP2A Demethylation Inhibitors
2.2. Gene Microarray Analysis of AGSE versus Commercial GSE
2.3. AGSE Possesses Potent Antioxidant Activity
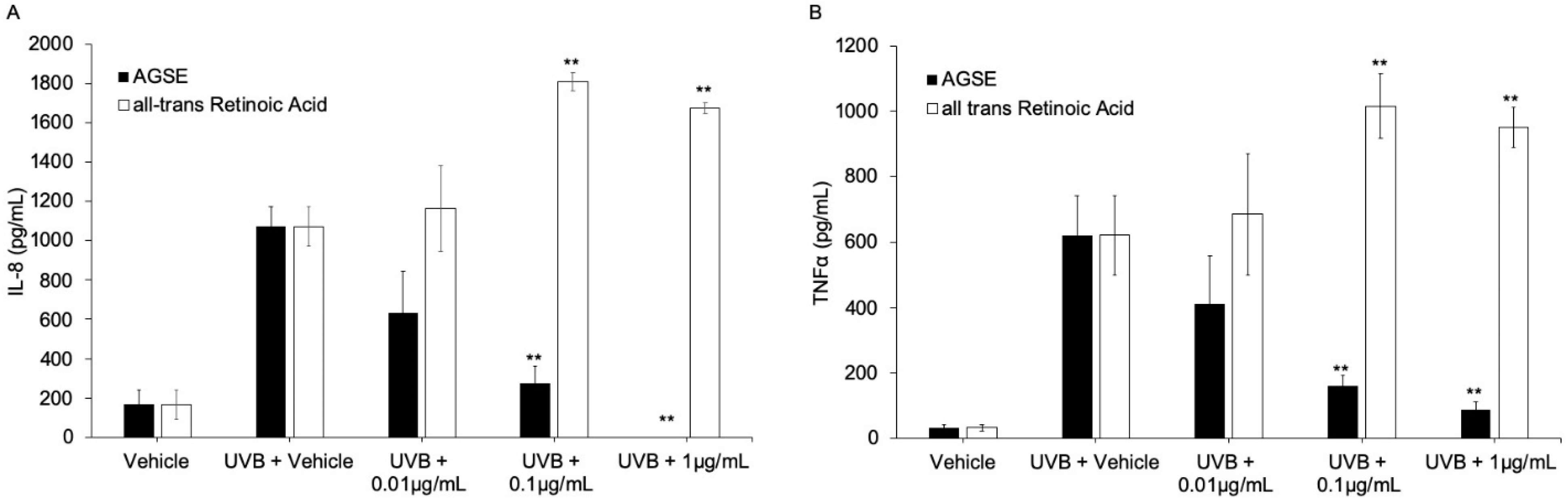
2.4. AGSE Inhibits UV Light and Chemical-Induced Pro-Inflammatory Cytokine Production
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Botanical Extraction, Fractionation and AGSE Production
3.3. Demethylation of PP2A by PME-1 (Protein Phosphatase Methylesterase 1)
3.4. Chemical Analysis
3.5. Gene Microarray
3.6. Antioxidant Assays
3.7. Cell Culture
3.8. Gene Expression Assays
3.9. Anti-Inflammatory Assays
3.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Mazhar, S.; Taylor, S.E.; Sangodkar, J.; Narla, G. Targeting PP2A in cancer: Combination therapies. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Taleski, G.; Sontag, E. Protein phosphatase 2A and tau: An orchestrated ‘Pas de Deux’. FEBS Lett. 2018, 592, 1079–1095. [Google Scholar] [CrossRef] [PubMed]
- Koh, E.M.; Lee, E.K.; Song, C.H.; Song, J.; Chung, H.Y.; Chae, C.H.; Jung, K.J. Ferulate, an Active Component of Wheat Germ, Ameliorates Oxidative Stress-Induced PTK/PTP Imbalance and PP2A Inactivation. Toxicol. Res. 2018, 34, 333–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Mhamdi, A.; Trotta, A.; Kangasjarvi, S.; Noctor, G. The protein phosphatase subunit PP2A-B’gamma is required to suppress day length-dependent pathogenesis responses triggered by intracellular oxidative stress. New Phytol. 2014, 202, 145–160. [Google Scholar] [CrossRef] [PubMed]
- Elgenaidi, I.S.; Spiers, J.P. Regulation of the phosphoprotein phosphatase 2A system and its modulation during oxidative stress: A potential therapeutic target? Pharmacol. Ther. 2019, 198, 68–89. [Google Scholar] [CrossRef] [PubMed]
- O’Shaughnessy, R.F.; Welti, J.C.; Sully, K.; Byrne, C. Akt-dependent Pp2a activity is required for epidermal barrier formation during late embryonic development. Development 2009, 136, 3423–3431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kam, E.; Resing, K.A.; Lim, S.K.; Dale, B.A. Identification of rat epidermal profilaggrin phosphatase as a member of the protein phosphatase 2A family. J. Cell Sci. 1993, 106, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Dobrowsky, R.T.; Kamibayashi, C.; Mumby, M.C.; Hannun, Y.A. Ceramide activates heterotrimeric protein phosphatase 2A. J. Biol. Chem. 1993, 268, 15523–15530. [Google Scholar] [CrossRef]
- Voronkov, M.; Braithwaite, S.P.; Stock, J.B. Phosphoprotein phosphatase 2A: A novel druggable target for Alzheimer’s disease. Future Med. Chem. 2011, 3, 821–833. [Google Scholar] [CrossRef] [Green Version]
- Huber, K.L.; Fernandez, J.R.; Webb, C.; Rouzard, K.; Healy, J.; Tamura, M.; Voronkov, M.; Stock, J.B.; Stock, M.; Perez, E. HYVIA: A novel, topical chia seed extract that improves skin hydration. J. Cosmet. Dermatol. 2020, 19, 2386–2393. [Google Scholar] [CrossRef] [PubMed]
- Asam, K.; Staniszewski, A.; Zhang, H.; Melideo, S.L.; Mazzeo, A.; Voronkov, M.; Huber, K.L.; Perez, E.; Stock, M.; Stock, J.B.; et al. Eicosanoyl-5-hydroxytryptamide (EHT) prevents Alzheimer’s disease-related cognitive and electrophysiological impairments in mice exposed to elevated concentrations of oligomeric beta-amyloid. PLoS ONE 2017, 12, e0189413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basurto-Islas, G.; Blanchard, J.; Tung, Y.C.; Fernandez, J.R.; Voronkov, M.; Stock, M.; Zhang, S.; Stock, J.B.; Iqbal, K. Therapeutic benefits of a component of coffee in a rat model of Alzheimer’s disease. Neurobiol. Aging 2014, 35, 2701–2712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.W.; Im, J.Y.; Woo, J.M.; Grosso, H.; Kim, Y.S.; Cristovao, A.C.; Sonsalla, P.K.; Schuster, D.S.; Jalbut, M.M.; Fernandez, J.R.; et al. Neuroprotective and anti-inflammatory properties of a coffee component in the MPTP model of Parkinson’s disease. Neurotherapeutics 2013, 10, 143–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, R.; Zhang, J.; Park, H.J.; Park, E.S.; Oh, S.; Zheng, H.; Junn, E.; Voronkov, M.; Stock, J.B.; Mouradian, M.M. Synergistic neuroprotection by coffee components eicosanoyl-5-hydroxytryptamide and caffeine in models of Parkinson’s disease and DLB. Proc. Natl. Acad. Sci. USA 2018, 115, E12053–E12062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.W.; Cheng, Y.C.; Hung, Y.C.; Lee, C.H.; Fang, J.Y.; Li, W.T.; Wu, Y.R.; Pan, T.L. Red Raspberry Extract Protects the Skin against UVB-Induced Damage with Antioxidative and Anti-inflammatory Properties. Oxidative Med. Cell Longev. 2019, 2019, 9529676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Z.; Park, S.Y.; Hwang, E.; Park, B.; Seo, S.A.; Cho, J.G.; Zhang, M.; Yi, T.H. Dietary Foeniculum vulgare Mill extract attenuated UVB irradiation-induced skin photoaging by activating of Nrf2 and inhibiting MAPK pathways. Phytomedicine 2016, 23, 1273–1284. [Google Scholar] [CrossRef] [PubMed]
- Yarovaya, L.; Waranuch, N.; Wisuitiprot, W.; Khunkitti, W. Effect of grape seed extract on skin fibroblasts exposed to UVA light and its photostability in sunscreen formulation. J. Cosmet. Dermatol. 2021, 20, 1271–1282. [Google Scholar] [CrossRef] [PubMed]
- Hemmati, A.A.; Foroozan, M.; Houshmand, G.; Moosavi, Z.B.; Bahadoram, M.; Maram, N.S. The topical effect of grape seed extract 2% cream on surgery wound healing. Glob. J. Health Sci. 2014, 7, 52–58. [Google Scholar] [CrossRef] [Green Version]
- Katiyar, S.K. Grape seed proanthocyanidines and skin cancer prevention: Inhibition of oxidative stress and protection of immune system. Mol. Nutr. Food Res. 2008, 52, S71–S76. [Google Scholar] [CrossRef] [Green Version]
- Xia, E.Q.; Deng, G.F.; Guo, Y.J.; Li, H.B. Biological activities of polyphenols from grapes. Int. J. Mol. Sci. 2010, 11, 622–646. [Google Scholar] [CrossRef] [PubMed]
- Nassiri-Asl, M.; Hosseinzadeh, H. Review of the pharmacological effects of Vitis vinifera (Grape) and its bioactive compounds. Phytother. Res. 2009, 23, 1197–1204. [Google Scholar] [CrossRef] [PubMed]
- Nassiri-Asl, M.; Hosseinzadeh, H. Review of the Pharmacological Effects of Vitis vinifera (Grape) and its Bioactive Constituents: An Update. Phytother. Res. 2016, 30, 1392–1403. [Google Scholar] [CrossRef] [PubMed]
- De Rosso, M.; Panighel, A.; Vedova, A.D.; Gardiman, M.; Flamini, R. Characterization of Non-Anthocyanic Flavonoids in Some Hybrid Red Grape Extracts Potentially Interesting for Industrial Uses. Molecules 2015, 20, 18095–18106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shati, A.A.; Alfaifi, M.Y. Trans-resveratrol Inhibits Tau Phosphorylation in the Brains of Control and Cadmium Chloride-Treated. A.Rats by Activating PP2A and PI3K/Akt Induced-Inhibition of GSK3beta. Neurochem. Res. 2019, 44, 357–373. [Google Scholar] [CrossRef] [PubMed]
- Schweiger, S.; Matthes, F.; Posey, K.; Kickstein, E.; Weber, S.; Hettich, M.M.; Pfurtscheller, S.; Ehninger, D.; Schneider, R.; Krauss, S. Resveratrol induces dephosphorylation of Tau by interfering with the MID1-PP2A complex. Sci. Rep. 2017, 7, 13753. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Li, Z.; Rizak, J.D.; Wu, S.; Wang, Z.; He, R.; Su, M.; Qin, D.; Wang, J.; Hu, X. Resveratrol Attenuates Formaldehyde Induced Hyperphosphorylation of Tau Protein and Cytotoxicity in N2a Cells. Front. Neurosci. 2016, 10, 598. [Google Scholar] [CrossRef] [Green Version]
- Lavker, R. Cutaneous aging: Chronologic versus photoaging. Camb. Blackwell Sci. 1995, 1, 123–125. [Google Scholar]
- Mays, P.K.; Bishop, J.E.; Laurent, G.J. Age-related changes in the proportion of types I and III collagen. Mech. Ageing Dev. 1988, 45, 203–212. [Google Scholar] [CrossRef]
- Poschl, E.; Schlotzer-Schrehardt, U.; Brachvogel, B.; Saito, K.; Ninomiya, Y.; Mayer, U. Collagen IV is essential for basement membrane stability but dispensable for initiation of its assembly during early development. Development 2004, 131, 1619–1628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lephart, E.D. Resveratrol, 4’ Acetoxy Resveratrol, R-equol, Racemic Equol or S-equol as Cosmeceuticals to Improve Dermal Health. Int. J. Mol. Sci. 2017, 18, 1193. [Google Scholar] [CrossRef] [Green Version]
- Daly, C.H.; Odland, G.F. Age-related changes in the mechanical properties of human skin. J. Investig. Dermatol. 1979, 73, 84–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.; Melichian, D.S.; de la Garza, M.; Gruner, K.; Bhattacharyya, S.; Barr, L.; Nair, A.; Shahrara, S.; Sporn, P.H.; Mustoe, T.A.; et al. Essential roles for early growth response transcription factor Egr-1 in tissue fibrosis and wound healing. Am. J. Pathol. 2009, 175, 1041–1055. [Google Scholar] [CrossRef] [Green Version]
- Clark, A.R.; Dean, J.L. The control of inflammation via the phosphorylation and dephosphorylation of tristetraprolin: A tale of two phosphatases. Biochem. Soc. Trans. 2016, 44, 1321–1337. [Google Scholar] [CrossRef] [Green Version]
- Patial, S.; Blackshear, P.J. Tristetraprolin as a Therapeutic Target in Inflammatory Disease. Trends Pharmacol. Sci. 2016, 37, 811–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapiere, C.M. The ageing dermis: The main cause for the appearance of ‘old’ skin. Br. J. Dermatol. 1990, 122 (Suppl. 35), 5–11. [Google Scholar] [CrossRef] [PubMed]
- Harman, D. Aging: A theory based on free radical and radiation chemistry. J. Gerontol. 1956, 11, 298–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zillich, O.V.; Schweiggert-Weisz, U.; Eisner, P.; Kerscher, M. Polyphenols as active ingredients for cosmetic products. Int. J. Cosmet. Sci. 2015, 37, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Bickers, D.R.; Athar, M. Oxidative stress in the pathogenesis of skin disease. J. Investig. Dermatol. 2006, 126, 2565–2575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strickland, I.; Rhodes, L.E.; Flanagan, B.F.; Friedmann, P.S. TNF-alpha and IL-8 are upregulated in the epidermis of normal human skin after UVB exposure: Correlation with neutrophil accumulation and E-selectin expression. J. Investig. Dermatol. 1997, 108, 763–768. [Google Scholar] [CrossRef] [Green Version]
- Wilmer, J.L.; Luster, M.I. Chemical induction of interleukin-8, a proinflammatory chemokine, in human epidermal keratinocyte cultures and its relation to cytogenetic toxicity. Cell Biol. Toxicol. 1995, 11, 37–50. [Google Scholar] [CrossRef]
- Redondo, P.; Garcia-Foncillas, J.; Espana, A.; Cuevillas, F.; Quintanilla, E. Differential modulation of IL-8 and TNF-alpha expression in human keratinocytes by buflomedil chlorhydrate and pentoxifylline. Exp. Dermatol. 1997, 6, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Cataisson, C.; Pearson, A.J.; Tsien, M.Z.; Mascia, F.; Gao, J.L.; Pastore, S.; Yuspa, S.H. CXCR2 ligands and G-CSF mediate PKCalpha-induced intraepidermal inflammation. J. Clin. Investig. 2006, 116, 2757–2766. [Google Scholar] [CrossRef]
- Schneider, L.A.; Raizner, K.; Wlaschek, M.; Brenneisen, P.; Gethoffer, K.; Scharffetter-Kochanek, K. UVA-1 exposure in vivo leads to an IL-6 surge within the skin. Exp. Dermatol. 2017, 26, 830–832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szymanski, L.; Skopek, R.; Palusinska, M.; Schenk, T.; Stengel, S.; Lewicki, S.; Kraj, L.; Kaminski, P.; Zelent, A. Retinoic Acid and Its Derivatives in Skin. Cells 2020, 9, 2660. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]


| Extract Material | Name | Source | IC50 (µg/mL) |
|---|---|---|---|
| Seeds | Almond | Blue Mountain Organics | >5 |
| Avocado | Farmers Market | 6 | |
| Black Raspberry | Fruitsmart/FruitBasics | 6 | |
| Blueberry | Fruitsmart/FruitBasics | 8 | |
| Celery | Triarco | 7 | |
| Coffee | Umalaxmi Organics | 6 | |
| Cranberry | Fruitsmart/FruitBasics | 10 | |
| Fennel | Triarco | 1 | |
| Grape | Fruitsmart | 0.4 | |
| Guarana | Pharma Resources International | 1.5 | |
| Hazelnut | Bob’s Red Mill | >10 | |
| Red Raspberry | Fruitsmart/FruitBasics | 1 | |
| Fruits | Avocado | Farmers Market | >25 |
| Blueberry | Wilderness Family Naturals | >50 | |
| Cranberry | Wilderness Family Naturals | >30 | |
| Grapefruit (Oil) | Organic Herbal Essence | 25 | |
| Juniper Berry | Triarco | 5 | |
| Maqui Berry | Sunfood Superfoods | >10 | |
| Mulberry | Z Natural Foods | >10 | |
| Promegranate | Navitas | >27 | |
| Schisandra | Triarco | 6 | |
| Strawberry | Wilderness Family Naturals | >5 | |
| Root, Bark or Leafs | Maca Root | Triarco | 3 |
| Goldenseal Root | Triarco | 4 | |
| Magnolia Bark | Triarco | 2 | |
| Pygeum Bark | Triarco | 3 | |
| Red Raspberry Leaf | Starwest Botanicals | 2 | |
| Other | Cocoa Butter | Gourmet Imports | 12.5 |
| Cocoa Powder | Gourmet Imports | 6.25 | |
| Echinacea Angustifolia | Triarco | 4 | |
| Echinacea | Triarco | 12 |
| Compound | MW (g/mol) | Molecular Formula | IC50 (µM) |
|---|---|---|---|
| 2-(acetyloxy)-4-[3,7-bis(acetyloxy)-5-hydroxy-4-oxo-3,4-dihydro-2H-chromen-2-yl]phenyl acetate | 472.41 | C23H20O11 | >15 |
| 2′,4-dihydroxy-4′,6′-dimethoxychalcone | 300.32 | C14H12O3 | >25 |
| 3,5,7-Trihydroxy-3′,4′,5′-Trimethoxyflavone (Myricetin trimethyl ether) | 360.36 | C18H16O8 | >15 |
| 4-methylcatechol | 124.14 | C7H8O2 | >15 |
| 5,7-dimethoxy-4′hydroxyflavanone | 300.31 | C17H16O5 | >25 |
| Apigenin | 270.25 | C15H10O5 | >15 |
| Azoxystrobin | 403.39 | C22H17N3O5 | >15 |
| Baicalein | 270.24 | C15H10O5 | 3.4 |
| Biochanin a | 284.27 | C16H12O5 | ~15 |
| Caffeic Acid | 180.16 | C9H8O4 | >15 |
| Catechin | 290.28 | C15H14O6 | >15 |
| Chrysoeriol | 300.27 | C16H12O6 | ~25 |
| Cyanidin chloride | 322.7 | C15H11O6Cl | 4.3 |
| Delphinidin | 303.246 | C15H11O7+ | 5.1 |
| Diadzein | 254.24 | C15H12O4 | >15 |
| Diadzin | 416.38 | C21H20O9 | >15 |
| Diosmin | 608.55 | C28H32O15 | ~25 |
| Disometin | 300.27 | C16H12O6 | >25 |
| Epicatechin gallate | 442.37 | C22H18O10 | 1.4 |
| Epigallocatechin | 306.27 | C15H14O7 | >15 |
| Epigallocatechin gallate | 458.37 | C22H18O11 | 2.6 |
| Eriodictyol | 288.26 | C15H12O6 | >25 |
| Eugenol | 164.2 | C10H12O2 | >15 |
| Ferulic acid | 194.19 | C10H10O4 | >15 |
| Formonoetin | 268.26 | C16H12O4 | >15 |
| Forskolin | 410.51 | C22H34O7 | >25 |
| Gallic acid | 170.13 | C7H6O5 | >15 |
| Gossypetin | 318.24 | C15H10O8 | 4.9 |
| Iprodione | 330.17 | C13H13Cl2N3O3 | >15 |
| Isoorientin | 448.38 | C21H20O11 | >25 |
| Isoquercetin | 464.379 | C21H20O12 | >15 |
| Isorhamnetin (3,5,7,4′-Tetrahydroxy-3′-methoxyflavone) | 316.27 | C16H12O7 | >15 |
| Kaempferide | 300.27 | C16H12O6 | 2 |
| Luteolin | 286.25 | C15H10O6 | 1.7 |
| Luteolin 7-O-B-glucoside | 448.38 | C21H20O11 | >25 |
| Mangostine | 410.47 | C24H26O6 | 1.7 |
| Methyl 6,7-dimethoxycoumarin-4-acetate | 278.26 | C14H14O6 | >25 |
| Morin | 302.24 | C15H10O7 | >15 |
| Myclobutanil | 288.78 | C15H17ClN4 | >15 |
| Myricetin | 318.25 | C15H10O8 | 0.99 |
| Naringin | 580.54 | C27H32O14 | >25 |
| Narirutin | 580.54 | C27H32O14 | >25 |
| Orientin | 448.38 | C21H20O11 | >25 |
| Phloroglucinol | 126.111 | C6H6O3 | >15 |
| Phosmet | 317.32 | C11H12NO4PS2 | >15 |
| Picrotin | 310.29 | C15H18O7 | >25 |
| Procyanidin B1 | 578.52 | C30H26O12 | >35 |
| Procyanidin B2 | 578.52 | C30H26O12 | >35 |
| Procyanidin B3 | 578.52 | C30H26O12 | >35 |
| Pyrogallol | 126.11 | C6H3(OH)3 | ~15 |
| Quercetagetin | 318.25 | C15H10O8 | ~0.5 |
| Quercetin | 302.24 | C15H10O7 | 2.5 |
| Resveratrol | 228.25 | C14H12O3 | >100 |
| Rutin | 610.52 | C27H30O16 | >15 |
| Sciadopitysin | 580.547 | C33H24O10 | 1.1 |
| Scutellarin | 462.36 | C21H18O12 | 4.9 |
| Syringic acid | 198.18 | C9H10O5 | >15 |
| Tebuconazole | 307.82 | C16H22ClN3O | >15 |
| Vanillic acid | 168.15 | C8H8O4 | >15 |
| Material | IC50 (µg/mL) * | |||
|---|---|---|---|---|
| NHDFs-UVA-IL-6 | NHEK-UVB-IL-8 | NHEK-UVB-TNFα | NHEK-TPA-IL-8 | |
| Vitamin C | >10 | >10 | >10 | - |
| Ferulic Acid | 0.01 | <0.01 | <0.01 | - |
| AGSE | 0.1 | 0.01 | 0.05 | 0.387 |
| ATRA | >1 | >1 | >1 | - |
| Clobetasol | - | - | - | 5 × 10−7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huber, K.L.; Fernández, J.R.; Webb, C.; Rouzard, K.; Healy, J.; Tamura, M.; Stock, J.B.; Stock, M.; Pérez, E. AGSE: A Novel Grape Seed Extract Enriched for PP2A Activating Flavonoids That Combats Oxidative Stress and Promotes Skin Health. Molecules 2021, 26, 6351. https://doi.org/10.3390/molecules26216351
Huber KL, Fernández JR, Webb C, Rouzard K, Healy J, Tamura M, Stock JB, Stock M, Pérez E. AGSE: A Novel Grape Seed Extract Enriched for PP2A Activating Flavonoids That Combats Oxidative Stress and Promotes Skin Health. Molecules. 2021; 26(21):6351. https://doi.org/10.3390/molecules26216351
Chicago/Turabian StyleHuber, Kristen L., José R. Fernández, Corey Webb, Karl Rouzard, Jason Healy, Masanori Tamura, Jeffry B. Stock, Maxwell Stock, and Eduardo Pérez. 2021. "AGSE: A Novel Grape Seed Extract Enriched for PP2A Activating Flavonoids That Combats Oxidative Stress and Promotes Skin Health" Molecules 26, no. 21: 6351. https://doi.org/10.3390/molecules26216351
APA StyleHuber, K. L., Fernández, J. R., Webb, C., Rouzard, K., Healy, J., Tamura, M., Stock, J. B., Stock, M., & Pérez, E. (2021). AGSE: A Novel Grape Seed Extract Enriched for PP2A Activating Flavonoids That Combats Oxidative Stress and Promotes Skin Health. Molecules, 26(21), 6351. https://doi.org/10.3390/molecules26216351







