Unravelling the Anticancer Mechanisms of Traditional Herbal Medicines with Metabolomics
Abstract
:1. Introduction
| FDA Approved Drug/Year | Initial Discovery | Plant Source | Cancer Type | Mechanism of Action | References |
|---|---|---|---|---|---|
| Paclitaxel/1992 | 1960s | Taxus brevifolia Nutt. (bark) | Breast, ovarian, lung, pancreatic | Stabilizes microtubule | [8] |
| Homoharringtonine/2012 | 1970s | Cephalotaxus harringtonii (Knight ex J.Forbes) K.Koch (bark) | Chronic myeloid leukaemia | Disables the elongation of peptide chain inhibiting protein synthesis | [9] |
| Camptothecin/1996 | 1960s | Camptotheca acuminata Decne (bark and stem) | Gastrointestinal, ovarian, small-cell lung | Inhibits deoxyribonucleic acid (DNA) re-ligation through interaction with topoisomerase-type I DNA complex causing DNA damage | [10] |
| Vincristine sulphate/1963 | 1950s | Catharanthus roseus (Linnaeus) G.Don (leaf) | Leukemia | Inhibits the formation of microtubules and interferes with nucleic acid and protein synthesis by blocking glutamic acid utilization | [11] |
| Vinblastine sulphate/1965 | 1950s | Catharanthus roseus (leaf) | Lymphoma, choriocarcinoma, breast | Inhibits microtubule formation resulting in cell cycle arrest | [11] |
| Teniposide (semisynthetic analogues of podophyllotoxin)/1990 | 1960s | Podophyllum peltatum Linnaeus (rhizome) | Leukaemia | Inhibits type II DNA topoisomerase complex | [12] |
| Etoposide (semisynthetic analogues of podophyllotoxin)/1983 | 1960s | Podophyllum peltatum Linnaeus (rhizome) | Testes, lung | Inhibits type II DNA topoisomerase complex | [13] |
2. Metabolomics: An Indispensable Tool for Anticancer Drug Discovery from Plants
- Genomics, epigenetics, proteomics, and transcriptomics ultimately converge to metabolomics, making metabolic profiling indispensable to uncover the molecular target of medicinal plants [15].
- Analyses of subtle changes in metabolite levels in many metabolic pathways aid in hypothesis generation, enabling a top-down approach in discovering mechanisms of drug action [16].
- In the investigation for anticancer efficacy of potential therapeutics, metabolite biomarkers are stable end products that may be more reliable as indices of the initiation or progression of cancers than mRNA or proteins [17].
- Metabolomics offers the possibility of conducting non-invasive, large-scale studies (using bio-fluids like plasma, serum, and urine samples) to determine the efficacy of medicinal plants and their chemopreventive potential in the malignant transformation of normal cells [18].
- Metabolomics offers the opportunity to decipher the pharmacokinetics of herbal medicines and possible interactions with conventional anticancer drugs [19]. It is a tool to assess potential drug toxicity and to avoid drug withdrawals in early-phase clinical trials due to toxicity concerns [20]. These pharmacological evaluations are essential during the long-term use of traditional herbal medicines.
- Metabolic profiling is useful for the authentication and standardization of traditional herbal medicines [21]. It can be a quick preliminary guideline to uncover the most dominant compound related to the anticancer activity or predict potential herb-drug interactions and toxicity. Besides, the seasonal variations in plant components can also be studied using metabolomics [22].
3. Cancer Metabolic Reprogramming
3.1. Addiction to Glucose, Glutamine, and Other Amino Acids
3.2. Enhanced Lipid Utilization
3.3. Oncogenic Activation of Metabolic Pathways
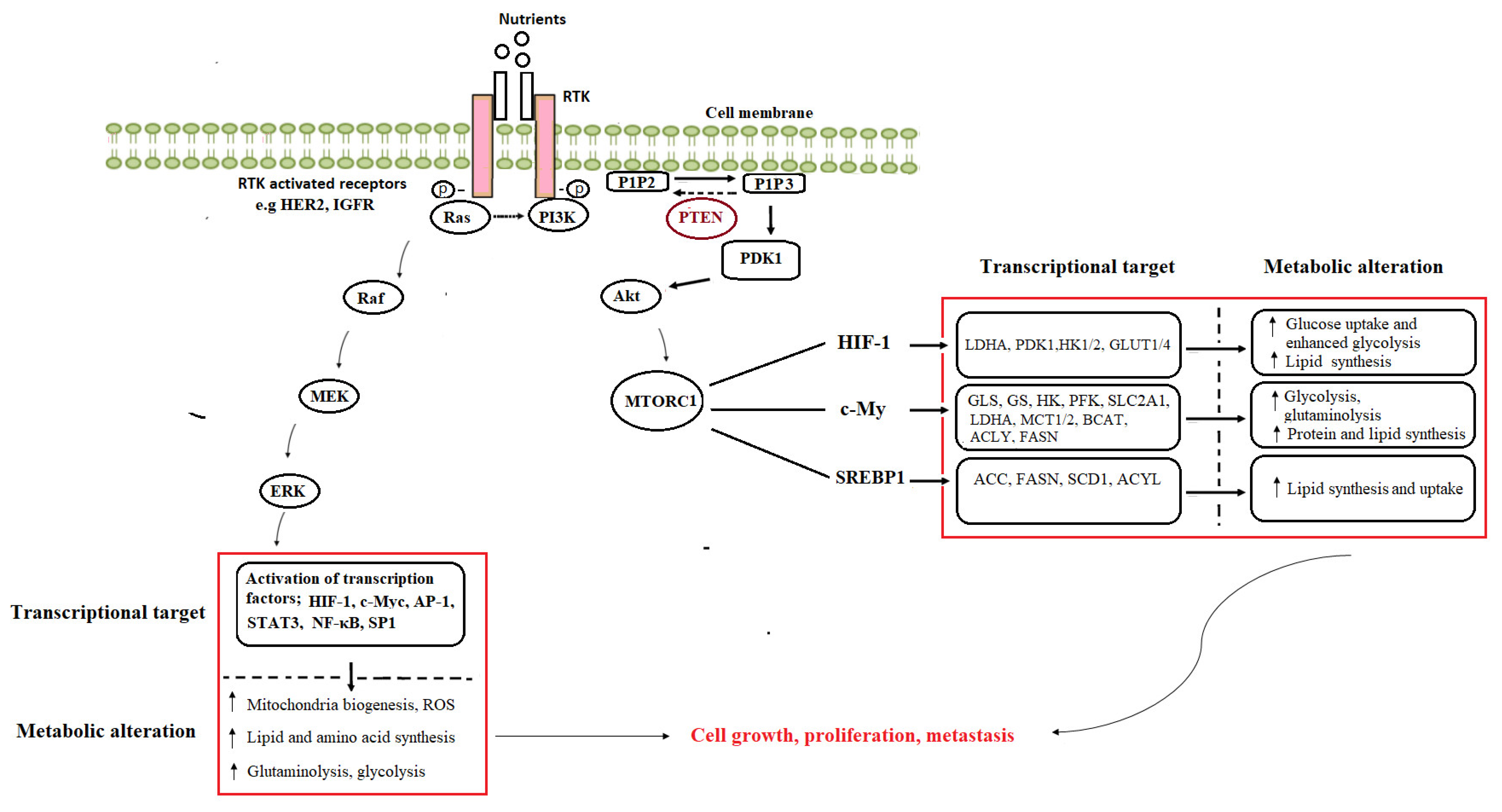
4. Medicinal Plants as Modulators of Cancer Metabolism
5. Deciphering the Anticancer Mechanisms of Traditional (African Chinese, Indian) Herbal Medicines with Metabolomics
5.1. Curcuma Longa Linnaeus
5.2. Zingiber Officinale Roscoe
5.3. Glycyrrhiza Species (G. glabra L, G. uralensis Fisch. and G. inflata Batalin)
5.4. Nigella sativa Linnaeus
5.5. Crithmum maritimum Linnaeus
5.6. Cyperus rotundus Linnaeus
6. Understudied Herbal Medicines as Potential Candidates for Cancer Metabolomics Studies
7. Metabolomics Approach in the Quality Control and Safety Evaluation of Herbal Exposure
8. Challenges
9. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Wei, Q.; Zhou, Y.; Wang, J.; Liu, Q.; Xu, H. A systematic analysis of FDA-approved anticancer drugs. BMC Syst. Biol. 2017, 11, 87. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasir, J.K.; Labhasetwar, V. Targeted drug delivery in cancer therapy. Technol. Cancer Res. Treat. 2005, 4, 363–374. [Google Scholar] [CrossRef]
- Langlois-Klassen, D.; Kipp, W.; Jhangri, G.S.; Rubaale, T. Use of traditional herbal medicine by AIDS patients in Kabarole District, western Uganda. Am. J. Trop. Med. Hyg. 2007, 77, 757–763. [Google Scholar] [CrossRef] [Green Version]
- Mwaka, A.D.; Abbo, C.; Kinengyere, A.A. Traditional and Complementary Medicine Use among Adult Cancer Patients Undergoing Conventional Treatment in Sub-Saharan Africa: A Scoping Review on the Use, Safety and Risks. Cancer Manag. Res. 2020, 12, 3699–3712. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.G.; Winson, M.K.; Kell, D.B.; Baganz, F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998, 16, 373–378. [Google Scholar] [CrossRef]
- Kampan, N.C.; Madondo, M.T.; McNally, O.M.; Quinn, M.; Plebanski, M. Paclitaxel and its evolving role in the management of ovarian cancer. BioMed. Res. Int. 2015, 2015, 413076. [Google Scholar] [CrossRef]
- Li, S.; Bo, Z.; Jiang, Y.; Song, X.; Wang, C.; Tong, Y. Homoharringtonine promotes BCR-ABL degradation through the p62-mediated autophagy pathway. Oncol. Rep. 2020, 43, 113–120. [Google Scholar] [CrossRef]
- Li, F.; Jiang, T.; Li, Q.; Ling, X. Camptothecin (CPT) and its derivatives are known to target topoisomerase I (Top1) as their mechanism of action: Did we miss something in CPT analogue molecular targets for treating human disease such as cancer? Am. J. Cancer Res. 2017, 7, 2350–2394. [Google Scholar]
- Martino, E.; Casamassima, G.; Castiglione, S.; Cellupica, E.; Pantalone, S.; Papagni, F.; Rui, M.; Siciliano, A.M.; Collina, S. Vinca alkaloids and analogues as anti-cancer agents: Looking back, peering ahead. Bioorg. Med. Chem. Lett. 2018, 28, 2816–2826. [Google Scholar] [CrossRef]
- Hirschfeld, S.; Ho, P.T.; Smith, M.; Pazdur, R. Regulatory approvals of pediatric oncology drugs: Previous experience and new initiatives. J. Clin. Oncol. 2003, 21, 1066–1073. [Google Scholar] [CrossRef]
- Montecucco, A.; Zanetta, F.; Biamonti, G. Molecular mechanisms of etoposide. EXCLI J. 2015, 14, 95. [Google Scholar] [PubMed]
- Wang, X.X.; Yu, P.C.; Li, J. High-Throughput Metabolomics for Identification of Metabolic Pathways and Deciphering the Effect Mechanism of Dioscin on Rectal Cancer From Cell Metabolic Profiles Coupled with Chemometrics Analysis. Front. Pharm. 2020, 11, 68. [Google Scholar] [CrossRef]
- Shah, N.J.; Sureshkumar, S.; Shewade, D.G. Metabolomics: A tool ahead for understanding molecular mechanisms of drugs and diseases. Indian J. Clin. Biochem. 2015, 30, 247–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prosser, G.A.; Larrouy-Maumus, G.; de Carvalho, L.P.S. Metabolomic strategies for the identification of new enzyme functions and metabolic pathways. EMBO Rep. 2014, 15, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Ahmed, S. Emerging field of metabolomics: Big promise for cancer biomarker identification and drug discovery. J. Pharm. Biomed. Anal. 2015, 107, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D. A review of applications of metabolomics in cancer. Metabolites 2013, 3, 552–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, K.; Xie, G.; Jia, W. Towards polypharmacokinetics: Pharmacokinetics of multicomponent drugs and herbal medicines using a metabolomics approach. Evid.-Based Complementary Altern. Med. 2013, 2013, 819147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, L.; Guo, L.; Wang, L.; Yin, Q.; Zhang, C.-M.; Zheng, Y.-G.; Liu, E.-H. Application of metabolomics in toxicity evaluation of traditional Chinese medicines. Chin. Med. 2018, 13, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.-M.; Jeon, J.-Y.; Lee, B.-J.; Lee, H.; Choi, H.-K. Application of metabolomics to quality control of natural product derived medicines. Biomol. Ther. 2017, 25, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramírez-Briones, E.; Rodríguez-Macías, R.; Salcedo-Pérez, E.; Ramírez-Chávez, E.; Molina-Torres, J.; Tiessen, A.; Ordaz-Ortiz, J.; Martínez-Gallardo, N.; Délano-Frier, J.P.; Zañudo-Hernández, J. Seasonal changes in the metabolic profiles and biological activity in leaves of Diospyros digyna and D. rekoi “Zapote” trees. Plants 2019, 8, 449. [Google Scholar] [CrossRef] [Green Version]
- Commisso, M.; Strazzer, P.; Toffali, K.; Stocchero, M.; Guzzo, F. Untargeted metabolomics: An emerging approach to determine the composition of herbal products. Comput. Struct. Biotechnol. J. 2013, 4, e201301007. [Google Scholar] [CrossRef] [Green Version]
- De Vos, R.C.; Moco, S.; Lommen, A.; Keurentjes, J.J.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778. [Google Scholar] [CrossRef]
- Kruger, N.J.; Troncoso-Ponce, M.A.; Ratcliffe, R.G. 1 H NMR metabolite fingerprinting and metabolomic analysis of perchloric acid extracts from plant tissues. Nat. Protoc. 2008, 3, 1001–1012. [Google Scholar] [CrossRef]
- Sumner, L.W.; Mendes, P.; Dixon, R.A. Plant metabolomics: Large-scale phytochemistry in the functional genomics era. Phytochemistry 2003, 62, 817–836. [Google Scholar] [CrossRef] [Green Version]
- Lu, W.; Su, X.; Klein, M.S.; Lewis, I.A.; Fiehn, O.; Rabinowitz, J.D. Metabolite Measurement: Pitfalls to Avoid and Practices to Follow. Annu. Rev. Biochem. 2017, 86, 277–304. [Google Scholar] [CrossRef]
- Warburg, O. On respiratory impairment in cancer cells. Science 1956, 124, 269–270. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Moreno-Sanchez, R.; Rodriguez-Enriquez, S.; Marin-Hernandez, A.; Saavedra, E. Energy metabolism in tumor cells. FEBS J. 2007, 274, 1393–1418. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg effect: How does it benefit cancer cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [Green Version]
- Nagao, A.; Kobayashi, M.; Koyasu, S.; Chow, C.C.T.; Harada, H. HIF-1-Dependent Reprogramming of Glucose Metabolic Pathway of Cancer Cells and Its Therapeutic Significance. Int. J. Mol. Sci. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bott, A.J.; Maimouni, S.; Zong, W.X. The Pleiotropic Effects of Glutamine Metabolism in Cancer. Cancers 2019, 11, 770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altman, B.J.; Stine, Z.E.; Dang, C.V. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer 2016, 16, 619–634. [Google Scholar] [CrossRef] [PubMed]
- Gaucher, C.; Boudier, A.; Bonetti, J.; Clarot, I.; Leroy, P.; Parent, M. Glutathione: Antioxidant Properties Dedicated to Nanotechnologies. Antioxidants 2018, 7, 62. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.K.; Park, K.G. Targeting Glutamine Metabolism for Cancer Treatment. Biomol. Ther. 2018, 26, 19–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koundouros, N.; Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef] [Green Version]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef] [Green Version]
- Baenke, F.; Peck, B.; Miess, H.; Schulze, A. Hooked on fat: The role of lipid synthesis in cancer metabolism and tumour development. Dis. Models Mech. 2013, 6, 1353–1363. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Huang, J. The expanded role of fatty acid metabolism in cancer: New aspects and targets. Precis. Clin. Med. 2019, 2, 183–191. [Google Scholar] [CrossRef] [Green Version]
- Santos, C.R.; Schulze, A. Lipid metabolism in cancer. FEBS J. 2012, 279, 2610–2623. [Google Scholar] [CrossRef]
- Hoxhaj, G.; Manning, B.D. The PI3K–AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 2020, 20, 74–88. [Google Scholar] [CrossRef]
- Jiang, N.; Dai, Q.; Su, X.; Fu, J.; Feng, X.; Peng, J. Role of PI3K/AKT pathway in cancer: The framework of malignant behavior. Mol. Biol. Rep. 2020, 47, 4587–4629. [Google Scholar] [CrossRef]
- Marbaniang, C.; Kma, L. Dysregulation of glucose metabolism by oncogenes and tumor suppressors in cancer cells. Asian Pac. J. Cancer Prev. APJCP 2018, 19, 2377. [Google Scholar] [PubMed]
- Manning, B.D.; Toker, A. AKT/PKB Signaling: Navigating the Network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef] [Green Version]
- Lukey, M.J.; Wilson, K.F.; Cerione, R.A. Therapeutic strategies impacting cancer cell glutamine metabolism. Future Med. Chem. 2013, 5, 1685–1700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Y.; Tu, R.; Liu, H.; Qing, G. Regulation of cancer cell metabolism: Oncogenic MYC in the driver’s seat. Signal Transduct. Target. Ther. 2020, 5, 124. [Google Scholar] [CrossRef]
- Jun, J.C.; Rathore, A.; Younas, H.; Gilkes, D.; Polotsky, V.Y. Hypoxia-inducible factors and cancer. Curr. Sleep Med. Rep. 2017, 3, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, D.; Bell, E.H.; Mischel, P.; Chakravarti, A. Targeting SREBP-1-driven lipid metabolism to treat cancer. Curr. Pharm. Des. 2014, 20, 2619–2626. [Google Scholar] [CrossRef] [Green Version]
- Aquila, S.; Santoro, M.; Caputo, A.; Panno, M.L.; Pezzi, V.; De Amicis, F. The tumor suppressor PTEN as molecular switch node regulating cell metabolism and autophagy: Implications in immune system and tumor microenvironment. Cells 2020, 9, 1725. [Google Scholar] [CrossRef]
- Qin, J.J.; Li, X.; Hunt, C.; Wang, W.; Wang, H.; Zhang, R. Natural products targeting the p53-MDM2 pathway and mutant p53: Recent advances and implications in cancer medicine. Genes Dis. 2018, 5, 204–219. [Google Scholar] [CrossRef]
- Papa, S.; Choy, P.M.; Bubici, C. The ERK and JNK pathways in the regulation of metabolic reprogramming. Oncogene 2019, 38, 2223–2240. [Google Scholar] [CrossRef] [Green Version]
- Dhillon, A.S.; Hagan, S.; Rath, O.; Kolch, W. MAP kinase signalling pathways in cancer. Oncogene 2007, 26, 3279–3290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raez, L.E.; Papadopoulos, K.; Ricart, A.D.; Chiorean, E.G.; Dipaola, R.S.; Stein, M.N.; Rocha Lima, C.M.; Schlesselman, J.J.; Tolba, K.; Langmuir, V.K.; et al. A phase I dose-escalation trial of 2-deoxy-D-glucose alone or combined with docetaxel in patients with advanced solid tumors. Cancer Chemother. Pharm. 2013, 71, 523–530. [Google Scholar] [CrossRef]
- Pajak, B.; Siwiak, E.; Soltyka, M.; Priebe, A.; Zielinski, R.; Fokt, I.; Ziemniak, M.; Jaskiewicz, A.; Borowski, R.; Domoradzki, T.; et al. 2-Deoxy-d-Glucose and Its Analogs: From Diagnostic to Therapeutic Agents. Int. J. Mol. Sci. 2019, 21, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, T.; Feng, Q.; Wang, Z.; Li, W.; Sun, Z.; Wilhelm, J.; Huang, G.; Vo, T.; Sumer, B.D.; Gao, J. Tumor-Targeted Inhibition of Monocarboxylate Transporter 1 Improves T-Cell Immunotherapy of Solid Tumors. Adv. Healthc. Mater. 2021, 10, 2000549. [Google Scholar] [CrossRef] [PubMed]
- Stacpoole, P.W. Therapeutic targeting of the pyruvate dehydrogenase complex/pyruvate dehydrogenase kinase (PDC/PDK) axis in cancer. JNCI J. Natl. Cancer Inst. 2017, 109, djx071. [Google Scholar] [CrossRef] [Green Version]
- Myers, R.A.; Wirth, S.; Williams, S.; Kiel, P.J. Enasidenib: An oral IDH2 inhibitor for the treatment of acute myeloid leukemia. J. Adv. Pract. Oncol. 2018, 9, 435. [Google Scholar] [PubMed]
- Schulte, M.L.; Fu, A.; Zhao, P.; Li, J.; Geng, L.; Smith, S.T.; Kondo, J.; Coffey, R.J.; Johnson, M.O.; Rathmell, J.C. Pharmacological blockade of ASCT2-dependent glutamine transport leads to antitumor efficacy in preclinical models. Nat. Med. 2018, 24, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Falchook, G.; Infante, J.; Arkenau, H.-T.; Patel, M.R.; Dean, E.; Borazanci, E.; Brenner, A.; Cook, N.; Lopez, J.; Pant, S. First-in-human study of the safety, pharmacokinetics, and pharmacodynamics of first-in-class fatty acid synthase inhibitor TVB-2640 alone and with a taxane in advanced tumors. EClinicalMedicine 2021, 34, 100797. [Google Scholar] [CrossRef]
- Menendez, J.A.; Vellon, L.; Mehmi, I.; Oza, B.P.; Ropero, S.; Colomer, R.; Lupu, R. Inhibition of fatty acid synthase (FAS) suppresses HER2/neu (erbB-2) oncogene overexpression in cancer cells. Proc. Natl. Acad. Sci. USA 2004, 101, 10715–10720. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.M.; Huang, C.S.; Hsu, T.N.; Huang, M.S.; Fong, I.H.; Lee, W.H.; Liu, S.C. Disruption of Cancer Metabolic SREBP1/miR-142-5p Suppresses Epithelial-Mesenchymal Transition and Stemness in Esophageal Carcinoma. Cells 2019, 9, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- She, K.; Fang, S.; Du, W.; Fan, X.; He, J.; Pan, H.; Huang, L.; He, P.; Huang, J. SCD1 is required for EGFR-targeting cancer therapy of lung cancer via re-activation of EGFR/PI3K/AKT signals. Cancer Cell Int. 2019, 19, 103. [Google Scholar] [CrossRef]
- Jiang, L.; Zawacka-Pankau, J. The p53/MDM2/MDMX-targeted therapies—A clinical synopsis. Cell Death Dis. 2020, 11, 237. [Google Scholar] [CrossRef]
- Gluck, W.L.; Gounder, M.M.; Frank, R.; Eskens, F.; Blay, J.Y.; Cassier, P.A.; Soria, J.-C.; Chawla, S.; de Weger, V.; Wagner, A.J. Phase 1 study of the MDM2 inhibitor AMG 232 in patients with advanced P53 wild-type solid tumors or multiple myeloma. Investig. New Drugs 2020, 38, 831–843. [Google Scholar] [CrossRef] [Green Version]
- Fendt, S.-M. Is there a therapeutic window for metabolism-based cancer therapies? Front. Endocrinol. 2017, 8, 150. [Google Scholar] [CrossRef] [Green Version]
- Torres, M.P.; Rachagani, S.; Purohit, V.; Pandey, P.; Joshi, S.; Moore, E.D.; Johansson, S.L.; Singh, P.K.; Ganti, A.K.; Batra, S.K. Graviola: A novel promising natural-derived drug that inhibits tumorigenicity and metastasis of pancreatic cancer cells in vitro and in vivo through altering cell metabolism. Cancer Lett. 2012, 323, 29–40. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Xu, Q.; Yang, W.; Wu, T.; Lu, X. Oleanolic acid reduces aerobic glycolysis-associated proliferation by inhibiting yes-associated protein in gastric cancer cells. Gene 2019, 712, 143956. [Google Scholar] [CrossRef] [PubMed]
- Chai, X.X.; Le, Y.F.; Wang, J.C.; Mei, C.X.; Feng, J.F.; Zhao, H.; Wang, C.; Lu, D.Z. Carpesium abrotanoides (L.) Root as a Potential Source of Natural Anticancer Compounds: Targeting Glucose Metabolism and PKM2/HIF-1α Axis of Breast Cancer Cells. J. Food Sci. 2019, 84, 3825–3832. [Google Scholar] [CrossRef]
- Saunier, E.; Antonio, S.; Regazzetti, A.; Auzeil, N.; Laprevote, O.; Shay, J.W.; Coumoul, X.; Barouki, R.; Benelli, C.; Huc, L.; et al. Resveratrol reverses the Warburg effect by targeting the pyruvate dehydrogenase complex in colon cancer cells. Sci. Rep. 2017, 7, 6945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.J.; Chung, T.W.; Lee, J.H.; Kim, B.S.; Kim, E.Y.; Lee, S.O.; Ha, K.T. Water-extracted branch of Cinnamomum cassia promotes lung cancer cell apoptosis by inhibiting pyruvate dehydrogenase kinase activity. J. Pharm. Sci. 2018, 138, 146–154. [Google Scholar] [CrossRef]
- Chung, T.-W.; Lee, J.H.; Choi, H.-J.; Park, M.-J.; Kim, E.-Y.; Han, J.H.; Jang, S.B.; Lee, S.-O.; Lee, S.W.; Hang, J. Anemone rivularis inhibits pyruvate dehydrogenase kinase activity and tumor growth. J. Ethnopharmacol. 2017, 203, 47–54. [Google Scholar] [CrossRef]
- Kwak, C.-H.; Lee, J.-H.; Kim, E.-Y.; Han, C.W.; Kim, K.-J.; Lee, H.; Cho, M.; Jang, S.B.; Kim, C.-H.; Chung, T.-W. Huzhangoside A Suppresses Tumor Growth through Inhibition of Pyruvate Dehydrogenase Kinase Activity. Cancers 2019, 11, 712. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.S.; Jeong, S.-J.; Kim, J.-H.; Lee, H.-J.; Song, H.S.; Kim, M.-S.; Ko, E.; Lee, H.-J.; Khil, J.-H.; Jang, H.-J. The genome-wide expression profile of 1, 2, 3, 4, 6-penta-O-galloyl-β-D-glucose-treated MDA-MB-231 breast cancer cells: Molecular target on cancer metabolism. Mol. Cells 2011, 32, 123. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, D.; Han, S.; Wang, N.; Mo, F.; Loo, T.Y.; Shen, J.; Huang, H.; Chen, J. Bioactivity-guided identification and cell signaling technology to delineate the lactate dehydrogenase A inhibition effects of Spatholobus suberectus on breast cancer. PLoS ONE 2013, 8, e56631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.Y.; Choi, H.J.; Park, M.J.; Jung, Y.S.; Lee, S.O.; Kim, K.J.; Choi, J.H.; Chung, T.W.; Ha, K.T. Myristica fragrans Suppresses Tumor Growth and Metabolism by Inhibiting Lactate Dehydrogenase A. Am. J. Chin. Med. 2016, 44, 1063–1079. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Islam, M.S.; Tian, J.; Lui, V.W.; Xiao, D. Inactivation of ATP citrate lyase by Cucurbitacin B: A bioactive compound from cucumber, inhibits prostate cancer growth. Cancer Lett. 2014, 349, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, J.; Yuan, L.; Lay, Y.; Wong, Y.K.; Lim, T.K.; Ong, C.S.; Lin, Q.; Wang, J.; Hua, Z. Andrographolide suppresses MV4-11 cell proliferation through the inhibition of FLT3 signaling, fatty acid synthesis and cellular iron uptake. Molecules 2017, 22, 1444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menendez, J.A.; Vazquez-Martin, A.; Oliveras-Ferraros, C.; Garcia-Villalba, R.; Carrasco-Pancorbo, A.; Fernandez-Gutierrez, A.; Segura-Carretero, A. Analyzing effects of extra-virgin olive oil polyphenols on breast cancer-associated fatty acid synthase protein expression using reverse-phase protein microarrays. Int. J. Mol. Med. 2008, 22, 433–439. [Google Scholar] [CrossRef] [Green Version]
- Messeha, S.; Zarmouh, N.; Taka, E.; Gendy, S.; Shokry, G.; Kolta, M.; Soliman, K. The role of monocarboxylate transporters and their chaperone cd147 in lactate efflux inhibition and the anticancer effects of Terminalia chebula in neuroblastoma cell line n2-a. Eur. J. Med. Plants 2016, 12, EJMP.23992. [Google Scholar] [CrossRef] [Green Version]
- Lodi, A.; Saha, A.; Lu, X.; Wang, B.; Sentandreu, E.; Collins, M.; Kolonin, M.G.; DiGiovanni, J.; Tiziani, S. Combinatorial treatment with natural compounds in prostate cancer inhibits prostate tumor growth and leads to key modulations of cancer cell metabolism. NPJ Precis. Oncol. 2017, 1, 18. [Google Scholar] [CrossRef]
- Hajrah, N.H.; Abdul, W.M.; Al-Garni, S.M.; Sheikh, A.; Ahmed, M.M.M.; Hall, N.; Saini, K.S.; Mohammad Sabir, J.S.; Bora, R.S. Gene expression profiling to elucidate the pharmacological and toxicological effects of Ricinus communis L. leaf extract in mammalian cells. Biotechnol. Biotechnol. Equip. 2019, 33, 397–407. [Google Scholar] [CrossRef] [Green Version]
- Othman, F.; Motalleb, G.; Peng, S.L.T.; Rahmat, A.; Basri, R.; Pei, C.P. Effect of neem leaf extract (Azadirachta indica) on c-Myc oncogene expression in 4T1 breast cancer cells of BALB/c mice. Cell J. Yakhteh 2012, 14, 53. [Google Scholar]
- Xu, T.; Pang, Q.; Wang, Y.; Yan, X. Betulinic acid induces apoptosis by regulating PI3K/Akt signaling and mitochondrial pathways in human cervical cancer cells. Int. J. Mol. Med. 2017, 40, 1669–1678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Lu, P.; Song, G.; Liu, Q.; Zhu, D.; Liu, X. Involvement of PI3K/Akt, ERK and p38 signaling pathways in emodin-mediated extrinsic and intrinsic human hepatoblastoma cell apoptosis. Food Chem. Toxicol. 2016, 92, 26–37. [Google Scholar] [CrossRef]
- Kim, A.; Im, M.; Ma, J.Y. Ethanol extract of Remotiflori radix induces endoplasmic reticulum stress-mediated cell death through AMPK/mTOR signaling in human prostate cancer cells. Sci. Rep. 2015, 5, 8394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddiqui, F.A.; Prakasam, G.; Chattopadhyay, S.; Rehman, A.U.; Padder, R.A.; Ansari, M.A.; Irshad, R.; Mangalhara, K.; Bamezai, R.N.; Husain, M. Curcumin decreases Warburg effect in cancer cells by down-regulating pyruvate kinase M2 via mTOR-HIF1α inhibition. Sci. Rep. 2018, 8, 8323. [Google Scholar] [CrossRef]
- Motadi, L.R.; Choene, M.S.; Mthembu, N.N. Anticancer properties of Tulbaghia violacea regulate the expression of p53-dependent mechanisms in cancer cell lines. Sci. Rep. 2020, 10, 12924. [Google Scholar] [CrossRef]
- Kong, Y.; Lu, Z.L.; Wang, J.J.; Zhou, R.; Guo, J.; Liu, J.; Sun, H.L.; Wang, H.; Song, W.; Yang, J.; et al. Platycodin D, a metabolite of Platycodin grandiflorum, inhibits highly metastatic MDA-MB-231 breast cancer growth in vitro and in vivo by targeting the MDM2 oncogene. Oncol. Rep. 2016, 36, 1447–1456. [Google Scholar] [CrossRef]
- Arafa el, S.A.; Zhu, Q.; Shah, Z.I.; Wani, G.; Barakat, B.M.; Racoma, I.; El-Mahdy, M.A.; Wani, A.A. Thymoquinone up-regulates PTEN expression and induces apoptosis in doxorubicin-resistant human breast cancer cells. Mutat. Res. 2011, 706, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Ghorbani, A.; Zand, H.; Jeddi-Tehrani, M.; Koohdani, F.; Shidfar, F.; Keshavarz, S.A. PTEN over-expression by resveratrol in acute lymphoblastic leukemia cells along with suppression of AKT/PKB and ERK1/2 in genotoxic stress. J. Nat. Med. 2015, 69, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Quan, H.Y.; Kim, S.J.; Jo, H.K.; Kim, G.W.; Chung, S.H. Betulinic acid alleviates non-alcoholic fatty liver by inhibiting SREBP1 activity via the AMPK–mTOR–SREBP signaling pathway. Biochem. Pharmacol. 2013, 85, 1330–1340. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, Z.; Zhang, S.; Ren, J.; Zhang, R.; Zeng, H.; Li, Q.; Wu, G. Inhibitory action of Celastrol on hypoxia-mediated angiogenesis and metastasis via the HIF-1α pathway. Int. J. Mol. Med. 2011, 27, 407–415. [Google Scholar] [PubMed]
- Li, Y.; Gan, C.; Zhang, Y.; Yu, Y.; Fan, C.; Deng, Y.; Zhang, Q.; Yu, X.; Zhang, Y.; Wang, L.; et al. Inhibition of Stat3 Signaling Pathway by Natural Product Pectolinarigenin Attenuates Breast Cancer Metastasis. Front Pharm. 2019, 10, 1195. [Google Scholar] [CrossRef] [Green Version]
- Che, C.-T.; George, V.; Ijinu, T.; Pushpangadan, P.; Andrae-Marobela, K. Traditional medicine. In Pharmacognosy; Elsevier: Amsterdam, The Netherlands, 2017; pp. 15–30. [Google Scholar]
- Xiang, Y.; Guo, Z.; Zhu, P.; Chen, J.; Huang, Y. Traditional Chinese medicine as a cancer treatment: Modern perspectives of ancient but advanced science. Cancer Med. 2019, 8, 1958–1975. [Google Scholar] [CrossRef]
- Sen, S.; Chakraborty, R. Herbal Medicine in India: Indigenous Knowledge, Practice, Innovation and Its Value; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Mahomoodally, M.F. Traditional medicines in Africa: An appraisal of ten potent African medicinal plants. Evid.-Based Complementary Altern. Med. 2013, 2013, 617459. [Google Scholar] [CrossRef] [Green Version]
- Kuttan, R.; Sudheeran, P.; Josph, C. Turmeric and curcumin as topical agents in cancer therapy. Tumori J. 1987, 73, 29–31. [Google Scholar] [CrossRef]
- Mansouri, K.; Rasoulpoor, S.; Daneshkhah, A.; Abolfathi, S.; Salari, N.; Mohammadi, M.; Rasoulpoor, S.; Shabani, S. Clinical effects of curcumin in enhancing cancer therapy: A systematic review. BMC Cancer 2020, 20, 791. [Google Scholar] [CrossRef]
- Sultana, S.; Munir, N.; Mahmood, Z.; Riaz, M.; Akram, M.; Rebezov, M.; Kuderinova, N.; Moldabayeva, Z.; Shariati, M.A.; Rauf, A. Molecular targets for the management of cancer using Curcuma longa Linn. phytoconstituents: A Review. Biomed. Pharmacother. 2021, 135, 111078. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.-L.; Zheng, J.-Y.; Cheng, X.-Q.; Xin, G.-Z.; Wang, S.-L.; Xie, T. Chemical markers’ knockout coupled with UHPLC-HRMS-based metabolomics reveals anti-cancer integration effects of the curcuminoids of turmeric (Curcuma longa L.) on lung cancer cell line. J. Pharm. Biomed. Anal. 2019, 175, 112738. [Google Scholar] [CrossRef]
- Kukula-Koch, W.; Grabarska, A.; Luszczki, J.; Czernicka, L.; Nowosadzka, E.; Gumbarewicz, E.; Jarzab, A.; Audo, G.; Upadhyay, S.; Glowniak, K.; et al. Superior anticancer activity is demonstrated by total extract of Curcuma longa L. as opposed to individual curcuminoids separated by centrifugal partition chromatography. Phytother. Res. 2018, 32, 933–942. [Google Scholar] [CrossRef] [Green Version]
- Bayet-Robert, M.; Morvan, D. Metabolomics reveals metabolic targets and biphasic responses in breast cancer cells treated by curcumin alone and in association with docetaxel. PLoS ONE 2013, 8, e57971. [Google Scholar] [CrossRef]
- Bhandari, M.; Ravipati, A.S.; Reddy, N.; Koyyalamudi, S.R. Traditional ayurvedic medicines: Pathway to develop anti-cancer drugs. J. Mol. Pharm. Org. Process Res. 2015, 3. [Google Scholar] [CrossRef]
- Zhu, L.; Li, L.; Li, Y.; Wang, J.; Wang, Q. Chinese herbal medicine as an adjunctive therapy for breast cancer: A systematic review and meta-analysis. Evid.-Based Complementary Altern. Med. 2016, 2016, 9469276. [Google Scholar] [CrossRef] [Green Version]
- Kabbaj, F.; Meddah, B.; Cherrah, Y.; Faouzi, E. Ethnopharmacological profile of traditional plants used in Morocco by cancer patients as herbal therapeutics. Phytopharmacology 2012, 2, 243–256. [Google Scholar]
- Manju, V.; Viswanathan, P.; Nalini, N. Hypolipidemic effect of ginger in 1, 2-dimethyl hydrazine-induced experimental colon carcinogenesis. Toxicol. Mech. Methods 2006, 16, 461–472. [Google Scholar] [CrossRef]
- Plengsuriyakarn, T.; Viyanant, V.; Eursitthichai, V.; Tesana, S.; Chaijaroenkul, W.; Itharat, A.; Na-Bangchang, K. Cytotoxicity, toxicity, and anticancer activity of Zingiber officinale Roscoe against cholangiocarcinoma. Asian Pac. J. Cancer Prev. 2012, 13, 4597–4606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Citronberg, J.; Bostick, R.; Ahearn, T.; Turgeon, D.K.; Ruffin, M.T.; Djuric, Z.; Sen, A.; Brenner, D.E.; Zick, S.M. Effects of ginger supplementation on cell-cycle biomarkers in the normal-appearing colonic mucosa of patients at increased risk for colorectal cancer: Results from a pilot, randomized, and controlled trial. Cancer Prev. Res. 2013, 6, 271–281. [Google Scholar] [CrossRef] [Green Version]
- Parvizzadeh, N.; Sadeghi, S.; Irani, S.; Iravani, A.; Kalayee, Z.; Rahimi, N.; Azadi, M.; Zamani, Z. A metabonomic study of the effect of methanol extract of ginger on Raji cells using 1HNMR spectroscopy. Biotechnol. Res. Int. 2014, 2014, 572534. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Y.; Gong, H.; Zhu, H. A systematic review of the comparison of three medicinal licorices, based on differences of the types and contents about their bioactive components. J. Chem. Biol. Pharm. Chem. 2018, 1, 1–6. [Google Scholar]
- Chebat, A.; Skalli, S.; Errihani, H.; Boulaâmane, L.; Mokrim, M.; Mahfoud, T.; Soulaymani, R.; Kahouadji, A. Étude de prévalence des effets indésirables liés à l’utilisation des plantes médicinales par les patients de l’Institut National d’Oncologie, Rabat. Phytothérapie 2014, 12, 25–32. [Google Scholar] [CrossRef]
- Lam, W.; Jiang, Z.; Guan, F.; Huang, X.; Hu, R.; Wang, J.; Bussom, S.; Liu, S.-H.; Zhao, H.; Yen, Y. PHY906 (KD018), an adjuvant based on a 1800-year-old Chinese medicine, enhanced the anti-tumor activity of Sorafenib by changing the tumor microenvironment. Sci. Rep. 2015, 5, 9384. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.-H.; Cheng, Y.-C. Old formula, new Rx: The journey of PHY906 as cancer adjuvant therapy. J. Ethnopharmacol. 2012, 140, 614–623. [Google Scholar] [CrossRef] [Green Version]
- Pastorino, G.; Cornara, L.; Soares, S.; Rodrigues, F.; Oliveira, M.B.P. Liquorice (Glycyrrhiza glabra): A phytochemical and pharmacological review. Phytother. Res. 2018, 32, 2323–2339. [Google Scholar] [CrossRef]
- Batiha, G.E.-S.; Beshbishy, A.M.; El-Mleeh, A.; Abdel-Daim, M.M.; Devkota, H.P. Traditional uses, bioactive chemical constituents, and pharmacological and toxicological activities of Glycyrrhiza glabra L. (Fabaceae). Biomolecules 2020, 10, 352. [Google Scholar] [CrossRef] [Green Version]
- Tang, F.; Xie, C.; Huang, D.; Wu, Y.; Zeng, M.; Yi, L.; Wang, Y.; Mei, W.; Cao, Y.; Sun, L. Novel potential markers of nasopharyngeal carcinoma for diagnosis and therapy. Clin. Biochem. 2011, 44, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Amelio, I.; Cutruzzola, F.; Antonov, A.; Agostini, M.; Melino, G. Serine and glycine metabolism in cancer. Trends Biochem. Sci. 2014, 39, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Sousa, C.M.; Biancur, D.E.; Wang, X.; Halbrook, C.J.; Sherman, M.H.; Zhang, L.; Kremer, D.; Hwang, R.F.; Witkiewicz, A.K.; Ying, H. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 2016, 536, 479–483. [Google Scholar] [CrossRef] [Green Version]
- Seo, M.; Crochet, R.; Lee, Y. Chapter 14—Targeting altered metabolism—emerging cancer therapeutic strategies. In Cancer Drug Design and Discovery, 2nd ed.; Neidle, S., Ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 427–448. [Google Scholar]
- Randhawa, M.A.; Alghamdi, M.S. Anticancer activity of Nigella sativa (black seed)—A review. Am. J. Chin. Med. 2011, 39, 1075–1091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baharetha, H.M.; Nassar, Z.D.; Aisha, A.F.; Ahamed, M.B.K.; Al-Suede, F.S.R.; Kadir, M.O.A.; Ismail, Z.; Majid, A.M.S.A. Proapoptotic and antimetastatic properties of supercritical CO2 extract of Nigella sativa Linn. against breast cancer cells. J. Med. Food 2013, 16, 1121–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agbaria, R.; Gabarin, A.; Dahan, A.; Ben-Shabat, S. Anticancer activity of Nigella sativa (black seed) and its relationship with the thermal processing and quinone composition of the seed. Drug Des. Dev. Ther. 2015, 9, 3119. [Google Scholar]
- Ait Mbarek, L.; Ait Mouse, H.; Elabbadi, N.; Bensalah, M.; Gamouh, A.; Aboufatima, R.; Benharref, A.; Chait, A.; Kamal, M.; Dalal, A. Anti-tumor properties of blackseed (Nigella sativa L.) extracts. Braz. J. Med. Biol. Res. 2007, 40, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Dogar, M.; Adnan, H.; Akhtar, M.S.; Sheikh, M.A. Prelimiary assessment of efficacy of Nigella sativa seeds in acute lymphoblastic leukemia local children. Pharmacologyonline 2009, 2, 769–777. [Google Scholar]
- Alghamdi, S.A. Effect of Nigella sativa and Foeniculum vulgare seeds extracts on male mice exposed to carbendazim. Saudi J. Biol. Sci. 2020, 27, 2521–2530. [Google Scholar] [CrossRef] [PubMed]
- Morad, S.A.; Cabot, M.C. Ceramide-orchestrated signalling in cancer cells. Nat. Rev. Cancer 2013, 13, 51–65. [Google Scholar] [CrossRef]
- Palma, C.D.; Perrotta, C. Ceramide as a target of chemotherapy: Its role in apoptosis and autophagy. Clin. Lipidol. 2012, 7, 111–119. [Google Scholar] [CrossRef]
- Gnocchi, D.; Cesari, G.; Calabrese, G.J.; Capone, R.; Sabbà, C.; Mazzocca, A. Inhibition of Hepatocellular Carcinoma Growth by Ethyl Acetate Extracts of Apulian Brassica oleracea L. and Crithmum maritimum L. Plant Food Hum. Nutr. 2020, 75, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Gnocchi, D.; Del Coco, L.; Girelli, C.R.; Castellaneta, F.; Cesari, G.; Sabbà, C.; Fanizzi, F.P.; Mazzocca, A. 1H-NMR metabolomics reveals a multitarget action of Crithmum maritimum ethyl acetate extract in inhibiting hepatocellular carcinoma cell growth. Sci. Rep. 2021, 11, 1259. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Song, X.; Ma, S.; Liu, C.; Sun, X.; Wang, X.; Liu, Z.; Liang, D.; Yu, Z. The treatment role of Cyperus rotundus L. to triple-negative breast cancer cells. Biosci. Rep. 2019, 39, BSR20190502. [Google Scholar] [CrossRef] [Green Version]
- Xiao, K.; Li, K.; Long, S.; Kong, C.; Zhu, S. Potential molecular mechanisms of Chaihu-Shugan-San in treatment of breast cancer based on network pharmacology. Evid.-Based Complementary Altern. Med. 2020, 2020, 3670309. [Google Scholar] [CrossRef]
- Ma, S.; Wang, F.; Zhang, C.; Wang, X.; Wang, X.; Yu, Z. Cell metabolomics to study the function mechanism of Cyperus rotundus L. on triple-negative breast cancer cells. BMC Complementary Med. Ther. 2020, 20, 262. [Google Scholar] [CrossRef]
- Mills, E.; Cooper, C.; Seely, D.; Kanfer, I. African herbal medicines in the treatment of HIV: Hypoxis and Sutherlandia. An overview of evidence and pharmacology. Nutr. J. 2005, 4, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Wyk, B.E. A broad review of commercially important southern African medicinal plants. J. Ethnopharmacol. 2008, 119, 342–355. [Google Scholar] [CrossRef]
- Aboyade, O.M.; Styger, G.; Gibson, D.; Hughes, G. Sutherlandia frutescens: The meeting of science and traditional knowledge. J. Altern. Complementary Med. 2014, 20, 71–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komakech, R.; Kang, Y.; Lee, J.-H.; Omujal, F. A review of the potential of phytochemicals from Prunus africana (Hook f.) Kalkman stem bark for chemoprevention and chemotherapy of prostate cancer. Evid.-Based Complementary Altern. Med. 2017, 2017, 3014019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tugume, P.; Kakudidi, E.K.; Buyinza, M.; Namaalwa, J.; Kamatenesi, M.; Mucunguzi, P.; Kalema, J. Ethnobotanical survey of medicinal plant species used by communities around Mabira Central Forest Reserve, Uganda. J. Ethnobiol. Ethnomed. 2016, 12, 5. [Google Scholar] [CrossRef] [Green Version]
- Anywar, G.; Kakudidi, E.; Byamukama, R.; Mukonzo, J.; Schubert, A.; Oryem-Origa, H. Indigenous traditional knowledge of medicinal plants used by herbalists in treating opportunistic infections among people living with HIV/AIDS in Uganda. J. Ethnopharmacol. 2020, 246, 112205. [Google Scholar] [CrossRef]
- Gouws, C.; Smit, T.; Willers, C.; Svitina, H.; Calitz, C.; Wrzesinski, K. Anticancer Potential of Sutherlandia frutescens and Xysmalobium undulatum in LS180 Colorectal Cancer Mini-Tumors. Molecules 2021, 26, 605. [Google Scholar] [CrossRef]
- Liu, J.; Liu, X.; Ma, J.; Li, K.; Xu, C. The clinical efficacy and safety of kanglaite adjuvant therapy in the treatment of advanced hepatocellular carcinoma: A PRISMA-compliant meta-analysis. Biosci. Rep. 2019, 39, BSR20193319. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Yu, L.; Ding, W. Efficacy and safety of Kanglaite injection combined with radiochemotherapy in the treatment of advanced pancreatic cancer: A PRISMA-compliant meta-analysis. Medicine 2019, 98, e16656. [Google Scholar] [CrossRef]
- Zhan, Y.-P.; Huang, X.-E.; Cao, J.; Lu, Y.-Y.; Wu, X.-Y.; Liu, J.; Xu, X.; Xiang, J.; Ye, L.-H. Clinical safety and efficacy of Kanglaite® (Coix Seed Oil) injection combined with chemotherapy in treating patients with gastric cancer. Asian Pac. J. Cancer Prev. 2012, 13, 5319–5321. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, W.; Wang, X.-J.; Liu, S. Antitumor effect of Kanglaite® injection in human pancreatic cancer xenografts. BMC Complementary Altern. Med. 2014, 14, 228. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Cai, X.; Zong, B.; Feng, R.; Ji, Y.; Chen, G.; Li, Z. Qianlie Xiaozheng decoction induces autophagy in human prostate cancer cells via inhibition of the Akt/mTOR pathway. Front. Pharmacol. 2018, 9, 234. [Google Scholar] [CrossRef] [Green Version]
- Trendafilova, A.; Moujir, L.M.; Sousa, P.; Seca, A.M. Research advances on health effects of edible Artemisia species and some sesquiterpene lactones constituents. Foods 2021, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Biswas, M.; Karan, T.; Bhattacharya, S.; Kumar, R.S.; Ghosh, A.; Haldar, P. Acute and sub-chronic toxicity study of Terminalia arjuna leaf in swiss albino mice. Pharmacologyonline 2011, 1, 366–371. [Google Scholar]
- Shyur, L.F.; Liu, C.P.; Chien, S.C. Metabolomics in herbal medicine research. Handb. Plant Metab. 2013, 155–174. [Google Scholar] [CrossRef]
- Oyenihi, A.B.; Smith, C. Are polyphenol antioxidants at the root of medicinal plant anti-cancer success? J. Ethnopharmacol. 2019, 229, 54–72. [Google Scholar] [CrossRef]
- Leenders, J.; Frederich, M.; de Tullio, P. Nuclear magnetic resonance: A key metabolomics platform in the drug discovery process. Drug Discov. Today Technol. 2015, 13, 39–46. [Google Scholar] [CrossRef]
- Nassar, A.F.; Wu, T.; Nassar, S.F.; Wisnewski, A.V. UPLC–MS for metabolomics: A giant step forward in support of pharmaceutical research. Drug Discov. Today 2017, 22, 463–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villate, A.; San Nicolas, M.; Gallastegi, M.; Aulas, P.-A.; Olivares, M.; Usobiaga, A.; Etxebarria, N.; Aizpurua-Olaizola, O. Review: Metabolomics as a prediction tool for plants performance under environmental stress. Plant Sci. 2021, 303, 110789. [Google Scholar] [CrossRef]
- Sun, S.; Wang, Y.; Wu, A.; Ding, Z.; Liu, X. Influence factors of the pharmacokinetics of herbal resourced compounds in clinical practice. Evid.-Based Complementary Altern. Med. 2019, 2019, 1983780. [Google Scholar] [CrossRef] [Green Version]
- Salem, M.A.; Perez de Souza, L.; Serag, A.; Fernie, A.R.; Farag, M.A.; Ezzat, S.M.; Alseekh, S. Metabolomics in the Context of Plant Natural Products Research: From Sample Preparation to Metabolite Analysis. Metabolites 2020, 10, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, G.; Zhao, A.; Zhao, L.; Chen, T.; Chen, H.; Qi, X.; Zheng, X.; Ni, Y.; Cheng, Y.; Lan, K.; et al. Metabolic fate of tea polyphenols in humans. J. Proteome Res. 2012, 11, 3449–3457. [Google Scholar] [CrossRef] [PubMed]
- Saigusa, D.; Matsukawa, N.; Hishinuma, E.; Koshiba, S. Identification of biomarkers to diagnose diseases and find adverse drug reactions by metabolomics. Drug Metab. Pharmacokinet. 2021, 37, 100373. [Google Scholar] [CrossRef]
- Chen, D.Q.; Chen, H.; Chen, L.; Tang, D.D.; Miao, H.; Zhao, Y.Y. Metabolomic application in toxicity evaluation and toxicological biomarker identification of natural product. Chem. Biol. Interact. 2016, 252, 114–130. [Google Scholar] [CrossRef]
- Zhang, P.-j.; Li, Y.-m.; Zhang, Y.-n.; Huang, W.; Li, Y.-b.; Zhang, Y.-j.; Liu, C.-x. Application and prospect of toxicity quality markers of Chinese materia medica based on metabolomics. Chin. Herb. Med. 2018, 10, 108–116. [Google Scholar] [CrossRef]
- Yan, Y.; Shi, N.; Han, X.; Li, G.; Wen, B.; Gao, J. UPLC/MS/MS-Based Metabolomics Study of the Hepatotoxicity and Nephrotoxicity in Rats Induced by Polygonum multiflorum Thunb. ACS Omega 2020, 5, 10489–10500. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.B.; Lee, B.M. Metabolomics, a New Promising Technology for Toxicological Research. Toxicol. Res. 2009, 25, 59–69. [Google Scholar] [CrossRef] [Green Version]
- Li, C.Y.; Tu, C.; Gao, D.; Wang, R.L.; Zhang, H.Z.; Niu, M.; Li, R.Y.; Zhang, C.E.; Li, R.S.; Xiao, X.H.; et al. Metabolomic Study on Idiosyncratic Liver Injury Induced by Different Extracts of Polygonum multiflorum in Rats Integrated with Pattern Recognition and Enriched Pathways Analysis. Front. Pharm. 2016, 7, 483. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.-N.; Liu, X.-M.; Fang, J.-H.; Zhu, X.; Yang, X.-W.; Xiao, X.-R.; Huang, J.-F.; Gonzalez, F.J.; Li, F. PPARα mediates the hepatoprotective effects of nutmeg. J. Proteome Res. 2018, 17, 1887–1897. [Google Scholar] [CrossRef]
- Li, F.; Yang, X.W.; Krausz, K.W.; Nichols, R.G.; Xu, W.; Patterson, A.D.; Gonzalez, F.J. Modulation of colon cancer by nutmeg. J. Proteome Res. 2015, 14, 1937–1946. [Google Scholar] [CrossRef]
- Robosky, L.C.; Robertson, D.G.; Baker, J.; Rane, S.; Reily, M.D. In Vivo toxicity screening programs using metabonomics. Comb. Chem. High Throughput Screen. 2002, 5, 651–662. [Google Scholar] [CrossRef]
- Robertson, D.G.; Watkins, P.B.; Reily, M.D. Metabolomics in toxicology: Preclinical and clinical applications. Toxicol. Sci. 2011, 120 (Suppl. 1), S146–S170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alseekh, S.; Fernie, A.R. Metabolomics 20 years on: What have we learned and what hurdles remain? Plant J. 2018, 94, 933–942. [Google Scholar] [CrossRef]
- Kumar, A.; Misra, B.B. Challenges and opportunities in cancer metabolomics. Proteomics 2019, 19, 1900042. [Google Scholar] [CrossRef]
- Kim, J.; DeBerardinis, R.J. Mechanisms and implications of metabolic heterogeneity in cancer. Cell Metab. 2019, 30, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R.; Goldansaz, S.A.; Jaine, J. Translational metabolomics: Current challenges and future opportunities. Metabolites 2019, 9, 108. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, D.R.; Patel, R.; Kirsch, D.G.; Lewis, C.A.; Vander Heiden, M.G.; Locasale, J.W. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J. Clin. 2021, 16, 760–763. [Google Scholar]
- Miggiels, P.; Wouters, B.; van Westen, G.J.; Dubbelman, A.-C.; Hankemeier, T. Novel technologies for metabolomics: More for less. TrAC Trends Anal. Chem. 2019, 120, 115323. [Google Scholar] [CrossRef]
- Sun, C.; Li, T.; Song, X.; Huang, L.; Zang, Q.; Xu, J.; Bi, N.; Jiao, G.; Hao, Y.; Chen, Y. Spatially resolved metabolomics to discover tumor-associated metabolic alterations. Proc. Natl. Acad. Sci. USA 2019, 116, 52–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, T.; Vergara, D.; Fanini, F.; Maffia, M.; Bravaccini, S.; Pirini, F. Microbiota-derived metabolites in tumor progression and metastasis. Int. J. Mol. Sci. 2020, 21, 5786. [Google Scholar] [CrossRef] [PubMed]
- Misra, B.B. New software tools, databases, and resources in metabolomics: Updates from 2020. Metabolomics 2021, 17, 49. [Google Scholar] [CrossRef] [PubMed]
- Schorn, M.A.; Verhoeven, S.; Ridder, L.; Huber, F.; Acharya, D.D.; Aksenov, A.A.; Aleti, G.; Moghaddam, J.A.; Aron, A.T.; Aziz, S. A community resource for paired genomic and metabolomic data mining. Nat. Chem. Biol. 2021, 17, 363–368. [Google Scholar] [CrossRef] [PubMed]
| Metabolic Target | Drug | Study Phase | Mode of Action | Reference |
|---|---|---|---|---|
| Glycolysis inhibitors | 2-deoxy-D-glucose | Phase III clinical trial | Competitively inhibits glucose uptake by interfering with HK | [54] |
| WP1122—Novel 2-DG analog) | Phase II clinical trial | Inhibits HK | [55] | |
| AZD3965 | Phase II clinical trial | Inhibits MCT 1 causing lactic acid accumulation and feedback inhibition of glycolysis | [56] | |
| Pyruvate dehydrogenase complex inhibitor | Dichloroacetate | Phase II clinical trial | Inhibits PDHK and reactivate the TCA cycle | [57] |
| Isocitrate dehydrogenase inhibitor | Enasidenib—novel IDH inhibitor | Phase III clinical trial | Inhibits mutant IDH2 variants and lowers serum levels of 2-HG in acute Myeloid Leukemia | [58] |
| Glutamine transport Inhibitor | V-9302 | Preclinical data | Selectively inhibits ASCT2 transporter | [59] |
| Fatty acid synthesis inhibitor | TVB-2640 | Phase 1 clinical trial | Inhibits FASN | [60] |
| Cerulenin | Preclinical data | Inhibits FASN | [61] | |
| Fatostatin | Preclinical data | inhibits SREBP activation | [62] | |
| A939572 | Preclinical data | Inhibits SCD-1 | [63] | |
| Mdm2 inhibitors | Idasanutlin (RG73388) | Phase III clinical trial | Inhibits MDM2-p53 interaction | [64] |
| AMG-232 | Phase I clinical trial | Blocks MDM2-p53 interaction | [65] |
| Target Classification | Metabolic Process | Target | Metabolic Effects of Plants and Derived Compounds on Cancer Cells and Tumour |
|---|---|---|---|
| Enzymes | Glycolysis | Hexokinase 2 (HK2) |
|
| Phosphofructokinase (PFK)-1 & 2 |
| ||
| Pyruvate kinase (PKM2) |
| ||
| Pyruvate metabolism Pyruvate metabolism | Pyruvate dehydrogenase (PDH) |
| |
| Pyruvate dehydrogenase kinase (PDHK) |
| ||
| Pyruvate dehydrogenase phosphatases (PDP)-1 & 2 |
| ||
| Pyruvate carboxylase (PC) |
| ||
| Lactate metabolism | Lactate dehydrogenase A (LDHA) |
| |
| Fatty acid synthesis | ATP citrate lyase (ACLY) |
| |
| Acetyl-CoA carboxylase (ACC) |
| ||
| Fatty acid synthase (FASN) |
| ||
| Transporters | Glucose transport | GLUT1/4 |
|
| Monocarboxylate transport | Monocarboxylate transporter (MCT) 1–4 |
| |
| Amino acid transport | Alanine-serine-cysteine transporter (ASCT) 2 |
| |
| Oncogenes and Tumor suppressors | Cell signalling and growth regulation | MAF |
|
| C-MYC |
| ||
| PI3K/AKT |
| ||
| MTORC1 | |||
| p53 |
| ||
| Phosphatase and tensin homolog (PTEN) |
| ||
| Transcription factors | Gene transcription | Sterol regulatory element-binding protein (SREBP) |
|
| Hypoxia-inducible factor (HIF)- 1 |
| ||
| Signal transducers and activators of transcription (STAT) 3 |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oyenihi, O.R.; Oyenihi, A.B.; Erhabor, J.O.; Matsabisa, M.G.; Oguntibeju, O.O. Unravelling the Anticancer Mechanisms of Traditional Herbal Medicines with Metabolomics. Molecules 2021, 26, 6541. https://doi.org/10.3390/molecules26216541
Oyenihi OR, Oyenihi AB, Erhabor JO, Matsabisa MG, Oguntibeju OO. Unravelling the Anticancer Mechanisms of Traditional Herbal Medicines with Metabolomics. Molecules. 2021; 26(21):6541. https://doi.org/10.3390/molecules26216541
Chicago/Turabian StyleOyenihi, Omolola R., Ayodeji B. Oyenihi, Joseph O. Erhabor, Motlalepula G. Matsabisa, and Oluwafemi O. Oguntibeju. 2021. "Unravelling the Anticancer Mechanisms of Traditional Herbal Medicines with Metabolomics" Molecules 26, no. 21: 6541. https://doi.org/10.3390/molecules26216541






