Towards Unravelling the Role of ERα-Targeting miRNAs in the Exosome-Mediated Transferring of the Hormone Resistance
Abstract
:1. Introduction
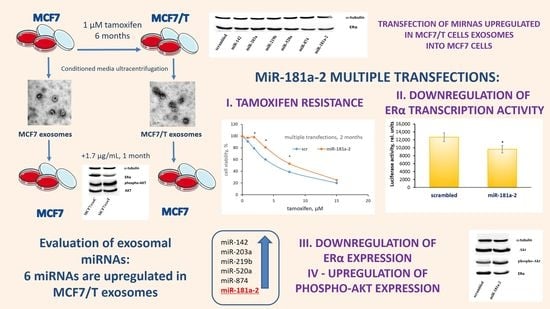
2. Results
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Cell Lines
5.2. Exosome Isolation and Visualization
5.3. MiRNA Analysis
5.4. RNA Isolation and Quantification by Quantitative RT–PCR
5.5. MiRNA Transfection
5.6. Immunoblotting, Reporter Analysis, and MTT-Test
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sever, B.; Altıntop, M.D.; Özdemir, A.; Akalın Çiftçi, G.; Ellakwa, D.E.; Tateishi, H.; Radwan, M.O.; Ibrahim, M.A.A.; Otsuka, M.; Fujita, M.; et al. In Vitro and In Silico Evaluation of Anticancer Activity of New Indole-Based 1,3,4-Oxadiazoles as EGFR and COX-2 Inhibitors. Molecules 2020, 25, 21. [Google Scholar] [CrossRef]
- Sever, B.; Akalın Çiftçi, G.; Altıntop, M.D. A new series of benzoxazole-based SIRT1 modulators for targeted therapy of non-small-cell lung cancer. Archiv Pharm. 2021, 354, e2000235. [Google Scholar] [CrossRef]
- Akalin Çiftçi, G.; Sever, B.; Altintop, M.D. Comprehensive Study on Thiadiazole-Based Anticancer Agents Inducing Cell Cycle Arrest and Apoptosis/Necrosis Through Suppression of Akt Activity in Lung Adenocarcinoma and Glioma Cells. Turk. J. Pharm. Sci. 2019, 16, 119–131. [Google Scholar] [CrossRef]
- Ciftci, H.I.; Bayrak, N.; Yıldız, M.; Yıldırım, H.; Sever, B.; Tateishi, H.; Otsuka, M.; Fujita, M.; Tuyun, A.F. Design, synthesis and investigation of the mechanism of action underlying anti-leukemic effects of the quinolinequinones as LY83583 analogs. Bioorganic Chem. 2021, 114, 105160. [Google Scholar] [CrossRef]
- Sever, B.; Altıntop, M.D.; Çiftçi, G.A.; Özdemir, A. A New Series of Triazolothiadiazines as Potential Anticancer Agents for Targeted Therapy of Non-small Cell Lung and Colorectal Cancers: Design, Synthesis, In Silico and In Vitro Studies Providing Mechanistic Insight into Their Anticancer Potencies. Med. Chem. 2020, 16. [Google Scholar] [CrossRef] [PubMed]
- Iftikhar, R.; Zahoor, A.F.; Irfan, M.; Rasul, A.; Rao, F. Synthetic molecules targeting yes associated protein activity as chemotherapeutics against cancer. Chem. Biol. Drug Des. 2021. [Google Scholar] [CrossRef]
- Dittmer, J. Nuclear Mechanisms Involved in Endocrine Resistance. Front. Oncol. 2021, 11, 736597. [Google Scholar] [CrossRef]
- Neven, P.; Sonke, G.S.; Jerusalem, G. Ribociclib plus fulvestrant in the treatment of breast cancer. Expert Rev. Anticancer. Ther. 2021, 21, 93–106. [Google Scholar] [CrossRef]
- McAndrew, N.P.; Finn, R.S. Management of ER positive metastatic breast cancer. Semin. Oncol. 2020, 47, 270–277. [Google Scholar] [CrossRef]
- Kowalczyk, W.; Waliszczak, G.; Jach, R.; Dulińska-Litewka, J. Steroid Receptors in Breast Cancer: Understanding of Molecular Function as a Basis for Effective Therapy Development. Cancers 2021, 13, 4779. [Google Scholar] [CrossRef]
- Andrahennadi, S.; Sami, A.; Haider, K.; Chalchal, H.I.; Le, D.; Ahmed, O.; Manna, M.; El-Gayed, A.; Wright, P.; Ahmed, S. Efficacy of Fulvestrant in Women with Hormone-Resistant Metastatic Breast Cancer (mBC): A Canadian Province Experience. Cancers 2021, 13, 4163. [Google Scholar] [CrossRef]
- Gründker, C.; Emons, G. Role of Gonadotropin-Releasing Hormone (GnRH) in Ovarian Cancer. Cells 2021, 10, 437. [Google Scholar] [CrossRef]
- Jillson, L.K.; Yette, G.A.; Laajala, T.D.; Tilley, W.D.; Costello, J.C.; Cramer, S.D. Androgen Receptor Signaling in Prostate Cancer Genomic Subtypes. Cancers 2021, 13, 3272. [Google Scholar] [CrossRef]
- Sasaki, T.; Nishikawa, K.; Kato, M.; Masui, S.; Yoshio, Y.; Sugimura, Y.; Inoue, T. Neoadjuvant Chemohormonal Therapy before Radical Prostatectomy for Japanese Patients with High-Risk Localized Prostate Cancer. Med. Sci. 2021, 9, 24. [Google Scholar] [CrossRef]
- Scherbakov, A.M.; Krasil’Nikov, M.A.; Kushlinskii, N.E. Molecular Mechanisms of Hormone Resistance of Breast Cancer. Bull. Exp. Biol. Med. 2013, 155, 384–395. [Google Scholar] [CrossRef]
- Hussein, S.; Khanna, P.; Yunus, N.; Gatza, M.L. Nuclear Receptor-Mediated Metabolic Reprogramming and the Impact on HR+ Breast Cancer. Cancers 2021, 13, 4808. [Google Scholar] [CrossRef]
- Dimauro, I.; Grazioli, E.; Antinozzi, C.; Duranti, G.; Arminio, A.; Mancini, A.; Greco, E.A.; Caporossi, D.; Parisi, A.; Di Luigi, L. Estrogen-Receptor-Positive Breast Cancer in Postmenopausal Women: The Role of Body Composition and Physical Exercise. Int. J. Environ. Res. Public Health 2021, 18, 18. [Google Scholar] [CrossRef] [PubMed]
- Stewart, H.J. Adjuvant endocrine therapy for operable breast cancer. Bull. Cancer 1991, 78, 379–384. [Google Scholar]
- Falkson, C.I.; Falkson, G.; Falkson, H.C. Postmenopausal breast cancer. Drug therapy in the 1990s. Drugs Aging 1993, 3, 106–121. [Google Scholar] [CrossRef] [PubMed]
- Jordan, V.C. 50th anniversary of the first clinical trial with ICI 46,474 (tamoxifen): Then what happened? Endocr. Relat. Cancer 2021, 28, R11–R30. [Google Scholar] [CrossRef] [PubMed]
- Yip, H.; Papa, A. Signaling Pathways in Cancer: Therapeutic Targets, Combinatorial Treatments, and New Developments. Cells 2021, 10, 659. [Google Scholar] [CrossRef]
- Murav’eva, N.I.; Kuz’mina, Z.V.; Smirnova, K.D.; Gershteĭn, E.S.; Ird, E.A. Action of tamoxifen on the sex organs of guinea pigs. Biulleten’ Eksperimental’noi Biol. Meditsiny 1982, 94, 77–80. [Google Scholar]
- Bogush, T.; Polezhaev, B.B.; Mamichev, I.; Bogush, E.A.; Polotsky, B.E.; Tjulandin, S.A.; Ryabov, A.B. Tamoxifen Never Ceases to Amaze: New Findings on Non-Estrogen Receptor Molecular Targets and Mediated Effects. Cancer Investig. 2018, 36, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Davies, C.; Pan, H.; Godwin, J.; Gray, R.; Arriagada, R.; Raina, V.; Abraham, M.; Alencar, V.H.M.; Badran, A.; Bonfill, X.; et al. Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trial. Lancet 2013, 381, 805–816. [Google Scholar] [CrossRef] [Green Version]
- Babyshkina, N.; Dronova, T.; Erdyneeva, D.; Gervas, P.; Cherdyntseva, N. Role of TGF-β signaling in the mechanisms of tamoxifen resistance. Cytokine Growth Factor Rev. 2021. [Google Scholar] [CrossRef]
- Xu, Y.; Huangyang, P.; Wang, Y.; Xue, L.; Devericks, E.; Nguyen, H.G.; Yu, X.; Oses-Prieto, J.A.; Burlingame, A.L.; Miglani, S.; et al. ERα is an RNA-binding protein sustaining tumor cell survival and drug resistance. Cell 2021, 184, 5215–5229.e17. [Google Scholar] [CrossRef] [PubMed]
- Brett, J.O.; Spring, L.M.; Bardia, A.; Wander, S.A. ESR1 mutation as an emerging clinical biomarker in metastatic hormone receptor-positive breast cancer. Breast Cancer Res. 2021, 23, 85. [Google Scholar] [CrossRef] [PubMed]
- Milani, A.; Geuna, E.; Mittica, G.; Valabrega, G. Overcoming endocrine resistance in metastatic breast cancer: Current evidence and future directions. World J. Clin. Oncol. 2014, 5, 990. [Google Scholar] [CrossRef] [Green Version]
- Viedma-Rodríguez, R.; Baiza-Gutman, L.; Salamanca-Gómez, F.; Diaz-Zaragoza, M.; Martínez-Hernández, G.; Ruiz Esparza-Garrido, R.; Velázquez-Flores, M.A.; Arenas-Aranda, D. Mechanisms associated with resistance to tamoxifen in estrogen receptor-positive breast cancer. Oncol. Rep. 2014, 32, 3–15. [Google Scholar]
- Scherbakov, A.M.; Sorokin, D.V.; Tatarskiy, V.V., Jr.; Prokhorov, N.S.; Semina, S.E.; Berstein, L.M.; Krasil’nikov, M.A. The phenomenon of acquired resistance to metformin in breast cancer cells: The interaction of growth pathways and estrogen receptor signaling. IUBMB Life 2016, 68, 281–292. [Google Scholar] [CrossRef]
- Suba, Z. DNA stabilization by the upregulation of estrogen signaling in BRCA gene mutation carriers. Drug Des. Dev. Ther. 2015, 9, 2663–2675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Camera, G.; Gelsomino, L.; Caruso, A.; Panza, S.; Barone, I.; Bonofiglio, D.; Andò, S.; Giordano, C.; Catalano, S. The Emerging Role of Extracellular Vesicles in Endocrine Resistant Breast Cancer. Cancers 2021, 13, 1160. [Google Scholar] [CrossRef]
- Augimeri, G.; La Camera, G.; Gelsomino, L.; Giordano, C.; Panza, S.; Sisci, D.; Morelli, C.; Győrffy, B.; Bonofiglio, D.; Andò, S.; et al. Evidence for Enhanced Exosome Production in Aromatase Inhibitor-Resistant Breast Cancer Cells. Int. J. Mol. Sci. 2020, 21, 5841. [Google Scholar] [CrossRef] [PubMed]
- Delort, L.; Bougaret, L.; Cholet, J.; Vermerie, M.; Billard, H.; Decombat, C.; Bourgne, C.; Berger, M.; Dumontet, C.; Caldefie-Chezet, F. Hormonal Therapy Resistance and Breast Cancer: Involvement of Adipocytes and Leptin. Nutrients 2019, 11, 2839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awan, A.; Esfahani, K. Endocrine Therapy for Breast Cancer in the Primary Care Setting. Curr. Oncol. 2018, 25, 285–291. [Google Scholar] [CrossRef] [Green Version]
- Clarke, R.; Tyson, J.J.; Dixon, J.M. Endocrine resistance in breast cancer-An overview and update. Mol. Cell. Endocrinol. 2015, 418, 220–234. [Google Scholar] [CrossRef] [Green Version]
- Petrelli, F.; Tomasello, G.; Barni, S.; Lonati, V.; Passalacqua, R.; Ghidini, M. Clinical and pathological characterization of HER2 mutations in human breast cancer: A systematic review of the literature. Breast Cancer Res. Treat. 2017, 166, 339–349. [Google Scholar] [CrossRef]
- Barchiesi, G.; Mazzotta, M.; Krasniqi, E.; Pizzuti, L.; Marinelli, D.; Capomolla, E.; Sergi, D.; Amodio, A.; Natoli, C.; Gamucci, T.; et al. Neoadjuvant Endocrine Therapy in Breast Cancer: Current Knowledge and Future Perspectives. Int. J. Mol. Sci. 2020, 21, 3528. [Google Scholar] [CrossRef]
- Zhou, Y.; Eppenberger-Castori, S.; Eppenberger, U.; Benz, C.C. The NFκB pathway and endocrine-resistant breast cancer. Endocr. Relat. Cancer 2005, 12, S37–S46. [Google Scholar] [CrossRef]
- Arpino, G.; DE Angelis, C.; Giuliano, M.; Giordano, A.; Falato, C.; De Laurentiis, M.; De Placido, S. Molecular Mechanism and Clinical Implications of Endocrine Therapy Resistance in Breast Cancer. Oncology 2009, 77 (Suppl. 1), 23–37. [Google Scholar] [CrossRef]
- Ghosh, A.; Awasthi, S.; Peterson, J.; Hamburger, A. Regulation of tamoxifen sensitivity by a PAK1–EBP1 signalling pathway in breast cancer. Br. J. Cancer 2013, 108, 557–563. [Google Scholar] [CrossRef] [Green Version]
- Poulard, C.; Jacquemetton, J.; Trédan, O.; Cohen, P.A.; Vendrell, J.; Ghayad, S.E.; Treilleux, I.; Marangoni, E.; Le Romancer, M. Oestrogen Non-Genomic Signalling is Activated in Tamoxifen-Resistant Breast Cancer. Int. J. Mol. Sci. 2019, 20, 2773. [Google Scholar] [CrossRef] [Green Version]
- Scherbakov, A.M.; Andreeva, O.E.; Shatskaya, V.; Krasil’Nikov, M. The relationships between snail1 and estrogen receptor signaling in breast cancer cells. J. Cell. Biochem. 2012, 113, 2147–2155. [Google Scholar] [CrossRef]
- Shi, X.-P.; Miao, S.; Wu, Y.; Zhang, W.; Zhang, X.-F.; Ma, H.-Z.; Xin, H.-L.; Feng, J.; Wen, A.-D.; Li, Y. Resveratrol Sensitizes Tamoxifen in Antiestrogen-Resistant Breast Cancer Cells with Epithelial-Mesenchymal Transition Features. Int. J. Mol. Sci. 2013, 14, 15655–15668. [Google Scholar] [CrossRef] [PubMed]
- Rascio, F.; Spadaccino, F.; Rocchetti, M.; Castellano, G.; Stallone, G.; Netti, G.; Ranieri, E. The Pathogenic Role of PI3K/AKT Pathway in Cancer Onset and Drug Resistance: An Updated Review. Cancers 2021, 13, 3949. [Google Scholar] [CrossRef] [PubMed]
- Neophytou, C.M.; Trougakos, I.P.; Erin, N.; Papageorgis, P. Apoptosis Deregulation and the Development of Cancer Multi-Drug Resistance. Cancers 2021, 13, 4363. [Google Scholar] [CrossRef]
- Dong, C.; Wu, J.; Chen, Y.; Nie, J.; Chen, C. Activation of PI3K/AKT/mTOR Pathway Causes Drug Resistance in Breast Cancer. Front. Pharmacol. 2021, 12, 628690. [Google Scholar] [CrossRef]
- Saatci, O.; Huynh-Dam, K.-T.; Sahin, O. Endocrine resistance in breast cancer: From molecular mechanisms to therapeutic strategies. J. Mol. Med. 2021, 1–20. [Google Scholar] [CrossRef]
- Chien, T.J. A review of the endocrine resistance in hormone-positive breast cancer. Am. J. Cancer Res. 2021, 11, 3813–3831. [Google Scholar]
- Miricescu, D.; Totan, A.; Stanescu-Spinu, I.-I.; Badoiu, S.C.; Stefani, C.; Greabu, M. PI3K/AKT/mTOR Signaling Pathway in Breast Cancer: From Molecular Landscape to Clinical Aspects. Int. J. Mol. Sci. 2020, 22, 173. [Google Scholar] [CrossRef] [PubMed]
- Nunnery, S.E.; Mayer, I.A. Targeting the PI3K/AKT/mTOR Pathway in Hormone-Positive Breast Cancer. Drugs 2020, 80, 1685–1697. [Google Scholar] [CrossRef] [PubMed]
- Di Leva, G.; Gasparini, P.; Piovan, C.; Ngankeu, A.; Garofalo, M.; Taccioli, C.; Iorio, M.V.; Li, M.; Volinia, S.; Alder, H.; et al. MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J. Natl. Cancer Inst. 2010, 102, 706–721. [Google Scholar] [CrossRef] [PubMed]
- He, Y.J.; Wu, J.Z.; Ji, M.H.; Ma, T.; Qiao, E.Q.; Ma, R.; Tang, J.H. miR-342 is associated with estrogen receptor-α expression and response to tamoxifen in breast cancer. Exp. Ther. Med. 2013, 5, 813–818. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Deng, C.; Lu, W.; Xiao, J.; Ma, D.; Guo, M.; Recker, R.R.; Gatalica, Z.; Wang, Z.; Xiao, G.G. let-7 microRNAs induce tamoxifen sensitivity by downregulation of estrogen receptor α signaling in breast cancer. Mol. Med. 2011, 17, 1233–1241. [Google Scholar] [CrossRef]
- Meng, D.; Li, Z.; Ma, X.; Fu, L.; Qin, G. MicroRNA-1280 modulates cell growth and invasion of thyroid carcinoma through targeting estrogen receptor α. Cell. Mol. Biol. 2016, 62, 1–6. [Google Scholar]
- Cui, J.; Yang, Y.; Li, H.; Leng, Y.; Qian, K.; Huang, Q.; Zhang, C.; Lu, Z.; Chen, J.; Sun, T.; et al. MiR-873 regulates ERalpha transcriptional activity and tamoxifen resistance via targeting CDK3 in breast cancer cells. Oncogene 2015, 34, 3895–3907. [Google Scholar] [CrossRef] [Green Version]
- Hossain, A.; Kuo, M.T.; Saunders, G.F. Mir-17-5p Regulates Breast Cancer Cell Proliferation by Inhibiting Translation of AIB1 mRNA. Mol. Cell. Biol. 2006, 26, 8191–8201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foley, N.H.; Bray, I.; Watters, K.M.; Das, S.; Bryan, K.; Bernas, T.; Prehn, J.; Stallings, R.L. MicroRNAs 10a and 10b are potent inducers of neuroblastoma cell differentiation through targeting of nuclear receptor corepressor 2. Cell Death Differ. 2011, 18, 1089–1098. [Google Scholar] [CrossRef] [Green Version]
- Bergamaschi, A.; Katzenellenbogen, B.S. Tamoxifen downregulation of miR-451 increases 14-3-3ζ and promotes breast cancer cell survival and endocrine resistance. Oncogene 2012, 31, 39–47. [Google Scholar] [CrossRef] [Green Version]
- Sachdeva, M.; Wu, H.; Ru, P.; Hwang, L.; Trieu, V.; Mo, Y. MicroRNA-101-mediated Akt activation and estrogen-independent growth. Oncogene 2011, 30, 822–831. [Google Scholar] [CrossRef]
- Chen, W.-X.; Liu, X.-M.; Lv, M.-M.; Chen, L.; Zhao, J.-H.; Zhong, S.; Ji, M.-H.; Hu, Q.; Luo, Z.; Wu, J.-Z.; et al. Exosomes from Drug-Resistant Breast Cancer Cells Transmit Chemoresistance by a Horizontal Transfer of MicroRNAs. PLoS ONE 2014, 9, e95240. [Google Scholar] [CrossRef] [PubMed]
- Phuong, N.T.T.; Kim, S.K.; Im, J.H.; Yang, J.W.; Choi, M.C.; Lim, S.C.; Lee, K.Y.; Kim, Y.-M.; Yoon, J.H.; Kang, K.W. Induction of methionine adenosyltransferase 2A in tamoxifen-resistant breast cancer cells. Oncotarget 2016, 7, 13902–13916. [Google Scholar] [CrossRef]
- Yu, X.; Li, R.; Shi, W.; Jiang, T.; Wang, Y.; Li, C.; Qu, X. Silencing of MicroRNA-21 confers the sensitivity to tamoxifen and fulvestrant by enhancing autophagic cell death through inhibition of the PI3K-AKT-mTOR pathway in breast cancer cells. Biomed. Pharmacother. 2016, 77, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-Y.; Park, M.K.; Park, J.-H.; Lee, H.J.; Shin, D.H.; Kang, Y.; Lee, C.H.; Kong, G. Loss of the polycomb protein Mel-18 enhances the epithelial–mesenchymal transition by ZEB1 and ZEB2 expression through the downregulation of miR-205 in breast cancer. Oncogene 2014, 33, 1325–1335. [Google Scholar] [CrossRef] [Green Version]
- Gajda, E.; Grzanka, M.; Godlewska, M.; Gawel, D. The Role of miRNA-7 in the Biology of Cancer and Modulation of Drug Resistance. Pharmaceuticals 2021, 14, 149. [Google Scholar] [CrossRef]
- Semina, S.E.; Scherbakov, A.M.; Vnukova, A.A.; Bagrov, D.V.; Evtushenko, E.G.; Safronova, V.M.; Golovina, D.A.; Lyubchenko, L.N.; Gudkova, M.V.; Krasil’nikov, M.A. Exosome-mediated transfer of cancer cell resistance to antiestrogen drugs. Molecules 2018, 23, 829. [Google Scholar] [CrossRef] [Green Version]
- Sansone, P.; Savini, C.; Kurelac, I.; Chang, Q.; Amato, L.B.; Strillacci, A.; Stepanova, A.; Iommarini, L.; Mastroleo, C.; Daly, L.; et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E9066–E9075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.X.; Vu, L.T.; Ismail, N.N.; Le, M.T.N.; Grimson, A. Landscape of extracellular vesicles in the tumour microenvironment: Interactions with stromal cells and with non-cell components, and impacts on metabolic reprogramming, horizontal transfer of neoplastic traits, and the emergence of therapeutic resistance. Semin. Cancer Biol. 2021, 74, 24–44. [Google Scholar] [CrossRef] [PubMed]
- Hertle, A.P.; Haberl, B.; Bock, R. Horizontal genome transfer by cell-to-cell travel of whole organelles. Sci. Adv. 2021, 7, 1. [Google Scholar] [CrossRef]
- Han, Z.; Li, Y.; Zhang, J.; Guo, C.; Li, Q.; Zhang, X.; Lan, Y.; Gu, W.; Xing, Z.; Liang, L.; et al. Tumor-derived circulating exosomal miR-342-5p and miR-574-5p as promising diagnostic biomarkers for early-stage Lung Adenocarcinoma. Int. J. Med. Sci. 2020, 17, 1428–1438. [Google Scholar] [CrossRef]
- Semina, S.; Bagrov, D.; Krasil’nikov, M. Intercellular interactions and progression of hormonal resistance of breast cancer cells. Adv. Mol. Oncol. 2015, 2, 50–55. [Google Scholar] [CrossRef] [Green Version]
- Mansoori, B.; Mohammadi, A.; Gjerstorff, M.F.; Shirjang, S.; Asadzadeh, Z.; Khaze, V.; Holmskov, U.; Kazemi, T.; Duijf, P.H.G.; Baradaran, B. miR-142-3p is a tumor suppressor that inhibits estrogen receptor expression in ER-positive breast cancer. J. Cell. Physiol. 2019, 234, 16043–16053. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.T.; Feng, C.X.; Zhao, J.C.; He, R.Q.; Ma, J.; Zhong, J.C. Upregulated miR-203a-3p and its potential molecular mechanism in breast cancer: A study based on bioinformatics analyses and a comprehensive metaanalysis. Mol. Med. Rep. 2018, 18, 4994–5008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leivonen, S.-K.; Mäkelä, R.; Östling, P.; Kohonen, P.; Haapa-Paananen, S.; Kleivi, K.; Enerly, E.; Aakula, A.; Hellström, K.; Sahlberg, K.K.; et al. Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene 2009, 28, 3926–3936. [Google Scholar] [CrossRef] [Green Version]
- Keklikoglou, I.; Koerner, C.; Schmidt, C.; Zhang, J.D.; Heckmann, D.; Shavinskaya, A.; Allgayer, H.; Guckel, B.; Fehm, T.; Schneeweiss, A.; et al. MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-kappaB and TGF-beta signaling pathways. Oncogene 2012, 31, 4150–4163. [Google Scholar] [CrossRef]
- Li, J.; Wei, J.; Mei, Z.; Yin, Y.; Li, Y.; Lu, M.; Jin, S. Suppressing role of miR-520a-3p in breast cancer through CCND1 and CD44. Am. J. Transl. Res. 2017, 9, 146–154. [Google Scholar]
- Wang, L.; Gao, W.; Hu, F.; Xu, Z.; Wang, F. MicroRNA-874 inhibits cell proliferation and induces apoptosis in human breast cancer by targeting CDK9. FEBS Lett. 2014, 588, 4527–4535. [Google Scholar] [CrossRef] [Green Version]
- Strotbek, M.; Schmid, S.; Sánchez-González, I.; Boerries, M.; Busch, H.; Olayioye, M.A. miR-181 elevates Akt signaling by co-targeting PHLPP2 and INPP4B phosphatases in luminal breast cancer. Int. J. Cancer 2017, 140, 2310–2320. [Google Scholar] [CrossRef]
- Haque, S.; Vaiselbuh, S.R. Silencing of Exosomal miR-181a Reverses Pediatric Acute Lymphocytic Leukemia Cell Proliferation. Pharmaceuticals 2020, 13, 9. [Google Scholar] [CrossRef]
- Nishimura, J.; Handa, R.; Yamamoto, H.; Tanaka, F.; Shibata, K.; Mimori, K.; Takemasa, I.; Mizushima, T.; Ikeda, M.; Sekimoto, M.; et al. microRNA-181a is associated with poor prognosis of colorectal cancer. Oncol. Rep. 2012, 28, 2221–2226. [Google Scholar] [CrossRef] [Green Version]
- Khan, I.A.; Rashid, S.; Singh, N.; Rashid, S.; Singh, V.; Gunjan, D.; Das, P.; Dash, N.R.; Pandey, R.M.; Chauhan, S.S.; et al. Panel of serum miRNAs as potential non-invasive biomarkers for pancreatic ductal adenocarcinoma. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Taylor, S.C.; Berkelman, T.; Yadav, G.; Hammond, M. A Defined Methodology for Reliable Quantification of Western Blot Data. Mol. Biotechnol. 2013, 55, 217–226. [Google Scholar] [CrossRef] [Green Version]
- Glover, H.R.; Barker, S.; Malouitre, S.D.M.; Puddefoot, J.R.; Vinson, G.P. Multiple Routes to Oestrogen Antagonism. Pharmaceuticals 2010, 3, 3417–3434. [Google Scholar] [CrossRef] [Green Version]
- McGowan, E.M.; Lin, Y.; Hatoum, D. Good Guy or Bad Guy? The Duality of Wild-Type p53 in Hormone-Dependent Breast Cancer Origin, Treatment, and Recurrence. Cancers 2018, 10, 172. [Google Scholar] [CrossRef] [Green Version]
- García-Becerra, R.; Santos, N.; Díaz, L.; Camacho, J. Mechanisms of Resistance to Endocrine Therapy in Breast Cancer: Focus on Signaling Pathways, miRNAs and Genetically Based Resistance. Int. J. Mol. Sci. 2013, 14, 108–145. [Google Scholar] [CrossRef] [Green Version]
- Chung, H.; Jung, Y.M.; Shin, D.H.; Lee, J.Y.; Oh, M.Y.; Kim, H.J.; Jang, K.S.; Jeon, S.J.; Son, K.H.; Kong, G. Anticancer effects of wogonin in both estrogen receptor-positive and -negative human breast cancer cell lines in vitro and in nude mice xenografts. Int. J. Cancer 2008, 122, 816–822. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Joo, H.S.; Won, H.Y.; Min, K.W.; Kim, H.Y.; Son, T.; Oh, Y.H.; Lee, J.Y.; Kong, G. Role of RBP2-Induced ER and IGF1R-ErbB Signaling in Tamoxifen Resistance in Breast Cancer. J. Natl. Cancer Inst. 2018, 110, 4. [Google Scholar] [CrossRef]
- Barazetti, J.; Jucoski, T.; Carvalho, T.; Veiga, R.; Kohler, A.; Baig, J.; Al Bizri, H.; Gradia, D.; Mader, S.; de Oliveira, J.C. From Micro to Long: Non-Coding RNAs in Tamoxifen Resistance of Breast Cancer Cells. Cancers 2021, 13, 3688. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.E.; Ghoshal, K.; Ramaswamy, B.; Roy, S.; Datta, J.; Shapiro, C.L.; Jacob, S.; Majumder, S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J. Biol. Chem. 2008, 283, 29897–29903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Zhang, W.; Liu, C.; Li, G. miR-200 affects tamoxifen resistance in breast cancer cells through regulation of MYB. Sci. Rep. 2019, 9, 1–6. [Google Scholar] [CrossRef]
- Dong, X.; Bai, X.; Ni, J.; Zhang, H.; Duan, W.; Graham, P.; Li, Y. Exosomes and breast cancer drug resistance. Cell Death Dis. 2020, 11, 987. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.J.; Sompel, K.M.; Elango, A.; Tennis, M.A. Non-Coding RNA and Frizzled Receptors in Cancer. Front. Mol. Biosci. 2021, 8, 712546. [Google Scholar] [CrossRef] [PubMed]
- Tabnak, P.; Masrouri, S.; Geraylow, K.R.; Zarei, M.; Esmailpoor, Z.H. Targeting miRNAs with anesthetics in cancer: Current understanding and future perspectives. Biomed. Pharmacother. 2021, 144, 112309. [Google Scholar] [CrossRef]
- Vlasov, V.V.; Rykova, E.I.; Ponomareva, A.A.; Zaporozhchenko, I.; Morozkin, E.S.; Cherdyntseva, N.V.; Laktionov, P.P. Circulating microRNAs in lung cancer: Prospects for diagnostics, prognosis and prediction of antitumor treatment efficiency. Mol. Biol. 2015, 49, 55–66. [Google Scholar] [CrossRef]
- Ponomareva, A.A.; Rykova, E.I.; Cherdyntseva, N.V.; Choĭnzonov, E.L.; Laktionov, P.P.; Vlasov, V.V. Molecular-genetic markers in lung cancer diagnostics. Mol. Biol. 2011, 45, 203–217. [Google Scholar]
- Sueta, A.; Fujiki, Y.; Goto-Yamaguchi, L.; Tomiguchi, M.; Yamamoto-Ibusuki, M.; Iwase, H.; Yamamoto, Y. Exosomal miRNA profiles of triple-negative breast cancer in neoadjuvant treatment. Oncol. Lett. 2021, 22, 1–10. [Google Scholar] [CrossRef]
- Isca, C.; Piacentini, F.; Mastrolia, I.; Masciale, V.; Caggia, F.; Toss, A.; Piombino, C.; Moscetti, L.; Barbolini, M.; Maur, M.; et al. Circulating and Intracellular miRNAs as Prognostic and Predictive Factors in HER2-Positive Early Breast Cancer Treated with Neoadjuvant Chemotherapy: A Review of the Literature. Cancers 2021, 13, 4894. [Google Scholar] [CrossRef]
- Monchusi, B.; Kaur, M. miRNAs as modulators of cholesterol in breast cancer stem cells: An approach to overcome drug resistance in cancer. Curr. Drug Targets 2021, 22, 1. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, S.; Tang, W.; Huang, Q.; Mei, Y.; Yang, H. Exosomes from tamoxifen-resistant breast cancer cells transmit drug resistance partly by delivering miR-9-5p. Cancer Cell Int. 2021, 21, 55. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Liu, X.; Li, Y.; Li, Q.; Xu, Y.; Zeng, W.; Zhong, G.; Yu, C. Exosomes Mediated Transfer of Circ_UBE2D2 Enhances the Resistance of Breast Cancer to Tamoxifen by Binding to MiR-200a-3p. Med Sci. Monit. 2020, 26, e922253. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, O.; Shchegolev, Y.; Scherbakov, A.; Mikhaevich, E.; Sorokin, D.; Gudkova, M.; Bure, I.; Kuznetsova, E.; Mikhaylenko, D.; Nemtsova, M.; et al. Secretion of Mutant DNA and mRNA by the Exosomes of Breast Cancer Cells. Molecules 2021, 26, 2499. [Google Scholar] [CrossRef]
- Tokar, T.; Pastrello, C.; Rossos, A.E.M.; Abovsky, M.; Hauschild, A.C.; Tsay, M.; Lu, R.; Jurisica, I. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018, 46, D360–D370. [Google Scholar] [CrossRef]
- Mruk, D.D.; Cheng, C.Y. Enhanced chemiluminescence (ECL) for routine immunoblotting: An inexpensive alternative to commercially available kits. Spermatogenesis 2011, 1, 121–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuznetsov, Y.V.; Levina, I.S.; Scherbakov, A.M.; Andreeva, O.; Fedyushkina, I.V.; Dmitrenok, A.S.; Shashkov, A.S.; Zavarzin, I.V. New estrogen receptor antagonists. 3,20-Dihydroxy-19-norpregna-1,3,5(10)-trienes: Synthesis, molecular modeling, and biological evaluation. Eur. J. Med. Chem. 2018, 143, 670–682. [Google Scholar] [CrossRef]
- Denizot, F.; Lang, R. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J. Immunol. Methods 1986, 89, 271–277. [Google Scholar] [CrossRef]





| MicroRNAs | Biological Activity | Reference |
|---|---|---|
| 142 | targets ESR1, reduces cell viability, induces apoptosis and decreases colony formation | [72] |
| 203a | is overexpressed in breast cancer and can influence ERα signalling by targeting ADCY5, IGF1, etc. | [73] |
| 219b | downregulates ERα | [74] |
| 520a | targets NF-κB and TGF-β signalling pathways; targets ESR1, inhibits CCND1 mRNA and cyclin D1 protein levels | [75,76] |
| 874 | targets ESR1, CDK9 | [77] |
| 181a | downregulates ERα; induces AKT signalling | [74,78] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andreeva, O.E.; Sorokin, D.V.; Mikhaevich, E.I.; Bure, I.V.; Shchegolev, Y.Y.; Nemtsova, M.V.; Gudkova, M.V.; Scherbakov, A.M.; Krasil’nikov, M.A. Towards Unravelling the Role of ERα-Targeting miRNAs in the Exosome-Mediated Transferring of the Hormone Resistance. Molecules 2021, 26, 6661. https://doi.org/10.3390/molecules26216661
Andreeva OE, Sorokin DV, Mikhaevich EI, Bure IV, Shchegolev YY, Nemtsova MV, Gudkova MV, Scherbakov AM, Krasil’nikov MA. Towards Unravelling the Role of ERα-Targeting miRNAs in the Exosome-Mediated Transferring of the Hormone Resistance. Molecules. 2021; 26(21):6661. https://doi.org/10.3390/molecules26216661
Chicago/Turabian StyleAndreeva, Olga E., Danila V. Sorokin, Ekaterina I. Mikhaevich, Irina V. Bure, Yuri Y. Shchegolev, Marina V. Nemtsova, Margarita V. Gudkova, Alexander M. Scherbakov, and Mikhail A. Krasil’nikov. 2021. "Towards Unravelling the Role of ERα-Targeting miRNAs in the Exosome-Mediated Transferring of the Hormone Resistance" Molecules 26, no. 21: 6661. https://doi.org/10.3390/molecules26216661