Interaction Study between ESIPT Fluorescent Lipophile-Based Benzazoles and BSA
Abstract
:1. Introduction
2. Results and Discussion
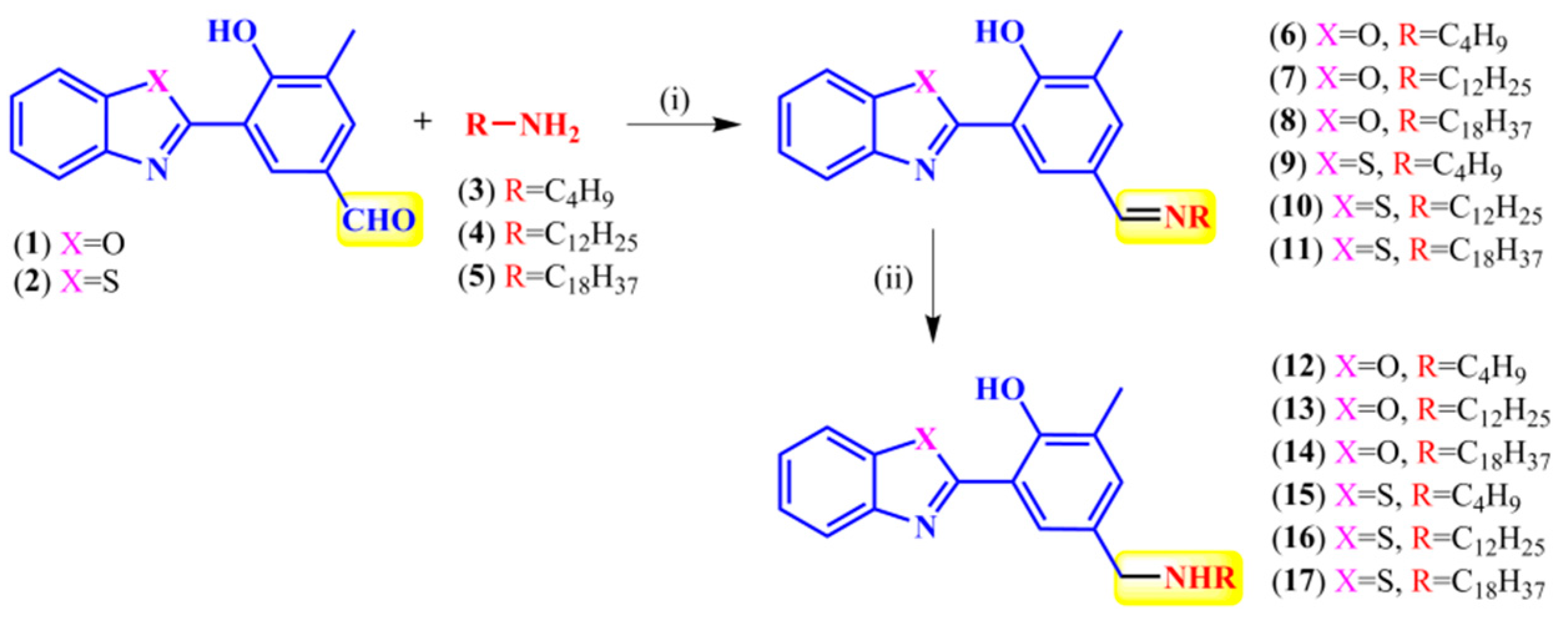
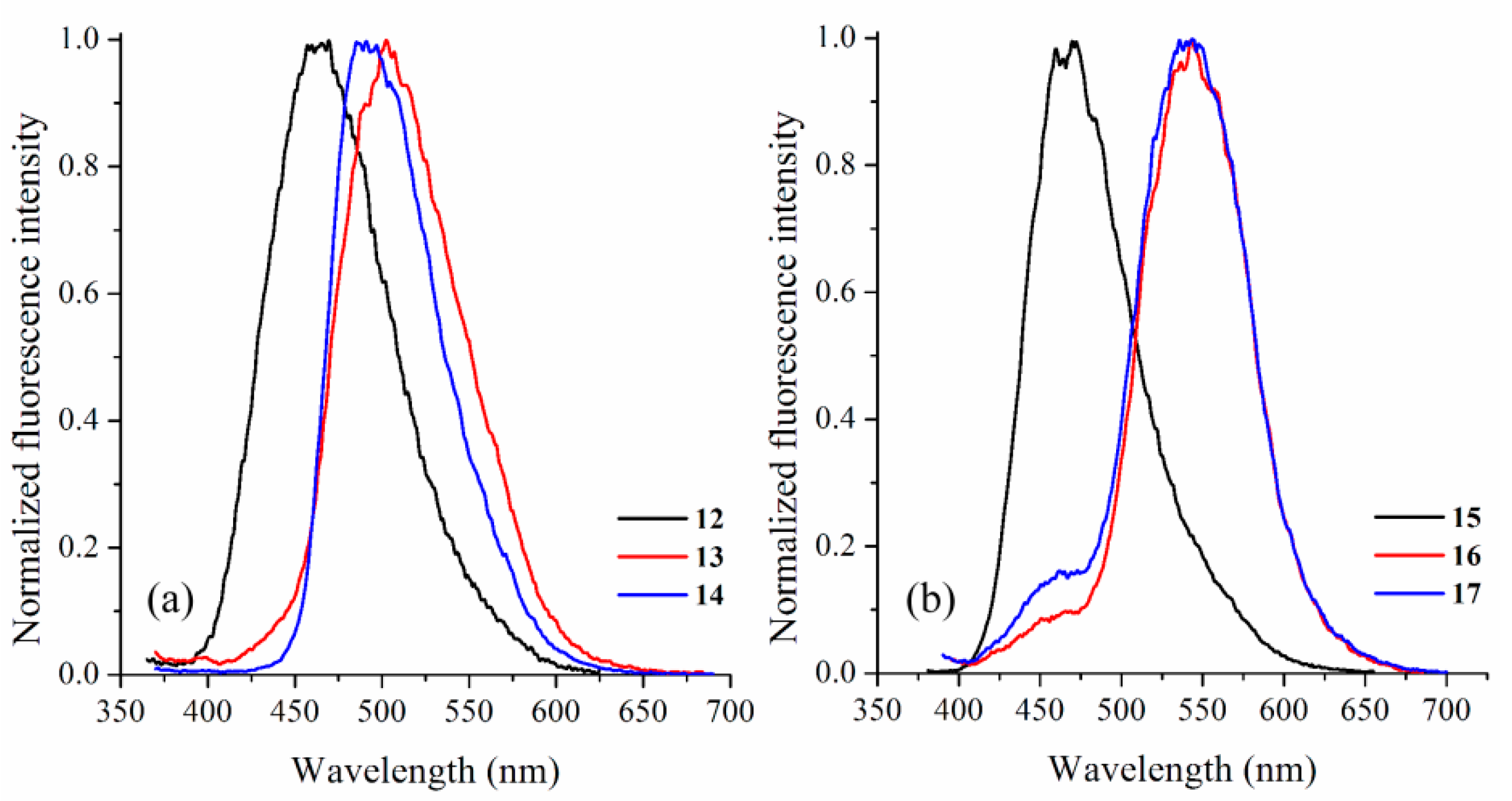
2.1. Synthesis and Photophysical Characterization
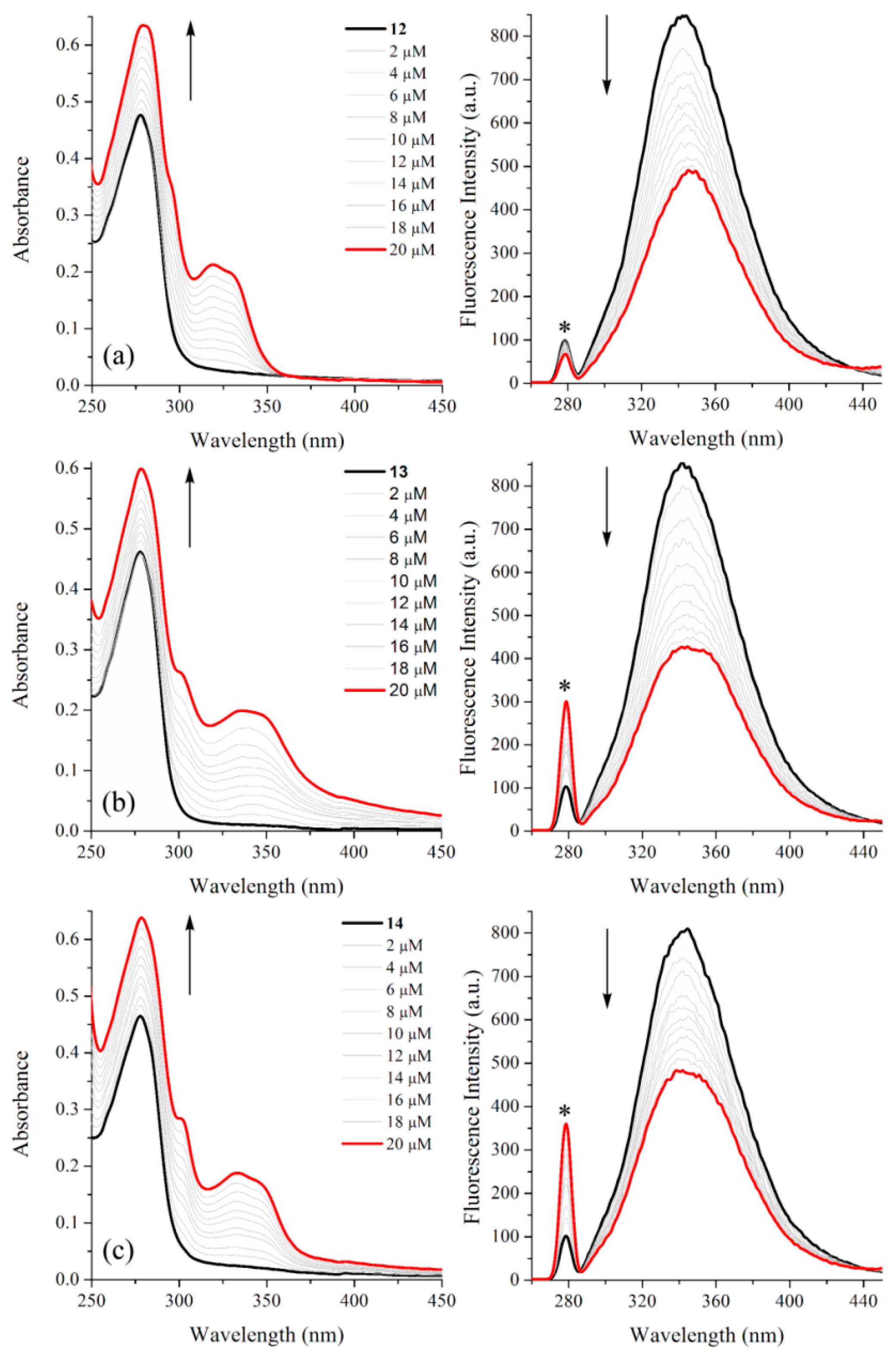
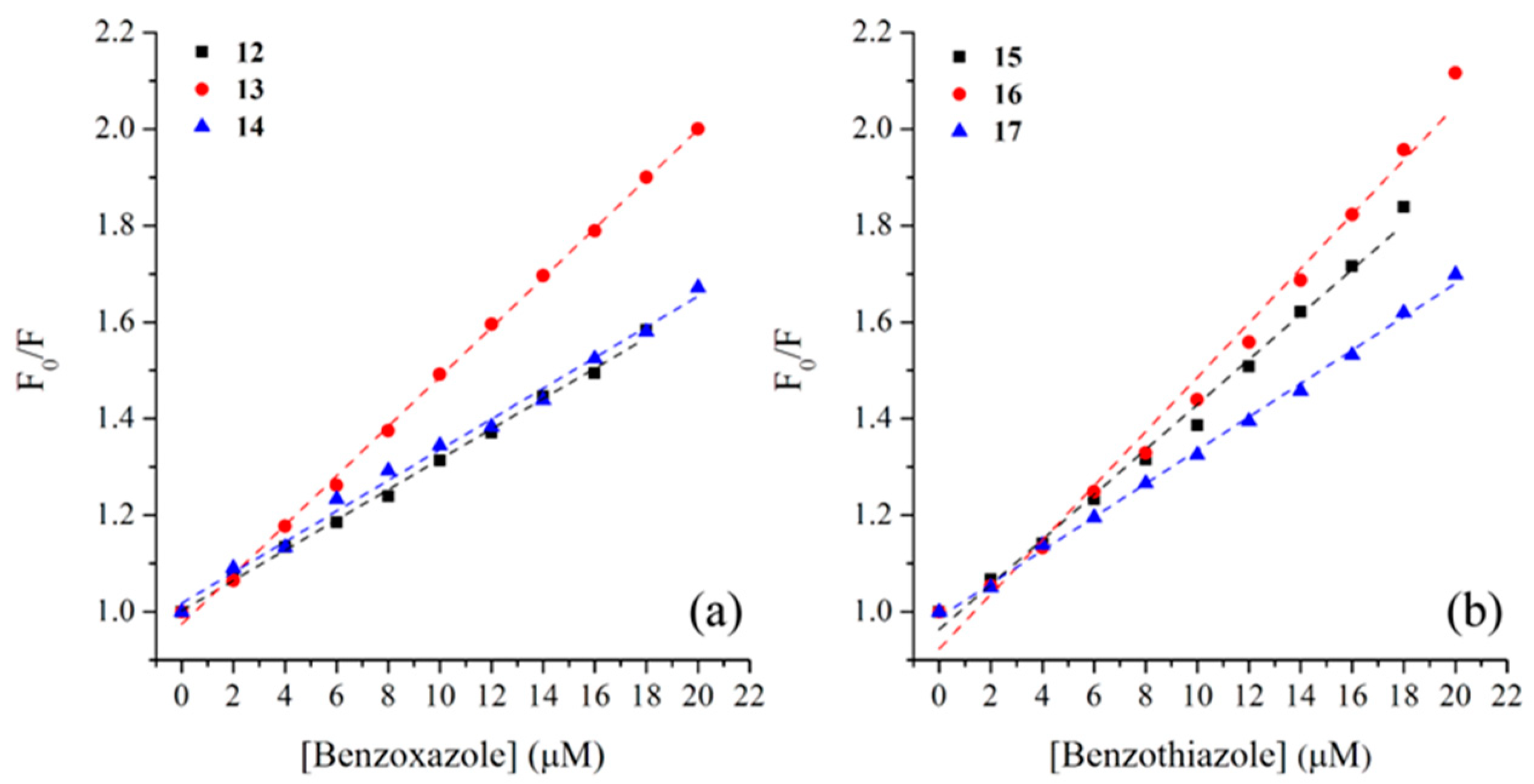
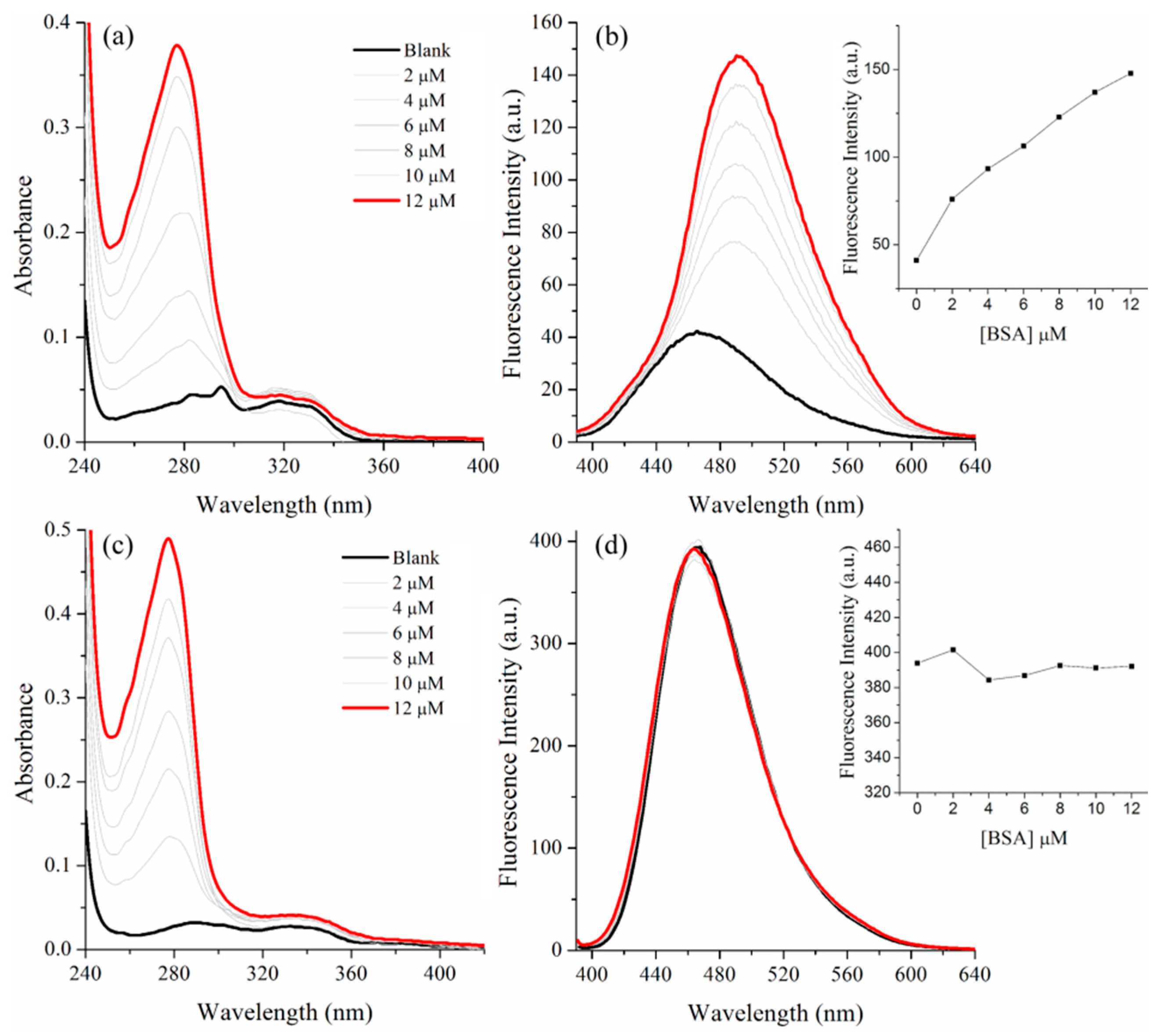
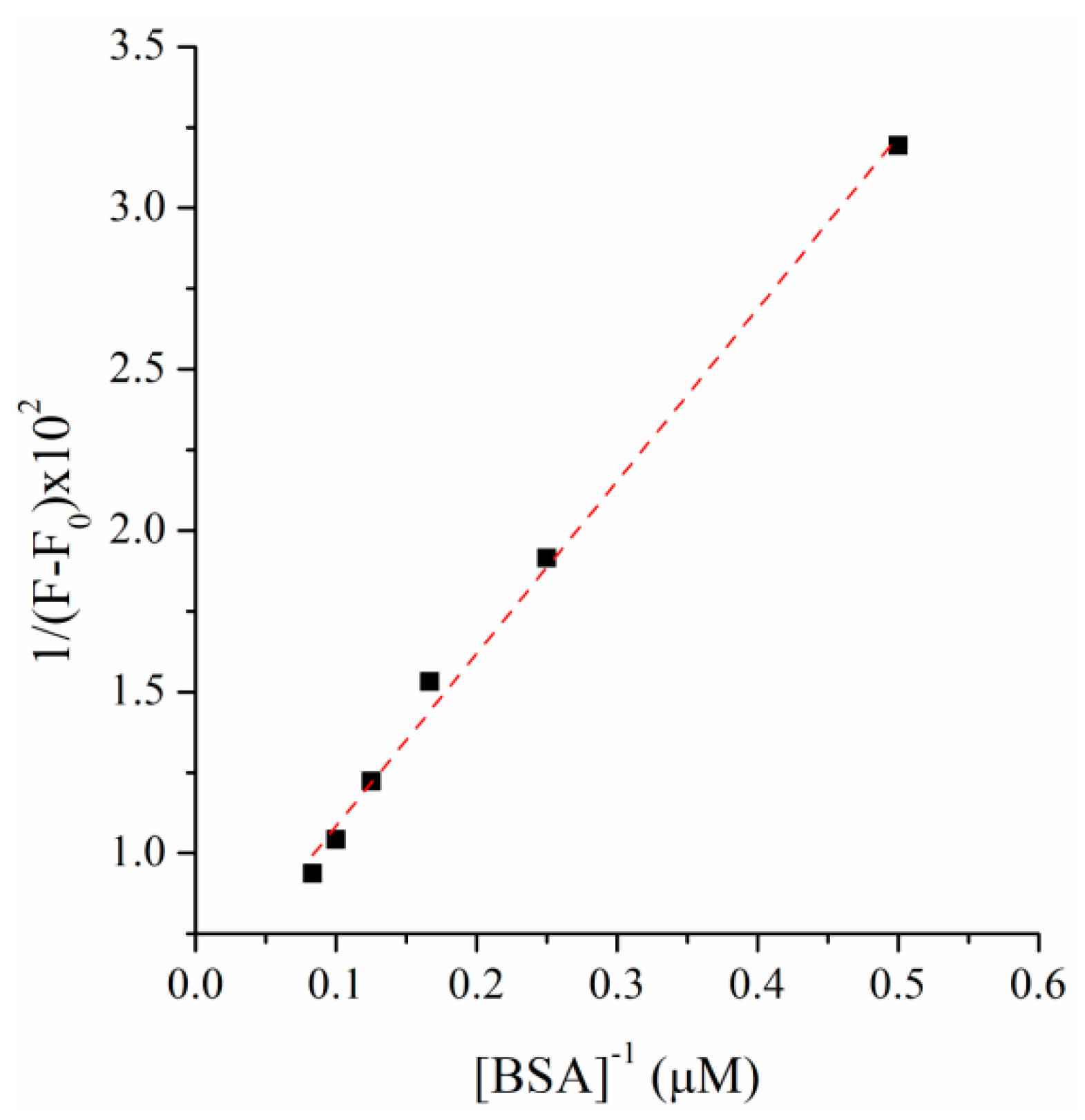
2.2. BSA Binding Study
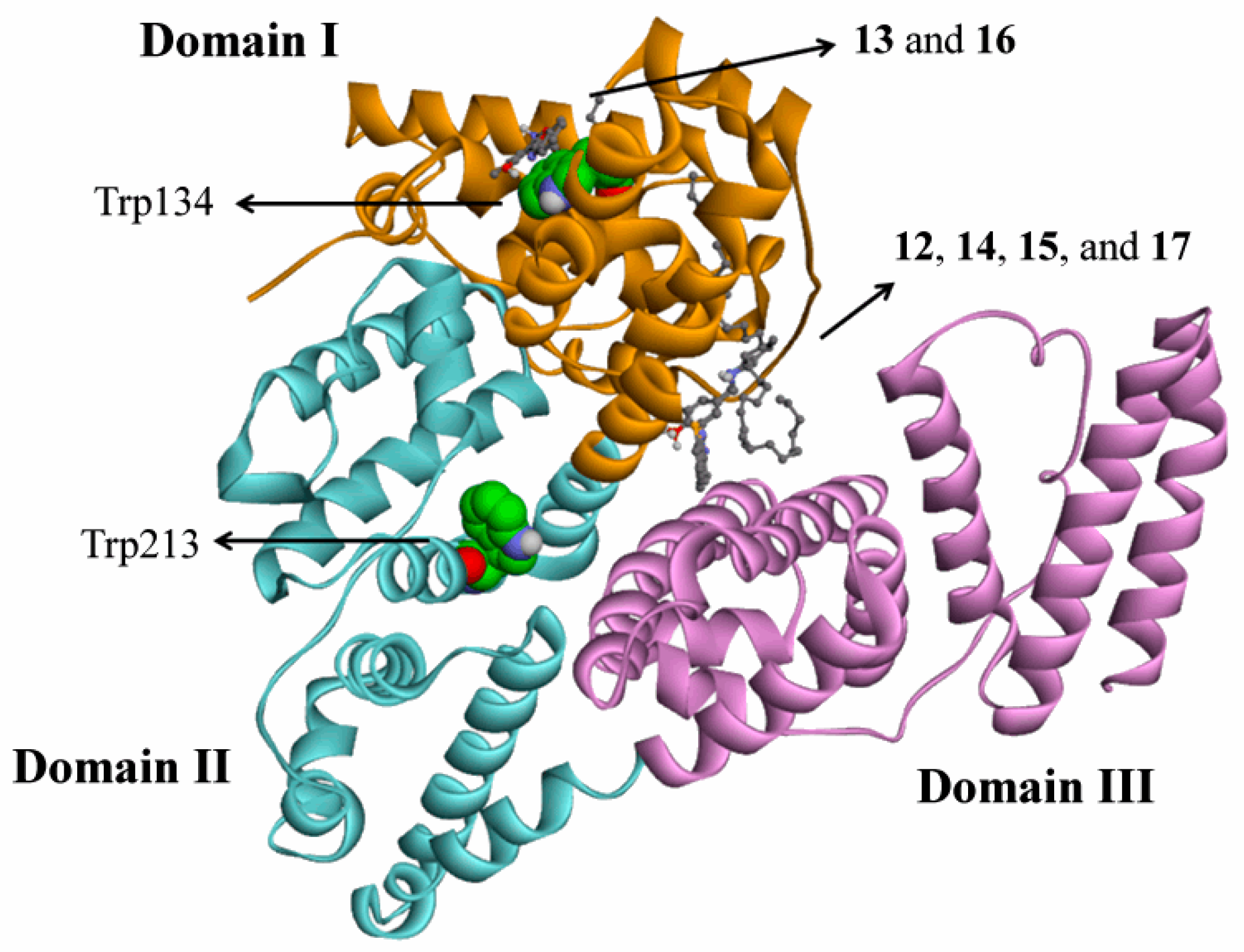
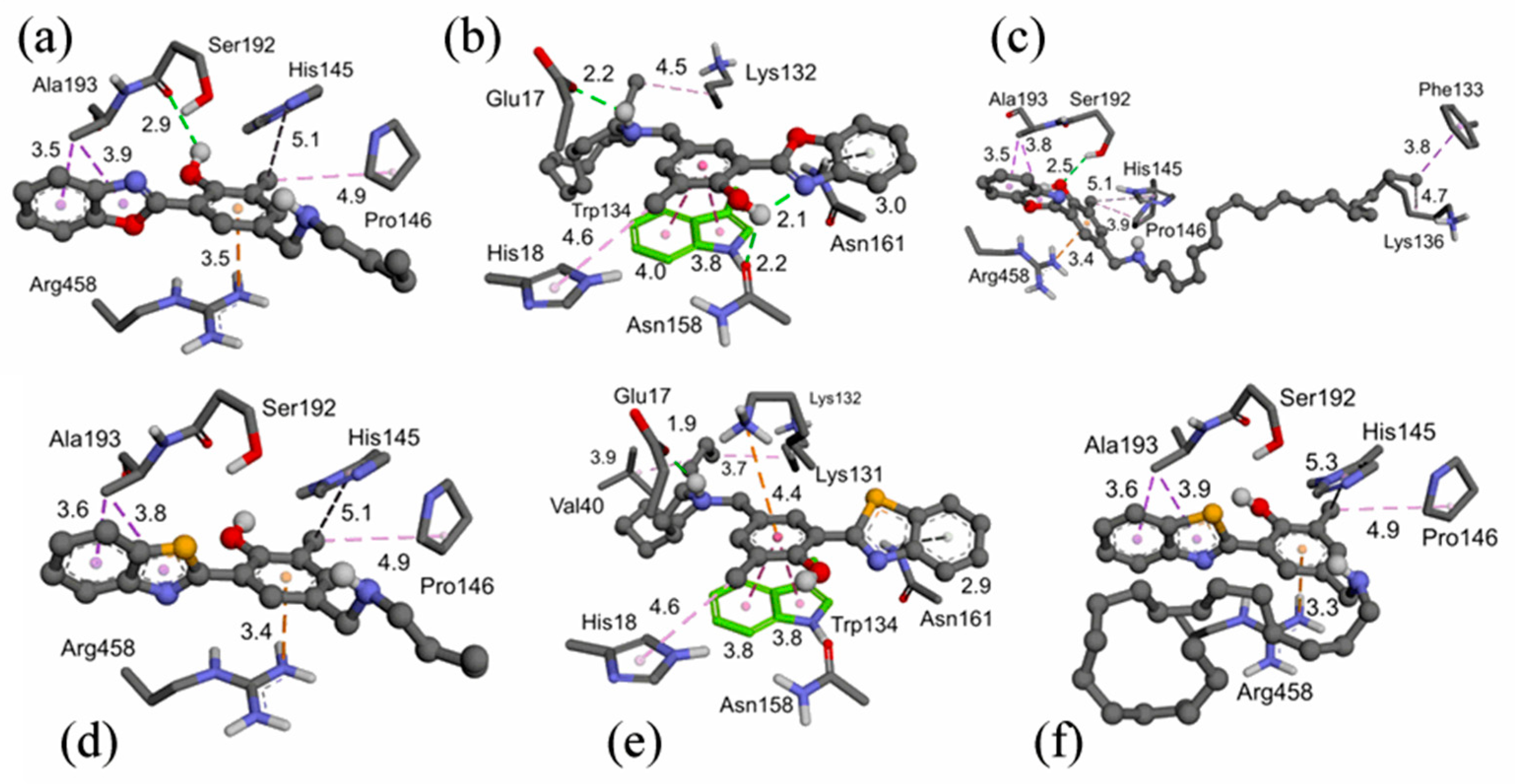
2.3. Molecular Docking
3. Experimental
3.1. Photophysical Characterization
3.2. BSA Interaction Studies
3.3. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bertucci, C.; Domenici, E. Reversible and covalent binding of drugs to human serum albumin: Methodological approaches and physiological relevance. Curr. Med. Chem. 2002, 9, 1463–1481. [Google Scholar] [CrossRef]
- Curry, S.; Mandelkow, H.; Brick, P.; Franks, N. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat. Struct. Biol. 1998, 5, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Y.Y.; Guo, Y.; Zhang, L.; Xu, R.; Xing, Z.Q.; Wang, S.X.; Zhang, X.D. Interaction of bovine serum albumin with Acridine Orange (CI Basic Orange 14) and its sonodynamic damage under ultrasonic irradiation. Dyes Pigments 2009, 80, 271–278. [Google Scholar] [CrossRef]
- Chen, L.B.; Wu, M.H.; Lin, X.C.; Xie, Z.H. Study on the interaction between human serum albumin and a novel bioactive acridine derivative using optical spectroscopy. Luminescence 2011, 26, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Gelamo, E.L.; Silva, C.; Imasato, H.; Tabak, M. Interaction of bovine (BSA) and human (HSA) serum albumins with ionic surfactants: Spectroscopy and modelling. BBA-Protein Struct. Mol. 2002, 1594, 84–99. [Google Scholar] [CrossRef]
- Sulkowska, A. Interaction of drugs with bovine and human serum albumin. J. Mol. Struct. 2002, 614, 227–232. [Google Scholar] [CrossRef]
- Abassi, P.; Abassi, F.; Yari, F.; Hashemi, M.; Nafisi, S. Study on the interaction of sulforaphane with human and bovine serum albumins. J. Photochem. Photobiol. B Biol. 2013, 122, 61–67. [Google Scholar] [CrossRef]
- Zhang, J.; Xiong, D.X.; Chen, L.N.; Kang, Q.L.; Zeng, B.R. Interaction of pyrrolizine derivatives with bovine serum albumin by fluorescence and UV-Vis spectroscopy. Spectrochim. Acta A 2012, 96, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.S.; Ramasamy, E.; Ramamurthy, V.; Rodembusch, F.S. Excited state behavior of benzoxazole derivatives in a confined environment afforded by a water soluble octaacid capsule. J. Photochem. Photobiol. A Chem. 2016, 317, 175–185. [Google Scholar] [CrossRef]
- Kwon, J.E.; Park, S.Y. Advanced organic optoelectronic materials: Harnessing excited-state intramolecular proton transfer (ESIPT) process. Adv. Mater. 2011, 23, 3615–3642. [Google Scholar] [CrossRef]
- Zhao, J.Z.; Ji, S.M.; Chen, Y.H.; Guo, H.M.; Yang, P. Excited state intramolecular proton transfer (ESIPT): From principal photophysics to the development of new chromophores and applications in fluorescent molecular probes and luminescent materials. Phys. Chem. Chem. Phys. 2012, 14, 8803–8817. [Google Scholar] [CrossRef] [PubMed]
- Rodembusch, F.S.; Campo, L.F.; Leusin, F.P.; Stefani, V. Excited state intramolecular proton transfer in amino 2-(2’-hydroxyphenyl)benzazole derivatives. Effects of the solvent and the amino group position. J. Lumin. 2007, 126, 728–734. [Google Scholar] [CrossRef]
- Lins, G.O.W.; Campo, L.F.; Rodembusch, F.S.; Stefani, V. Novel ESIPT fluorescent benzazolyl-4-quinolones: Synthesis, spectroscopic characterization and photophysical properties. Dyes Pigments 2010, 84, 114–120. [Google Scholar] [CrossRef]
- Santos, F.S.; Medeiros, N.G.; Affeldt, R.F.; Duarte, R.D.; Moura, S.; Rodembusch, F.S. Small heterocycles as highly luminescent building blocks in the solid state for organic synthesis. New J. Chem. 2016, 40, 2785–2791. [Google Scholar] [CrossRef]
- Berbigier, J.F.; Duarte, L.G.T.A.; Zawacki, M.; de Araújo, B.; Santos, C.; Atvars, T.D.Z.; Gonçalves, P.F.B.; Petzhold, C.L.; Rodembusch, F.S. ATRP initiators based on proton transfer benzazole dyes: Solid state photoactive polymer with very large Stokes shift. ACS Appl. Polym. Mater. 2020, 2, 1406–1416. [Google Scholar] [CrossRef]
- Chen, L.N.; Kuo, C.C.; Chiu, Y.C.; Chen, W.C. Ultra metal ions and pH sensing characteristics of thermoresponsive luminescent electrospun nanofibers prepared from poly(HPBO-co-NIPAAm-co-SA). RSC Adv. 2014, 4, 45345–45353. [Google Scholar] [CrossRef]
- Santos, F.S.; Duarte, L.G.T.A.; Germino, J.C.; Barboza, C.A.; Atvars, T.D.Z.; Rodembusch, F.S. Photoacidity as tool to rationalize excited state intramolecular proton transfer reactivity in flavonols. Photochem. Photobiol. Sci. 2018, 17, 231–238. [Google Scholar]
- Hong, K.I.; Park, S.H.; Lee, S.M.; Shin, I.; Jang, W.D. A pH-sensitive excited state intramolecular proton transfer fluorescent probe for imaging mitochondria and Helicobacter pylori. Sens. Actuators B Chem. 2019, 286, 148–153. [Google Scholar] [CrossRef]
- Coelho, F.L.; Santos, F.S.; Duarte, R.C.; Braga, C.A.; Toldo, J.M.; Gonçalves, P.F.B.; Rodembusch, F.S. Benzothiazole merocyanine dyes as middle pH optical sensors. Dyes Pigments 2020, 176, 108193. [Google Scholar] [CrossRef]
- Moraes, E.S.; Duarte, L.G.T.A.; Germino, J.C.; Atvars, T.D.Z. Near attack conformation as strategy for ESIPT modulation for white-light generation. J. Phys. Chem. C 2020, 124, 22406–22415. [Google Scholar] [CrossRef]
- Tang, K.C.; Chang, M.J.; Lin, T.Y.; Pan, H.A.; Fang, T.C.; Chen, K.Y.; Hung, W.Y.; Hsu, Y.H.; Chou, P.T. Fine tuning the energetics of excited-state intramolecular proton transfer (ESIPT): White light generation in a single ESIPT system. J. Am. Chem. Soc. 2011, 133, 17738–17745. [Google Scholar] [CrossRef] [PubMed]
- Duarte, L.G.T.A.; Germino, J.C.; Berbigier, J.F.; Mendes, R.A.; Faleiros, M.M.; Rodembusch, F.S.; Atvars, T.D.Z. The role of a simple and effective salicylidene derivative. Spectral broadening and performance improvement of PFO-based all-solution processed OLEDs. Dyes Pigments 2019, 171, 107671. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Y.A.; Hung, W.Y.; Tang, W.F.; Hsu, Y.H.; Chen, C.L.; Meng, F.Y.; Chou, P.T. Control of the reversibility of excited-state intramolecular proton transfer (ESIPT) reaction: Host-polarity tuning white organic light emitting diode on a new thiazolo[5,4-d]thiazole ESIPT system. Chem. Mater. 2016, 28, 8815–8824. [Google Scholar] [CrossRef]
- Chen, Y.; Fang, Y.; Gu, H.; Qiang, J.; Li, H.; Fan, J.; Cao, J.; Wang, F.; Lu, S.; Chen, X. Color-tunable and ESIPT-inspired solid fluorophores based on benzothiazole derivatives: Aggregation-induced emission, strong solvatochromic effect, and white light emission. ACS Appl. Mater. Interfaces 2020, 12, 55094–55106. [Google Scholar] [CrossRef]
- Duarte, L.G.T.A.; Germino, J.C.; Berbigier, J.F.; Barboza, C.A.; Faleiros, M.M.; Simoni, D.A.; Galante, M.T.; de Holanda, M.S.; Rodembusch, F.S.; Atvars, T.D.Z. White-light generation on all-solution-processed OLEDs using a benzothiazole-salophen derivative reactive to the ESIPT process. Phys. Chem. Chem. Phys. 2019, 21, 1172–1182. [Google Scholar] [CrossRef]
- Chen, L.; Fu, P.Y.; Wang, H.P.; Pan, M. Excited-state intramolecular proton transfer (ESIPT) for optical sensing in solid state. Adv. Opt. Mater. 2021. [Google Scholar] [CrossRef]
- Sedgwick, A.C.; Wu, L.; Han, H.H.; Bull, S.D.; He, X.P.; James, T.D.; Sessler, J.L.; Tang, B.Z.; Tian, H.; Yoon, J. Excited-state intramolecular proton-transfer (ESIPT) based fluorescence sensors and imaging agents. Chem. Soc. Rev. 2018, 47, 8842–8880. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Dahal, D.; Abeywickrama, C.S.; Pang, Y. Progress in tuning emission of the excited-state intramolecular proton transfer (ESIPT)-based fluorescent probes. ACS Omega 2021, 6, 6547–6553. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Ding, F.; Hu, X.; Feng, J.; Shen, J.; He, X. ESIPT-based fluorescent probe for bioimaging and identification of group IIIA ions in live cells and zebrafish. Bioorg. Chem. 2021, 109, 104746. [Google Scholar] [CrossRef]
- Singla, N.; Ahmad, M.; Dhiman, S.; Kumar, G.; Singh, S.; Verma, S.; Kaur, S.; Rashid, M.; Kaur, S.; Luxami, V.; et al. An ESIPT based versatile fluorescent probe for bioimaging strong acidic conditions in live-cells and E. coli. New J. Chem. 2021, 45, 19145–19153. [Google Scholar] [CrossRef]
- da Silva, C.B.; Gil, E.S.; Santos, F.S.; Morás, A.M.; Steffens, L.; Gonçalves, P.F.B.; Moura, D.J.; Lüdtke, D.S.; Rodembusch, F.S. Proton-transfer based azides with fluorescence off-on response for detection of hydrogen sulfide. An experimental, theoretical and bioimaging study. J. Org. Chem. 2018, 83, 15210–15224. [Google Scholar] [CrossRef] [PubMed]
- Tu, Z.; Liu, M.; Qian, Y.; Yang, G.; Cai, M.; Wang, L.; Huang, W. Easily fixed simple small ESIPT molecule with aggregation induced emission for fast and photostable “turn-on” bioimaging. RSC Adv. 2015, 5, 7789–7793. [Google Scholar] [CrossRef]
- Zhou, X.; Jiang, Y.; Zhao, X.; Guo, D. ESIPT-based photoactivatable fluorescent probe for ratiometric spatiotemporal bioimaging. Sensors 2016, 16, 1684. [Google Scholar] [CrossRef] [Green Version]
- De Souza, V.P.; Santos, F.S.; Rodembusch, F.S.; Braga, C.B.; Ornelas, C.; Pilli, R.A.; Russowsky, D. Hybrid dihydropyrimidinones as dual-functional bioactive molecules: Fluorescent probes and cytotoxic agents to cancer cells. New J. Chem. 2020, 44, 12440–12451. [Google Scholar] [CrossRef]
- Lv, H.M.; Yuan, D.H.; Liu, W.; Chen, Y.; Au, C.T.; Yin, S.F. A highly selective ESIPT-based fluorescent probe for cysteine sensing and its bioimaging application in living cells. Sens. Actuators B Chem. 2016, 233, 173–179. [Google Scholar] [CrossRef]
- Sulaiman, S.A.J.; Al-Rasbi, G.S.; Abou-Zied, O.K. Photophysical properties of hydroxyphenyl benzazoles and their applications as fluorescent probes to study local environment in DNA, protein, and lipid. Luminescence 2016, 31, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Abou-Zied, O.K.; Zahid, N.I.; Khyasudeen, M.F.; Giera, D.S.; Thimm, J.C.; Hashim, R. Detecting local heterogeneity and ionization ability in the head group region of different lipidic phases using modified fluorescent probes. Sci. Rep. 2015, 5, 8699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dick, P.F.; Coelho, F.L.; Rodembusch, F.S.; Campo, L.F. Amphiphilic ESIPT benzoxazole derivatives as prospective fluorescent membrane probes. Tetrahedron Lett. 2014, 55, 3024–3029. [Google Scholar] [CrossRef]
- Jiang, N.; Yang, C.; Dong, X.; Sun, X.; Zhang, D.; Liu, C. An ESIPT fluorescent probe sensitive to protein α-helix structures. Org. Biomol. Chem. 2014, 12, 5250–5259. [Google Scholar] [CrossRef] [PubMed]
- Coelho, F.L.; Rodembusch, F.S.; Campo, L.F. Synthesis, characterization and photophysics of new photoactive ESIPT lipophilic dyes. Partition experiments with different composed liposomes. Dyes Pigments 2014, 110, 134–142. [Google Scholar] [CrossRef]
- Kroetz, T.; dos Santos, M.C.; Beal, R.; Zanotto, G.M.; Santos, F.S.; Giacomelli, F.C.; Gonçalves, P.F.B.; de Lima, V.R.; Dal-Bó, A.G.; Rodembusch, F.S. Proton transfer fluorescent secondary amines. Synthesis, photophysics, theoretical calculation and preparation of photoactive phosphatidylcholine-based liposomes. Photochem. Photobiol. Sci. 2019, 18, 1171–1184. [Google Scholar] [CrossRef]
- Duarte, L.G.T.A.; Rodembusch, F.S.; Atvars, T.D.Z.; Weiss, R.G. Experimental and theoretical investigation of excited-state intramolecular proton transfer processes of benzothiazole derivatives in amino-polydimethylsiloxanes before and after crosslinking by CO2. J. Phys. Chem. A 2020, 124, 288–299. [Google Scholar] [CrossRef]
- Santos, F.S.; Ramasamy, E.; Ramamurthy, V.; Rodembusch, F.S. Photoinduced electron transfer across an organic molecular wall: Octaacid encapsulated ESIPT dyes as electron donors. Photochem. Photobiol. Sci. 2017, 16, 840–844. [Google Scholar] [CrossRef] [PubMed]
- Strickler, S.J.; Berg, R.A. Relationship between absorption intensity and fluorescence lifetime of molecules. J. Phys. Chem. 1962, 37, 814–822. [Google Scholar] [CrossRef] [Green Version]
- Turro, N.J.; Scaiano, J.C.; Ramamurthy, V. Principles of Molecular Photochemistry: An Introduction, 1st ed.; University Science Books: Sausalito, CA, USA, 2008. [Google Scholar]
- Hirayama, K.; Akashi, S.; Furuya, M.; Fukuhara, K. Rapid confirmation and revision of the primary structure of bovine serum albumin by ESIMS and Frit-FAB LC/MS. Biochem. Biophys. Res. Commun. 1990, 173, 639–646. [Google Scholar] [CrossRef]
- Kaur, A.; Banipal, P.; Banipal, T. Local anesthetic-bovine serum albumin interactional behaviour: Characterization by volumetric, calorimetric, and spectroscopic methods. J. Mol. Liq. 2017, 243, 91–101. [Google Scholar] [CrossRef]
- Thakur, A.; Patwa, J.; Pant, S.; Sharma, A.; Flora, S.J.S. Interaction study of monoisoamyl dimercaptosuccinic acid with bovine serum albumin using biophysical and molecular docking approaches. Sci. Rep. 2021, 11, 4068. [Google Scholar] [CrossRef]
- Majorek, K.A.; Porebski, P.J.; Dayal, A.; Zimmerman, M.D.; Jablonska, K.; Stewart, A.J.; Chruszcz, M.; Minor, W. Structural and immunologic characterization of bovine, horse, and rabbit serum albumins. Mol. Immunol. 2012, 52, 174–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roufegarinejad, L.; Jahanban-Esfahlan, A.; Sajed-Amin, S.; Panahi-Azar, V.; Tabibiazar, M. Molecular interactions of thymol with bovine serum albumin: Spectroscopic and molecular docking studies. J. Mol. Recognit. 2018, 31, e2704. [Google Scholar] [CrossRef]
- Borba, L.C.; Griebeler, C.H.; Bach, M.F.; Barboza, C.A.; Nogara, P.A.; da Rocha, J.B.T.; Amaral, S.S.; Rodembusch, F.S.; Schneider, P.H. Non-traditional intrinsic luminescence of amphiphilic-based ionic liquids from oxazolidines: Interaction studies in phosphatidylcholine composed liposomes and BSA optical sensing in solution. J. Mol. Liq. 2020, 313, 113525. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, NY, USA, 2006. [Google Scholar]
- Gehlen, M.H. The centenary of the Stern-Volmer equation of fluorescence quenching: From the single line plot to the SV quenching map. J. Photochem. Photobiol. C Photochem. Rev. 2020, 42, 100338. [Google Scholar] [CrossRef]
- Chaves, O.A.; Cesarin-Sobrinho, D.; Sant’Anna, C.M.R.; de Carvalho, M.G.; Suzart, L.R.; Catunda, F.E.A.; Netto-Ferreira, J.C.; Ferreira, A.B.B. Probing the interaction between 7-O-beta-D-glucopyranosyl-6-(3-methylbut-2-enyl)-5,4’-dihydroxyflavonol with bovine serum albumin (BSA). J. Photochem. Photobiol. A Chem. 2017, 336, 32–41. [Google Scholar] [CrossRef]
- Naveenraj, S.; Anandan, S. Binding of serum albumins with bioactive substances—Nanoparticles to drugs. J. Photochem. Photobiol. C 2013, 14, 53–71. [Google Scholar] [CrossRef]
- Montalti, M.; Credi, A.; Prodi, L.; Gandolfi, M.T. Handbook of Photochemistry, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Wang, C.; Wu, Q.H.; Wang, Z.; Zhao, J. Study of the interaction of Carbamazepine with bovine serum albumin by fluorescence quenching method. Anal. Sci. 2006, 22, 435–438. [Google Scholar] [CrossRef] [Green Version]
- Pisoni, D.S.; Todeschini, L.; Borges, A.C.A.; Petzhold, C.L.; Rodembusch, F.S.; Campo, L.F. Symmetrical and asymmetrical cyanine dyes. Synthesis, spectral properties and BSA association study. J. Org. Chem. 2014, 79, 5511–5522. [Google Scholar] [CrossRef]
- Rajamohan, R.; Nayaki, S.K.; Swaminathan, M. Inclusion complexation and photoprototropic behaviour of 3-amino-5-nitrobenzisothiazole with β-cyclodextrin. Spectrochim. Acta A 2008, 69, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Kragh-Hansen, U. Molecular aspects of ligand binding to serum albumin. Pharmacol. Rev. 1981, 33, 17–53. [Google Scholar]
- Moriyama, Y.; Ohta, D.; Hachiya, K.; Mitsui, Y.; Takeda, K. Fluorescence behavior of tryptophan residues of bovine and human serum albumins in ionic surfactant solutions: A comparative study of the two and one tryptophan(s) of bovine and human albumins. J. Protein Chem. 1996, 15, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, G.K.; Suthar, M.K.; Verma, N.; Asthana, S.; Tripathi, A.; Gupta, S.K.; Saxena, J.K.; Raisuddin, S.; Das, M. Investigation of the interaction of anthraquinones of Cassia occidentalis seeds with bovine serum albumin by molecular docking and spectroscopic analysis: Correlation to their in vitro cytotoxic potential. Food Res. Int. 2015, 77, 368–377. [Google Scholar] [CrossRef]
- Karami, K.; Rafiee, M.; Lighvan, Z.M.; Zakariazadeh, M.; Faal, A.Y.; Esmaeili, S.A.; Momtazi-Borojeni, A.A. Synthesis, spectroscopic characterization and in vitro cytotoxicities of new organometallic palladium complexes with biologically active β-diketones; Biological evaluation probing of the interaction mechanism with DNA/Protein and molecular docking. J. Mol. Struct. 2018, 1154, 480–495. [Google Scholar] [CrossRef]
- Wani, T.A.; Bakheit, A.H.; Abounassif, M.A.; Zargar, S. Study of Interactions of an anticancer drug Neratinib with bovine serum albumin: Spectroscopic and molecular docking approach. Front. Chem. 2018, 6, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chekalyuk, A.; Fadeev, V.; Georgiev, G.; Kalkanjjev, T.; Nickolov, Z. Determination of fluorescence quantum yields using a spontaneous Raman scattering line of the solvent as internal standard. Spectrosc. Lett. 1982, 15, 355–365. [Google Scholar] [CrossRef]
- Bujacz, A. Structures of bovine, equine and leporine serum albumin. Acta Crystallogr. Sect. D 2012, 68, 1278–1289. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1162. [Google Scholar] [CrossRef] [Green Version]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, J.J.P. Optimization of parameters for semiempirical methods V: Modification of NDDO approximations and application to 70 elements. J. Mol. Mod. 2007, 13, 1173–1213. [Google Scholar] [CrossRef] [Green Version]
- Stewart, J.J.P. MOPAC2012; Stewart Computational Chemistry: Colorado Springs, CO, USA, 2012; Available online: http://OpenMOPAC.net (accessed on 15 November 2019).
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Dassault Systèmes BIOVIA. Discovery Studio Modeling Environment, Release 2017; Dassault Systèmes: San Diego, CA, USA, 2017. [Google Scholar]


| Benzazole | λabs (nm) | ε (104 M−1·cm−1) | fe | ke0 (108 s−1) | τ0a (ns) | λem (nm) | ΔλST (cm−1) | ΦFL (%) |
|---|---|---|---|---|---|---|---|---|
| 12 | 330 | 1.53 | 0.895 | 8.22 | 1.22 | 464 | 8751 | 0.069 |
| 13 | 350 | 1.78 | 0.906 | 7.39 | 1.35 | 502 | 8651 | 0.094 |
| 14 | 350 | 2.02 | 0.943 | 7.70 | 1.30 | 492 | 8246 | 0.144 |
| 15 | 332 | 1.40 | 0.860 | 7.80 | 1.28 | 467 | 8707 | 0.087 |
| 16 | 367 | 1.39 | 0.773 | 5.74 | 1.74 | 542 | 8798 | 0.099 |
| 17 | 365 | 1.36 | 0.789 | 5.92 | 1.69 | 542 | 8947 | 0.148 |
| Benzazole | Q[a] (%) | KSV (×104 M−1) | kq (×1012 M−1·s−1) | KA (×104 M−1) | n | ΔG0[b] (kcal·mol−1) |
|---|---|---|---|---|---|---|
| 12 | 42 | 3.14 | 5.18 | 0.61 | 0.92 | −5.16 |
| 13 | 50 | 5.12 | 8.45 | 0.74 | 1.16 | −5.28 |
| 14 | 41 | 3.18 | 5.25 | 0.59 | 0.88 | −5.14 |
| 15 | 48 | 4.66 | 7.69 | 0.73 | 1.15 | −5.27 |
| 16 | 53 | 5.62 | 9.27 | 0.79 | 1.31 | −5.31 |
| 17 | 41 | 3.46 | 5.71 | 0.69 | 1.09 | −5.23 |
| Compound | 12 | 13 | 14 | 15 | 16 | 17 |
|---|---|---|---|---|---|---|
| ∆G bind (kcal·mol−1) | −8.0 | −6.8 | −6.5 | −8.0 | −6.9 | −6.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kroetz, T.; Nogara, P.A.; da Silveira Santos, F.; da Luz, L.C.; Câmara, V.S.; da Rocha, J.B.T.; Gonçalves Dal-Bó, A.; Rodembusch, F.S. Interaction Study between ESIPT Fluorescent Lipophile-Based Benzazoles and BSA. Molecules 2021, 26, 6728. https://doi.org/10.3390/molecules26216728
Kroetz T, Nogara PA, da Silveira Santos F, da Luz LC, Câmara VS, da Rocha JBT, Gonçalves Dal-Bó A, Rodembusch FS. Interaction Study between ESIPT Fluorescent Lipophile-Based Benzazoles and BSA. Molecules. 2021; 26(21):6728. https://doi.org/10.3390/molecules26216728
Chicago/Turabian StyleKroetz, Thais, Pablo Andrei Nogara, Fabiano da Silveira Santos, Lilian Camargo da Luz, Viktor Saraiva Câmara, João Batista Teixeira da Rocha, Alexandre Gonçalves Dal-Bó, and Fabiano Severo Rodembusch. 2021. "Interaction Study between ESIPT Fluorescent Lipophile-Based Benzazoles and BSA" Molecules 26, no. 21: 6728. https://doi.org/10.3390/molecules26216728







