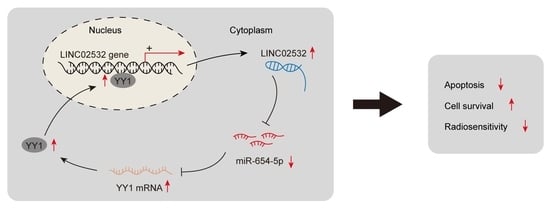
LINC02532 Contributes to Radiosensitivity in Clear Cell Renal Cell Carcinoma through the miR-654-5p/YY1 Axis
Abstract
:1. Background
2. Methods
2.1. Cell Lines and Cell Culture
2.2. Cell Transfection
2.3. Quantitative Real-Time PCR (qRT-PCR)
2.4. Subcellular Fractionation
2.5. CCK-8 Assay
2.6. Radiation Treatment
2.7. Cell Apoptosis Detection
2.8. Immunofluorescence Staining
2.9. Fluorescence In Situ Hybridization (FISH)
2.10. Western Blotting
2.11. Luciferase Reporter Assay
2.12. Chromatin Immunoprecipitation (ChIP)
2.13. Xenograft Assay
2.14. Statistical Analysis
3. Results
3.1. Knockdown of LINC02532 Suppresses Cell Viability in ccRCC Cells
3.2. LINC02532 Knockdown Accelerates Radiosensitivity in ccRCC
3.3. YY1 Transcriptionally Activates LINC02532 in ccRCC Cells
3.4. LINC02532 Sponges miR-654-5p to Regulate YY1 Expression in ccRCC Cells
3.5. miR-654-5p Overexpression Restores the Effect of LINC02532 Overexpression on Radiosensitivity in ccRCC Cells
3.6. YY1 Knockdown Abolishes the Effect of LINC02532 Overexpression on Radiosensitivity in ccRCC Cells
3.7. LINC02532 Knockdown Enhances Radiosensitivity of ccRCC Cells In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
| ATCC | American Type Culture Collection |
| CCK-8 | cell counting kit-8 |
| ccRCC | clear cell renal cell carcinoma |
| ChIP | chromatin immunoprecipitation |
| DAPI | 4′,6-diamidino-2-phenylindol |
| DSB | double-strand break |
| FBS | fetal bovine serum |
| FISH | fluorescence in situ hybridization |
| IR | ionizing radiation |
| lncRNA | long non-coding RNA |
| miRNA | microRNA |
| ncRNA | non-coding RNA |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| RCC | renal cell carcinoma |
| siRNA | small interfering RNA |
| TCGA | The Cancer Genome Atlas |
| UTR | untranslated region |
| YY1 | Yin Yang 1 |
References
- Linehan, W.M.; Schmidt, L.S.; Crooks, D.R.; Wei, D.; Srinivasan, R.; Lang, M.; Ricketts, C.J. The Metabolic Basis of Kidney Cancer. Cancer Discov. 2019, 9, 1006–1021. [Google Scholar] [CrossRef] [Green Version]
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ficarra, V.; Guille, F.; Schips, L.; de la Taille, A.; Prayer Galetti, T.; Tostain, J.; Cindolo, L.; Novara, G.; Zigeuner, R.; Bratti, E.; et al. Proposal for revision of the TNM classification system for renal cell carcinoma. Cancer 2005, 104, 2116–2123. [Google Scholar] [CrossRef]
- Dabestani, S.; Marconi, L.; Hofmann, F.; Stewart, F.; Lam, T.B.; Canfield, S.E.; Staehler, M.; Powles, T.; Ljungberg, B.; Bex, A. Local treatments for metastases of renal cell carcinoma: A systematic review. Lancet Oncol. 2014, 15, e549–e561. [Google Scholar] [CrossRef]
- Lichter, A.S.; Lawrence, T.S. Recent advances in radiation oncology. N. Engl. J. Med. 1995, 332, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Khanna, K.K.; Jackson, S.P. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat. Genet. 2001, 27, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Santivasi, W.L.; Xia, F. Ionizing radiation-induced DNA damage, response, and repair. Antioxid. Redox Signal. 2014, 21, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapman, J.R.; Taylor, M.R.; Boulton, S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 2012, 47, 497–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thapar, R. Regulation of DNA Double-Strand Break Repair by Non-Coding RNAs. Molecules 2018, 23, 2789. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, D.; Choi, Y.E.; Brault, M.E. Charity begins at home: Non-coding RNA functions in DNA repair. Nat. Rev. Mol. Cell Biol. 2013, 14, 181–189. [Google Scholar] [CrossRef]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Chen, S.; Yang, B.; Mao, W.; Yang, X.; Cai, J. Molecular mechanisms of lncRNAs in regulating cancer cell radiosensitivity. Biosci. Rep. 2019, 39, BSR20190590. [Google Scholar] [CrossRef] [PubMed]
- Michelini, F.; Pitchiaya, S.; Vitelli, V.; Sharma, S.; Gioia, U.; Pessina, F.; Cabrini, M.; Wang, Y.; Capozzo, I.; Iannelli, F.; et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 2017, 19, 1400–1411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Chen, X.; Chen, X.; Liu, J.; Gu, H.; Fan, R.; Ge, H. Long non-coding RNA HOTAIR knockdown enhances radiosensitivity through regulating microRNA-93/ATG12 axis in colorectal cancer. Cell Death Dis. 2020, 11, 175. [Google Scholar] [CrossRef]
- Zhong, Q.; Chen, Y.; Chen, Z. LncRNA MINCR regulates irradiation resistance in nasopharyngeal carcinoma cells via the microRNA-223/ZEB1 axis. Cell Cycle 2020, 19, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wang, H.; Yang, J.; Long, W.; Zhang, B.; Liu, J.; Yu, B. Down-regulated circPAPPA suppresses the proliferation and invasion of trophoblast cells via the miR-384/STAT3 pathway. Biosci. Rep. 2019, 39, BSR20191965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Amini Khorasgani, M.; Mohammady Nejad, P.; Moghani Bashi, M.M.; Hedayati, M. Evaluation of mir-377-3p Expression in Patients with Multiple Sclerosis. Sci. Med. J. 2019, 1, 48–54. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Croce, C.M. microRNA involvement in human cancer. Carcinogenesis 2012, 33, 1126–1133. [Google Scholar] [CrossRef] [Green Version]
- Gandellini, P.; Rancati, T.; Valdagni, R.; Zaffaroni, N. miRNAs in tumor radiation response: Bystanders or participants? Trends Mol. Med. 2014, 20, 529–539. [Google Scholar] [CrossRef]
- Chen, X.; Xu, Y.; Jiang, L.; Tan, Q. miRNA-218-5p increases cell sensitivity by inhibiting PRKDC activity in radiation-resistant lung carcinoma cells. Thorac. Cancer 2021, 12, 1549–1557. [Google Scholar] [CrossRef]
- Hu, P.; Zhou, L.; Wang, C.; Cao, G.; Chang, Y. MiR-181a reduces radiosensitivity of non-small cell lung cancer via inhibiting PTEN. Panminerva Med. 2020. [Google Scholar] [CrossRef]
- Lu, M.; Wang, C.; Chen, W.; Mao, C.; Wang, J. miR-654-5p Targets GRAP to Promote Proliferation, Metastasis, and Chemoresistance of Oral Squamous Cell Carcinoma Through Ras/MAPK Signaling. DNA Cell Biol. 2018, 37, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.Y.; Xu, X.Y.; Wang, J.F.; Zhang, C.W.; Zhang, S.C. MiR-654-5p attenuates breast cancer progression by targeting EPSTI1. Am. J. Cancer Res. 2016, 6, 522–532. [Google Scholar] [PubMed]
- Huang, F.; Wu, X.; Wei, M.; Guo, H.; Li, H.; Shao, Z.; Wu, Y.; Pu, J. miR-654-5p Targets HAX-1 to Regulate the Malignancy Behaviors of Colorectal Cancer Cells. Biomed. Res. Int. 2020, 2020, 4914707. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.Z.; Song, H.; Zhao, Y.; Zhang, L. MiR-654-5p regulated cell progression and tumor growth through targeting SIRT6 in osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 3517–3525. [Google Scholar] [CrossRef]
- Wu, S.; Wang, H.; Li, Y.; Xie, Y.; Huang, C.; Zhao, H.; Miyagishi, M.; Kasim, V. Transcription Factor YY1 Promotes Cell Proliferation by Directly Activating the Pentose Phosphate Pathway. Cancer Res. 2018, 78, 4549–4562. [Google Scholar] [CrossRef] [Green Version]
- Gordon, S.; Akopyan, G.; Garban, H.; Bonavida, B. Transcription factor YY1: Structure, function, and therapeutic implications in cancer biology. Oncogene 2006, 25, 1125–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figiel, M.; Gorecki, A. Physical Interaction of Human Yin Yang 1 Protein with DNA. Crit. Rev. Oncog. 2017, 22, 75–97. [Google Scholar] [CrossRef]
- Zaravinos, A.; Spandidos, D.A. Yin yang 1 expression in human tumors. Cell Cycle 2010, 9, 512–522. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.; Wang, G.; Yang, L.; Peng, B.; Wen, Y.; Ding, G.; Wang, Z. Transcription Factor YY1 Modulates Lung Cancer Progression by Activating lncRNA-PVT1. DNA Cell Biol. 2017, 36, 947–958. [Google Scholar] [CrossRef]
- Knauss, J.L.; Miao, N.; Kim, S.N.; Nie, Y.; Shi, Y.; Wu, T.; Pinto, H.B.; Donohoe, M.E.; Sun, T. Long noncoding RNA Sox2ot and transcription factor YY1 co-regulate the differentiation of cortical neural progenitors by repressing Sox2. Cell Death Dis. 2018, 9, 799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Li, R.; Qiu, J.Z.; Yu, J.B.; Cao, Y.; Yuan, R.T. YY1-mediated PTEN dephosphorylation antagonizes IR-induced DNA repair contributing to tongue squamous cell carcinoma radiosensitization. Mol. Cell Probes 2020, 53, 101577. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhou, X.; Ge, X.; Liu, P.; Cao, J.; Lu, X.; Ling, Y.; Zhang, S. Upregulation of Ying Yang 1 (YY1) suppresses esophageal squamous cell carcinoma development through heme oxygenase-1. Cancer Sci. 2013, 104, 1544–1551. [Google Scholar] [CrossRef]
- Shen, B.; Li, Y.; Ye, Q.; Qin, Y. YY1-mediated long non-coding RNA Kcnq1ot1 promotes the tumor progression by regulating PTEN via DNMT1 in triple negative breast cancer. Cancer Gene Ther. 2020, 28, 1099–1112. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Guan, C.; Hu, Z.; Liu, L.; Su, Z.; Kang, P.; Jiang, X.; Cui, Y. Yin Yang 1-induced LINC00667 up-regulates pyruvate dehydrogenase kinase 1 to promote proliferation, migration and invasion of cholangiocarcinoma cells by sponging miR-200c-3p. Hum. Cell 2021, 34, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Shen, Z.Z.; Xiao, C.M.; Sha, Q.K. YY1-mediated up-regulation of lncRNA LINC00466 facilitates glioma progression via miR-508/CHEK1. J. Gene Med. 2021, 23, e3287. [Google Scholar] [CrossRef] [PubMed]
- You, Q.; Yao, Y.; Wu, J.; Cheng, C.; Li, Y.; Yuan, H. YY1-induced lncRNA DSCR8 promotes the progression of ovarian cancer via miR-3192-5p/YY1 axis. Biomed. Pharm. 2020, 129, 110339. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, J.J.; Tay, Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef] [Green Version]
- Oosterwijk, E.; Rathmell, W.K.; Junker, K.; Brannon, A.R.; Pouliot, F.; Finley, D.S.; Mulders, P.F.; Kirkali, Z.; Uemura, H.; Belldegrun, A. Basic research in kidney cancer. Eur. Urol. 2011, 60, 622–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Meerleer, G.; Khoo, V.; Escudier, B.; Joniau, S.; Bossi, A.; Ost, P.; Briganti, A.; Fonteyne, V.; Van Vulpen, M.; Lumen, N.; et al. Radiotherapy for renal-cell carcinoma. Lancet Oncol. 2014, 15, e170–e177. [Google Scholar] [CrossRef]
- De Felice, F.; Tombolini, V. Radiation therapy in renal cell carcinoma. Crit. Rev. Oncol. Hematol. 2018, 128, 82–87. [Google Scholar] [CrossRef]
- Kwak, C.; Park, Y.H.; Jeong, C.W.; Lee, S.E.; Ku, J.H. Metastasectomy without systemic therapy in metastatic renal cell carcinoma: Comparison with conservative treatment. Urol. Int. 2007, 79, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Siva, S.; Kothari, G.; Muacevic, A.; Louie, A.V.; Slotman, B.J.; Teh, B.S.; Lo, S.S. Radiotherapy for renal cell carcinoma: Renaissance of an overlooked approach. Nat. Rev. Urol. 2017, 14, 549–563. [Google Scholar] [CrossRef] [PubMed]
- Agsalda-Garcia, M.; Shieh, T.; Souza, R.; Kamada, N.; Loi, N.; Oda, R.; Acosta-Maeda, T.; Choi, S.Y.; Lim, E.; Misra, A.; et al. Raman-Enhanced Spectroscopy (RESpect) Probe for Childhood Non-Hodgkin Lymphoma. Sci. Med. J. 2020, 2, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosvyra, A.; Maramis, C.; Chouvarda, I. Developing an Integrated Genomic Profile for Cancer Patients with the Use of NGS Data. Emerg. Sci. J. 2019, 3, 157–167. [Google Scholar] [CrossRef]
- Sekar, D.; Thirugnanasambantham, K.; Hairul Islam, V.I.; Saravanan, S. Sequencing approaches in cancer treatment. Cell Prolif. 2014, 47, 391–395. [Google Scholar] [CrossRef]
- Tuna, M.; Amos, C.I. Genomic sequencing in cancer. Cancer Lett. 2013, 340, 161–170. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Wang, C.; Zhang, J.; Zhou, Y.; Hu, W.; Guo, T. New insights into long noncoding RNAs and pseudogenes in prognosis of renal cell carcinoma. Cancer Cell Int. 2018, 18, 157. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, W.; Yan, Y.; Yao, X.; Gu, W.; Zhang, H. LINC01094 triggers radio-resistance in clear cell renal cell carcinoma via miR-577/CHEK2/FOXM1 axis. Cancer Cell Int. 2020, 20, 274. [Google Scholar] [CrossRef]
- Zhang, C.; Ma, M.H.; Liang, Y.; Wu, K.Z.; Dai, D.Q. Novel long non-coding RNA LINC02532 promotes gastric cancer cell proliferation, migration, and invasion in vitro. World J. Gastrointest. Oncol. 2019, 11, 91–101. [Google Scholar] [CrossRef]
- Zhang, H.; Hua, Y.; Jiang, Z.; Yue, J.; Shi, M.; Zhen, X.; Zhang, X.; Yang, L.; Zhou, R.; Wu, S. Cancer-associated Fibroblast-promoted LncRNA DNM3OS Confers Radioresistance by Regulating DNA Damage Response in Esophageal Squamous Cell Carcinoma. Clin. Cancer Res. 2019, 25, 1989–2000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Ding, L.; Sun, G.; Ning, H.; Huang, R. Suppression of LINC00460 mediated the sensitization of HCT116 cells to ionizing radiation by inhibiting epithelial-mesenchymal transition. Toxicol. Res. (Camb) 2020, 9, 107–116. [Google Scholar] [CrossRef]
- Thomas, M.J.; Seto, E. Unlocking the mechanisms of transcription factor YY1: Are chromatin modifying enzymes the key? Gene 1999, 236, 197–208. [Google Scholar] [CrossRef]
- Zhang, Y.; Jin, F.; Li, X.C.; Shen, F.J.; Ma, X.L.; Wu, F.; Zhang, S.M.; Zeng, W.H.; Liu, X.R.; Fan, J.X.; et al. The YY1-HOTAIR-MMP2 Signaling Axis Controls Trophoblast Invasion at the Maternal-Fetal Interface. Mol. Ther. 2017, 25, 2394–2403. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Tie, Y.; Lv, G.; Zhu, J.; Fu, H.; Zheng, X. Transcriptional activation of miR-320a by ATF2, ELK1 and YY1 induces cancer cell apoptosis under ionizing radiation conditions. Int. J. Oncol. 2018, 53, 1691–1702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moran, V.A.; Perera, R.J.; Khalil, A.M. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012, 40, 6391–6400. [Google Scholar] [CrossRef]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013, 14, 699–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majem, B.; Parrilla, A.; Jimenez, C.; Suarez-Cabrera, L.; Barber, M.; Marin, A.; Castellvi, J.; Tamayo, G.; Moreno-Bueno, G.; Ponce, J.; et al. MicroRNA-654-5p suppresses ovarian cancer development impacting on MYC, WNT and AKT pathways. Oncogene 2019, 38, 6035–6050. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.Y.; Wang, X.L.; Dang, Y.; Zhu, X.Z.; Zhang, Y.H.; Cai, B.X.; Zheng, L. Long non-coding RNA UCA1 promotes the progression of paclitaxel resistance in ovarian cancer by regulating the miR-654-5p/SIK2 axis. Eur. Rev. Med. Pharm. Sci. 2020, 24, 591–603. [Google Scholar] [CrossRef]
- Kong, R. Circular RNA hsa_circ_0085131 is involved in cisplatin-resistance of non-small-cell lung cancer cells by regulating autophagy. Cell Biol. Int. 2020, 44, 1945–1956. [Google Scholar] [CrossRef] [PubMed]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Zeng, B.; Li, Y.; Wang, H.; Zhang, X. LINC02532 Contributes to Radiosensitivity in Clear Cell Renal Cell Carcinoma through the miR-654-5p/YY1 Axis. Molecules 2021, 26, 7040. https://doi.org/10.3390/molecules26227040
Zhou X, Zeng B, Li Y, Wang H, Zhang X. LINC02532 Contributes to Radiosensitivity in Clear Cell Renal Cell Carcinoma through the miR-654-5p/YY1 Axis. Molecules. 2021; 26(22):7040. https://doi.org/10.3390/molecules26227040
Chicago/Turabian StyleZhou, Xiaoguang, Bowen Zeng, Yansheng Li, Haozhou Wang, and Xiaodong Zhang. 2021. "LINC02532 Contributes to Radiosensitivity in Clear Cell Renal Cell Carcinoma through the miR-654-5p/YY1 Axis" Molecules 26, no. 22: 7040. https://doi.org/10.3390/molecules26227040
APA StyleZhou, X., Zeng, B., Li, Y., Wang, H., & Zhang, X. (2021). LINC02532 Contributes to Radiosensitivity in Clear Cell Renal Cell Carcinoma through the miR-654-5p/YY1 Axis. Molecules, 26(22), 7040. https://doi.org/10.3390/molecules26227040