Molecular Cloning, Expression and Macrophage Activation of an Immunoregulatory Protein from Cordyceps militaris
Abstract
:1. Introduction
2. Results
2.1. Cloning, Expression and Purification of Recombinant CMIP
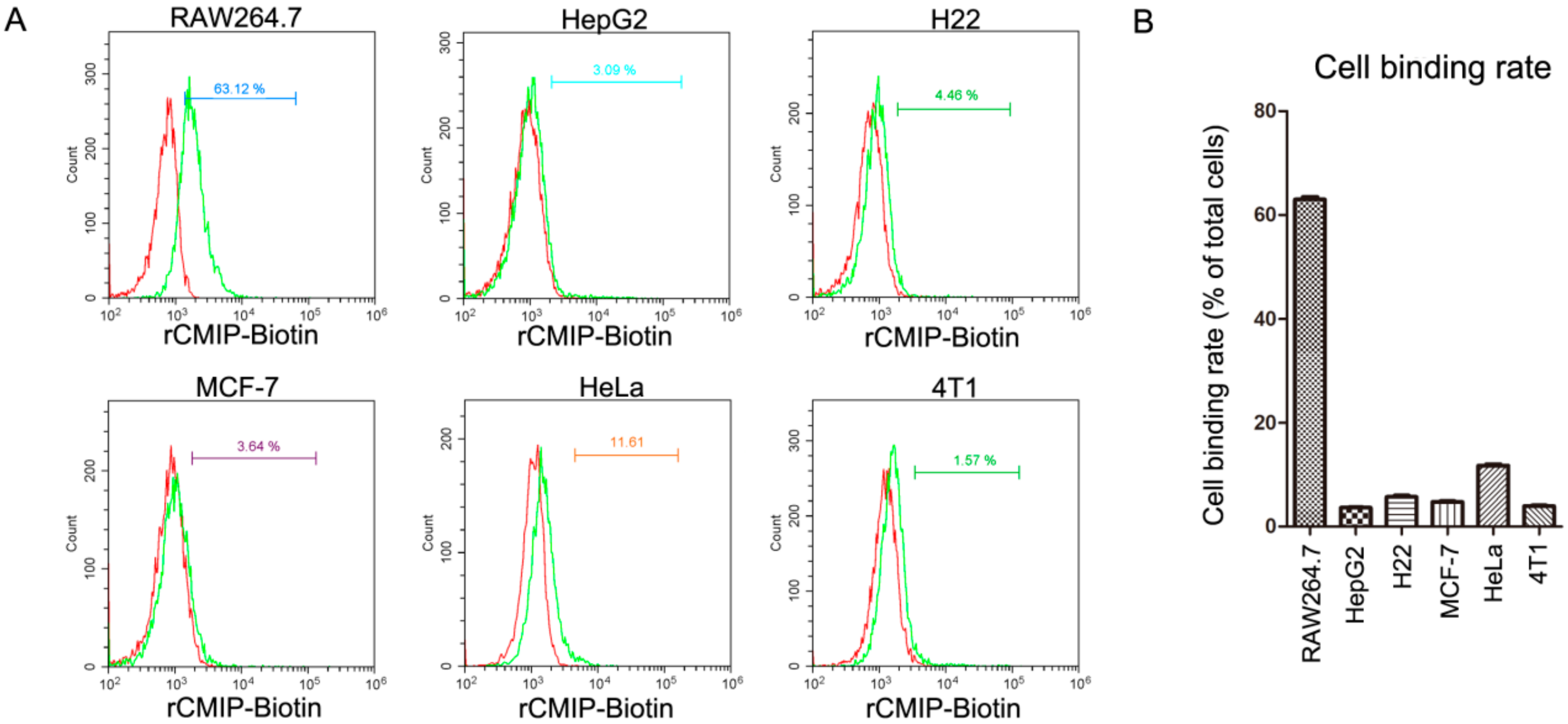
2.2. Cell Binding Specificity of Recombinant CMIP
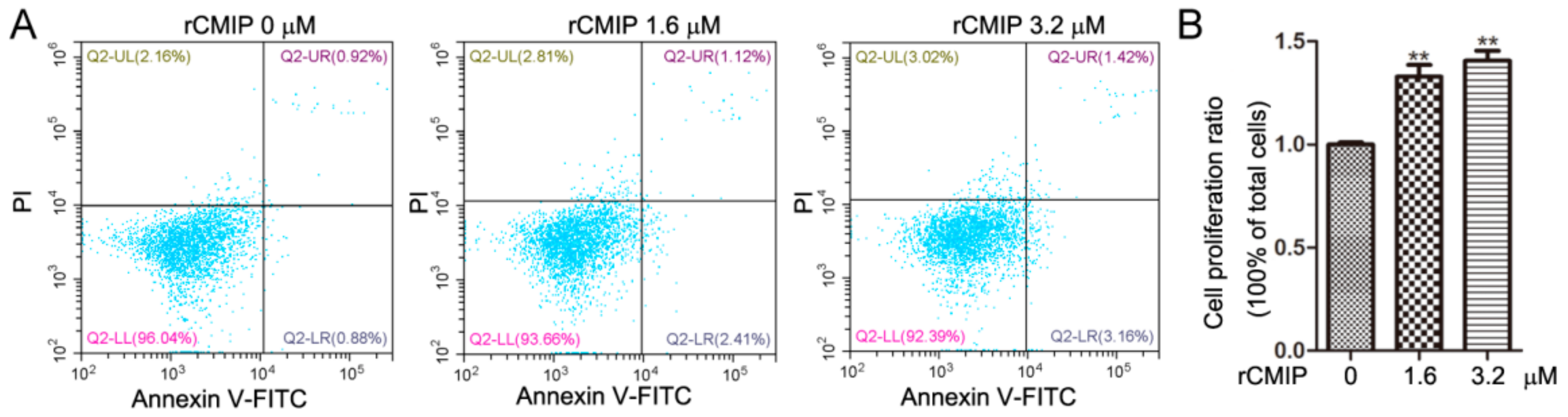
2.3. Effect of Recombinant CMIP on Cell Growth of Macrophages
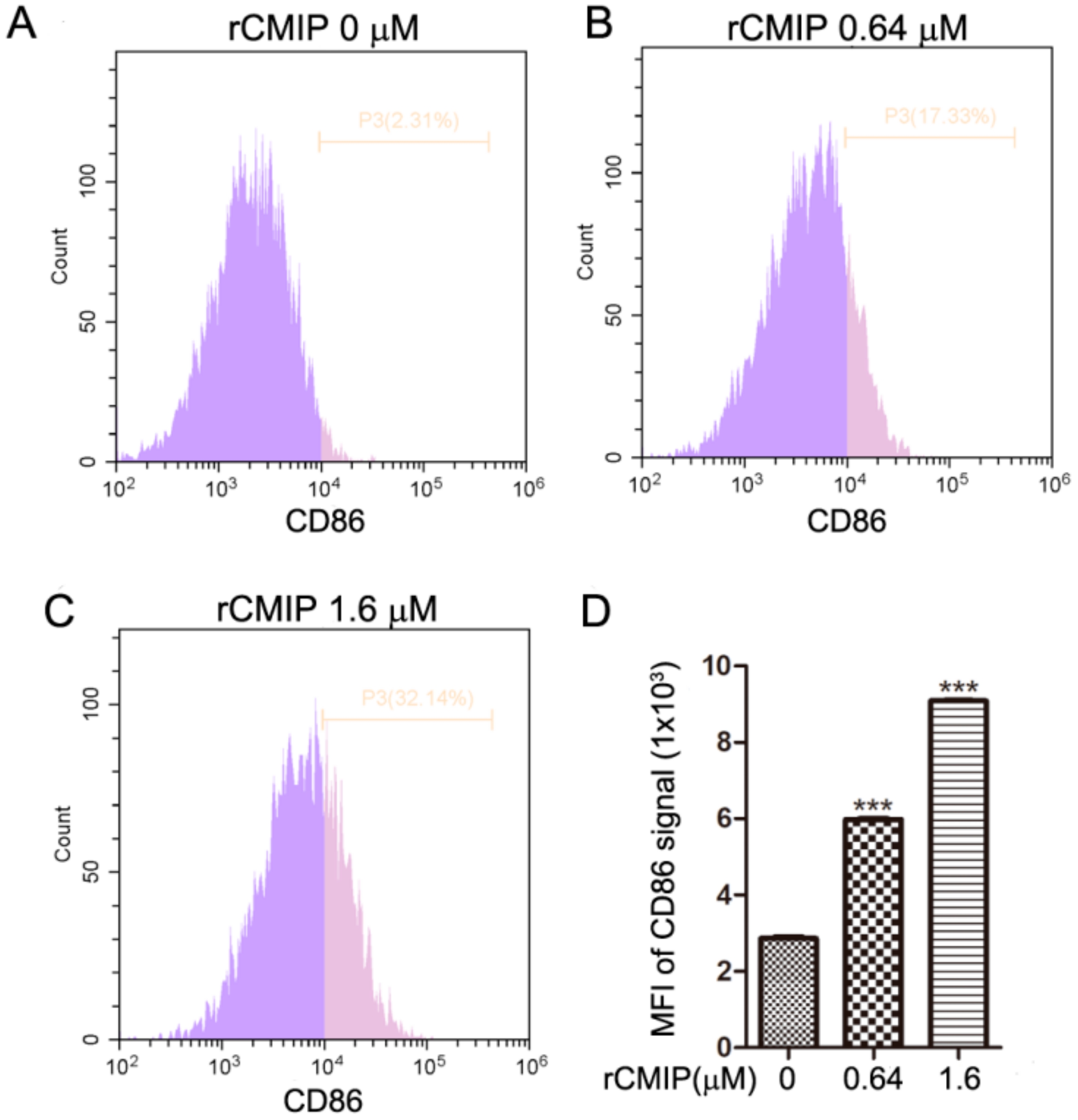
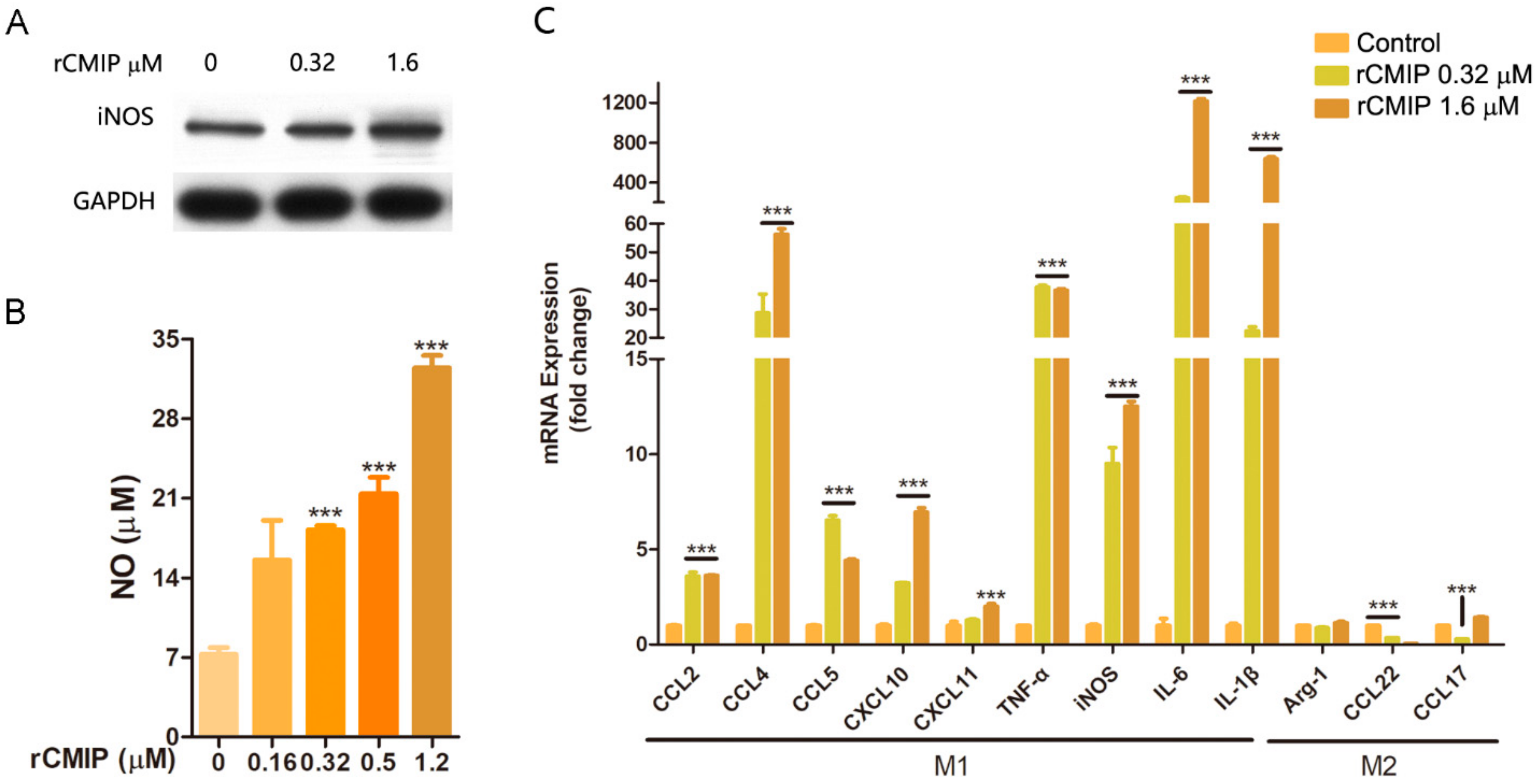
2.4. rCMIP-Induced Macrophages Polarized to M1 Phenotype
3. Discussion
4. Materials and Methods
4.1. Strains and Vectors
4.2. Materials and Chemicals
4.3. Cloning of CMIP Gene
4.4. Expression and Purification of Recombinant CMIP
4.5. Biotinylation of rCMIP
4.6. Cell Culture and Treatment
4.7. Cell-Adhesion Assay, Annexin V/PI Staining and FACS Analysis
4.8. Cell Proliferation Assay
4.9. Quantitative Real-Time PCR for Cytokine mRNAs
4.10. Flow Cytometry Detection of CD86 Expression
4.11. iNOS Expression and NO Production Detection
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Sample Availability
References
- Roncero-Ramos, I.; Delgado-Andrade, C. The beneficial role of edible mushrooms in human health. Curr. Opin. Food Sci. 2017, 14, 122–128. [Google Scholar] [CrossRef]
- Singh, R.S.; Walia, A.K.; Kennedy, J.F. Mushroom lectins in biomedical research and development. Int. J. Biol. Macromol. 2020, 151, 1340–1350. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Ruiz Beguerie, J.; Sze, D.M.-Y.; Chan, G.C.F. Ganoderma lucidum (Reishi mushroom) for cancer treatment. Cochrane Database Syst. Rev. 2016, 4, CD007731. [Google Scholar] [CrossRef]
- Motta, F.; Gershwin, M.E.; Selmi, C. Mushrooms and immunity. J. Autoimmun. 2021, 117, 102576. [Google Scholar] [CrossRef]
- Li, Q.-Z.; Zheng, Y.-Z.; Zhou, X.-W. Fungal immunomodulatory proteins: Characteristic, potential antitumor activities and their molecular mechanisms. Drug Discov. Today 2019, 24, 307–314. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, M.; Fang, Z. Efficient physical extraction of active constituents from edible fungi and their potential bioactivities: A review. Trends Food Sci. Technol. 2020, 105, 468–482. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, F.; Xu, Z.; Ding, Z. Bioactive Mushroom Polysaccharides: A Review on Monosaccharide Composition, Biosynthesis and Regulation. Molecules 2017, 22, 955. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Duan, Y.; Yang, W.; Zhang, H.; Li, C.; Zhang, J. Structural elucidation and immunostimulatory activity of polysaccharide isolated by subcritical water extraction from Cordyceps militaris. Carbohydr. Polym. 2017, 157, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-T.; Huang, K.-S.; Shaw, J.-F.; Chen, J.-R.; Kuo, W.-S.; Shen, G.; Grumezescu, A.M.; Holban, A.M.; Wang, Y.-T.; Wang, J.-S.; et al. Trends in the Immunomodulatory Effects of Cordyceps militaris: Total Extracts, Polysaccharides and Cordycepin. Front. Pharmacol. 2020, 11, 1824. [Google Scholar] [CrossRef]
- Joshi, M.; Sagar, A.; Kanwar, S.S.; Singh, S. Anticancer, antibacterial and antioxidant activities of Cordyceps militaris. Indian J. Exp. Biol. 2019, 57, 15–20. [Google Scholar]
- Zhu, S.-j.; Pan, J.; Zhao, B.; Liang, J.; Ze-yu, W.; Yang, J.-j. Comparisons on enhancing the immunity of fresh and dry Cordyceps militaris in vivo and in vitro. J. Ethnopharmacol. 2013, 149, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Ng, T.B.; Wang, H.; Sze, S.C.W.; Zhang, K.Y.; Li, Q.; Lu, X. Cordymin, an antifungal peptide from the medicinal fungus Cordyceps militaris. Phytomedicine 2011, 18, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Park, B.T.; Na, K.H.; Jung, E.C.; Park, J.W.; Kim, H.H. Antifungal and anticancer activities of a protein from the mushroom Cordyceps militaris. Korean J. Physiol. Pharmacol. 2009, 13, 49–54. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Yin, Y.; Yu, G.; Jin, Y.; Ye, X.; Shrestha, A.; Liu, W.; Yu, W.; Sun, H. A novel protein with anti-metastasis activity on 4T1 carcinoma from medicinal fungus Cordyceps militaris. Int. J. Biol. Macromol. 2015, 80, 385–391. [Google Scholar] [CrossRef]
- Gordon, S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003, 3, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Italiani, P.; Boraschi, D. From monocytes to M1/M2 macrophages: Phenotypical vs. functional differentiation. Front. Immunol. 2014, 5, 514. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Liang, H.; Zen, K. Molecular mechanisms that influence the macrophage M1–M2 polarization balance. Front. Immunol. 2014, 5, 614. [Google Scholar] [CrossRef] [Green Version]
- Mills, C. M1 and M2 macrophages: Oracles of health and disease. Crit. Rev. Immunol. 2012, 32, 463–488. [Google Scholar] [CrossRef] [Green Version]
- Jayasingam, S.D.; Citartan, M.; Thang, T.H.; Zin, A.A.M.; Ang, K.C.; Ch’ng, E.S. Evaluating the polarization of tumor-associated macrophages into M1 and M2 phenotypes in human cancer tissue: Technicalities and challenges in routine clinical practice. Front. Oncol. 2020, 9, 1512. [Google Scholar] [CrossRef] [Green Version]
- Bronte, V.; Murray, P.J. Understanding Local Macrophage Phenotypes In Disease: Modulating macrophage function to treat cancer. Nat. Med. 2015, 21, 117–119. [Google Scholar] [CrossRef]
- DeNardo, D.G.; Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 2019, 19, 369–382. [Google Scholar] [CrossRef]
- Cui, J.D. Biotechnological production and applications of Cordyceps militaris, a valued traditional Chinese medicine. Crit. Rev. Biotechnol. 2015, 35, 475–484. [Google Scholar] [CrossRef]
- Jung, E.C.; Kim, K.D.; Bae, C.H.; Kim, J.C.; Kim, D.K.; Kim, H.H. A mushroom lectin from ascomycete Cordyceps militaris. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2007, 1770, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Wang, H.; Ng, T.B. A haemagglutinin from the medicinal fungus Cordyceps militaris. Biosci. Rep. 2009, 29, 321–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, G.-m.; Pan, G.-F.; Guo, J.-Y. Anti-inflammatory and antinociceptive effects of cordymin, a peptide purified from the medicinal mushroom Cordyceps sinensis. Nat. Prod. Res. 2012, 26, 2358–2362. [Google Scholar] [CrossRef]
- Schmidt, F. Recombinant expression systems in the pharmaceutical industry. Appl. Microbiol. Biotechnol. 2004, 65, 363–372. [Google Scholar] [CrossRef]
- Sørensen, H.P.; Mortensen, K.K. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J. Biotechnol. 2005, 115, 113–128. [Google Scholar] [CrossRef] [PubMed]
- You, R.-I.; Wu, W.-S.; Cheng, C.-C.; Wu, J.-R.; Pan, S.-M.; Chen, C.-W.; Hu, C.-T. Involvement of N-glycan in multiple receptor tyrosine kinases targeted by Ling-Zhi-8 for suppressing HCC413 tumor progression. Cancers 2019, 11, 9. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.Y.; Hsu, H.Y.; Sun, W.H.; Wu, T.H.; Tsao, S.M. Induction of Cbl-dependent epidermal growth factor receptor degradation in Ling Zhi-8 suppressed lung cancer. Int. J. Cancer 2017, 140, 2596–2607. [Google Scholar] [CrossRef] [Green Version]
- Ejike, U.C.; Chan, C.J.; Okechukwu, P.N.; Lim, R.L.H. New advances and potentials of fungal immunomodulatory proteins for therapeutic purposes. Crit. Rev. Biotechnol. 2020, 40, 1172–1190. [Google Scholar] [CrossRef]
- Chen, J.N.; Ma, C.Y.; Tsai, P.F.; Wang, Y.T.; Wu, J.S. In vitro antitumor and immunomodulatory effects of the protein PCP-3A from mushroom Pleurotus citrinopileatus. J. Agric. Food Chem. 2010, 58, 12117–12122. [Google Scholar] [CrossRef]
- Sun, L.X.; Lin, Z.B.; Lu, J.; Li, W.D.; Niu, Y.D.; Sun, Y.; Hu, C.Y.; Zhang, G.Q.; Duan, X.S. The improvement of M1 polarization in macrophages by glycopeptide derived from Ganoderma lucidum. Immunol. Res. 2017, 65, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Nonnenmacher, Y.; Hiller, K. Biochemistry of proinflammatory macrophage activation. Cell. Mol. Life Sci. 2018, 75, 2093–2109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ditamo, Y.; Rupil, L.L.; Sendra, V.G.; Nores, G.A.; Roth, G.A.; Irazoqui, F.J. In vivo immunomodulatory effect of the lectin from edible mushroom Agaricus bisporus. Food Funct. 2016, 7, 262–269. [Google Scholar] [CrossRef]
- Meng, Y.; Qu, Y.; Wu, W.; Chen, L.; Sun, L.; Tai, G.; Zhou, Y.; Cheng, H. Galactan isolated from Cantharellus cibarius modulates antitumor immune response by converting tumor-associated macrophages toward M1-like phenotype. Carbohydr. Polym. 2019, 226, 115295. [Google Scholar] [CrossRef] [PubMed]
- van Horssen, R.; Ten Hagen, T.L.; Eggermont, A.M. TNF-α in cancer treatment: Molecular insights, antitumor effects, and clinical utility. Oncologist 2006, 11, 397–408. [Google Scholar] [CrossRef]
- Matsukawa, A.; Hogaboam, C.M.; Lukacs, N.W.; Lincoln, P.M.; Evanoff, H.L.; Kunkel, S.L. Pivotal role of the CC chemokine, macrophage-derived chemokine, in the innate immune response. J. Immunol. 2000, 164, 5362–5368. [Google Scholar] [CrossRef]
- Fauskanger, M.; Haabeth, O.A.W.; Skjeldal, F.M.; Bogen, B.; Tveita, A.A. Tumor Killing by CD4(+) T Cells Is Mediated via Induction of Inducible Nitric Oxide Synthase-Dependent Macrophage Cytotoxicity. Front. Immunol. 2018, 9, 1684. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Q.; Jia, B.; Liu, X.; Fang, J.; Zhao, L.; Xu, L.; Fang, M.; Gong, Z.; Sun, H. Molecular Cloning, Expression and Macrophage Activation of an Immunoregulatory Protein from Cordyceps militaris. Molecules 2021, 26, 7107. https://doi.org/10.3390/molecules26237107
Yang Q, Jia B, Liu X, Fang J, Zhao L, Xu L, Fang M, Gong Z, Sun H. Molecular Cloning, Expression and Macrophage Activation of an Immunoregulatory Protein from Cordyceps militaris. Molecules. 2021; 26(23):7107. https://doi.org/10.3390/molecules26237107
Chicago/Turabian StyleYang, Qing, Binmei Jia, Xiaomei Liu, Jialing Fang, Luyang Zhao, Lin Xu, Min Fang, Zhiyong Gong, and Hui Sun. 2021. "Molecular Cloning, Expression and Macrophage Activation of an Immunoregulatory Protein from Cordyceps militaris" Molecules 26, no. 23: 7107. https://doi.org/10.3390/molecules26237107






