A New Approach for Determination of the Botanical Origin of Monofloral Bee Honey, Combining Mineral Content, Physicochemical Parameters, and Self-Organizing Maps
Abstract
1. Introduction
2. Results
2.1. Samples
2.2. Basic Statistics
2.3. Set of Descriptors
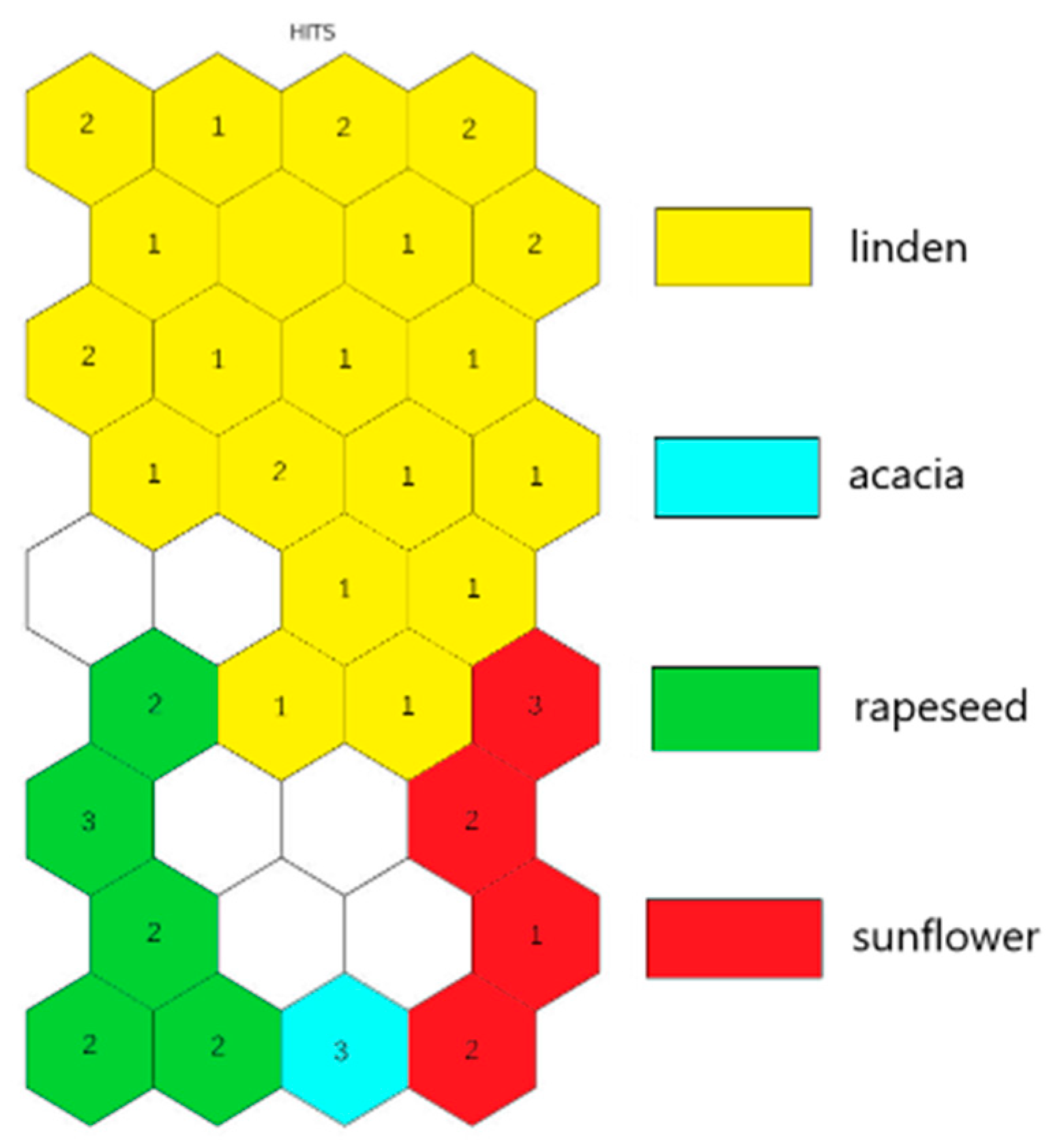
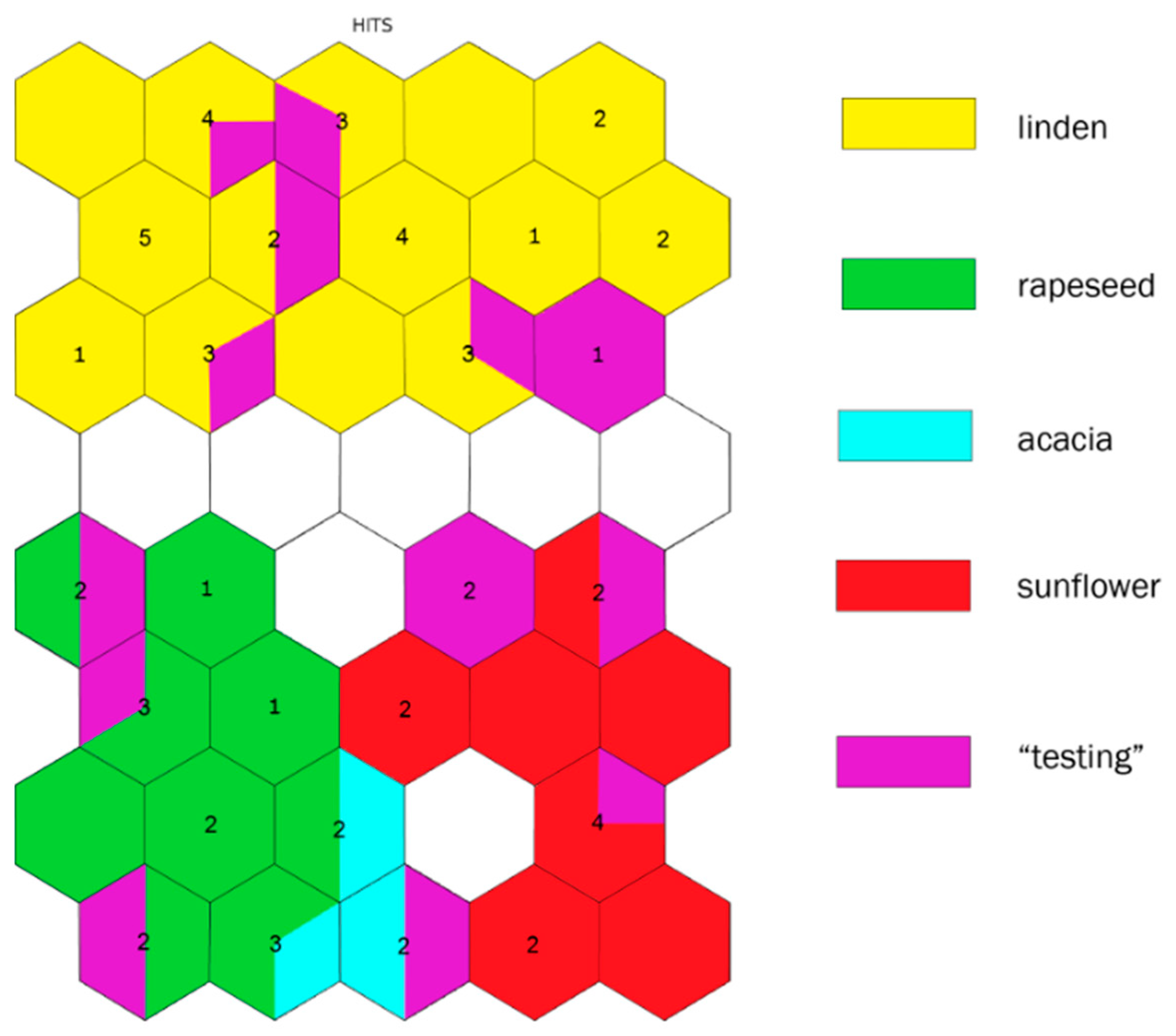
2.4. Botanical Origin Classification
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Melissopalynological Analysis
4.3. Physicochemical Analysis
4.4. Analysis of Chemical Elements
4.4.1. Sample Preparation Procedure
4.4.2. Apparatus
4.5. Chemometrics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bogdanov, S.; Haldimann, M.; Luginbühl, W.; Gallmann, P. Minerals in honey: Environmental, geographical and botanical aspects. J. Apic. Res. 2007, 46, 269–275. [Google Scholar] [CrossRef]
- Saitta, M.; Di Bella, G.; Fede, M.R.; Lo Turco, V.; Potortì, A.G.; Rando, R.; Russo, M.T.; Dugo, G. Gas chromatography-tandem mass spectrometry multi-residual analysis of contaminants in Italian honey samples. Food Addit. Contam. A 2017, 34, 800–808. [Google Scholar]
- Ramón-Sierra, J.M.; Ruiz-Ruiz, J.C.; de la Luz Ortiz-Vázquez, E. Electrophoresis characterisation of protein as a method to establish the entomological origin of stingless bee honeys. Food Chem. 2015, 183, 43–48. [Google Scholar] [CrossRef]
- Oddo, L.P.; Piro, R.R. Extra issue of European unifloral honeys. Apidologie 2004, 35, S38–S81. [Google Scholar]
- Fodor, P.; Molnar, E. Honey as an environmental indicator: Effect of sample preparation on trace element determination by ICP-AES. Mikrochim. Acta 1993, 112, 113–118. [Google Scholar] [CrossRef]
- Anklam, E. A review of the analytical methods to determine the geographical and botanical origin of honey. Food Chem. 1998, 63, 549–562. [Google Scholar] [CrossRef]
- Council Directive 2001/110/EC of the 20 December 2001 relating to honey. Off. J. Eur. Commun. 2002, 10, 47–52. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32001L0110&from=EN (accessed on 26 November 2021).
- Louveaux, J.; Maurizio, A.; Vorwohl, G. Methods of melissopalynology. Bee World 1978, 59, 139–157. [Google Scholar] [CrossRef]
- Bulgarian State Standard—BDS 2673, Bee Honey, 1989. Available online: https://bds-bg.org/en/project/show/bds:proj:22569 (accessed on 26 November 2021).
- Zhang, X.; Zhang, S.; Qing, X.; Lu, Z. A new strategy for rapid classification of honeys by simple cluster analysis method based on combination of various physicochemical parameters. Chem. J. Chin. Univ. 2019, 35, 390–394. [Google Scholar] [CrossRef]
- Dżugan, M.; Tomczyk, M.; Sowa, P.; Grabek-Lejko, D. Antioxidant activity as biomarker of honey variety. Molecules 2018, 23, 2069. [Google Scholar] [CrossRef]
- Roshan, A.-R.A.; Gad, H.A.; El-Ahmady, S.H.; Abou-Shoer, M.I.; Khanbash, M.S.; Al-Azizi, M.M. Characterization and discrimination of the floral origin of sidr honey by physicochemical data combined with multivariate analysis. Food Anal. Method 2017, 10, 137–146. [Google Scholar] [CrossRef]
- Adgaba, N.; Al-Ghamdi, A.A.; Getachew, A.; Tadesse, Y.; Belay, A.; Ansari, M.J.; Radloff, S.E.; Sharm, D. Characterization of honeys by their botanical and geographical origins based on physico-chemical properties and chemo-metrics analysis. J. Food Meas. Charact. 2017, 11, 1106–1117. [Google Scholar] [CrossRef]
- Sogut, E.; Seydim, A.C. Classification of honeys collected from different regions of Anatolia by chemometric methods. J. Food Process. Preserv. 2020, 44, e14960. [Google Scholar] [CrossRef]
- Rosiak, E.; Madras-Majewska, B.; Teper, D.; Łepecka, A.; Zielińska, D. Cluster analysis classification of honey from two different climatic zones based on selected physicochemical and of microbiological parameters. Molecules 2021, 26, 2361. [Google Scholar] [CrossRef] [PubMed]
- Majewska, E.; Druzyńska, B.; Wołosiak, R. Determination of the botanical origin of honeybee honeys based on the analysis of their selected physicochemical parameters coupled with chemometric assays. Food Sci. Biotechnol. 2019, 28, 1307–1314. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Jin, L.; Chang, Q.; Peng, T.; Hu, X.; Fan, C.; Pang, G.; Lu, M.; Wang, W. Discrimination of botanical origins for Chinese honey according to free amino acids content by HPLC-FLD with chemometric approaches. J. Sci. Food Agric. 2016, 97, 2042–2049. [Google Scholar] [CrossRef]
- Silva, B.; Gonzaga, L.V.; Maltez, H.F.; Samochvalov, K.B.; Fett, R.; Costa, A.C.O. Elemental profiling by ICP-MS as a tool for geographical discrimination: The case of bracatinga honeydew honey. J. Food Compos. Anal. 2021, 96, 103727–103735. [Google Scholar] [CrossRef]
- Castiglioni, S.; Stefano, M.; Pisani, M.; Carloni, P. Geographical characterisation of multifloral honeys from the Marche region (Italy) according to their antioxidant activity and colour using a chemometric approach. Int. J. Food Sci. Technol. 2018, 53, 571–581. [Google Scholar] [CrossRef]
- Drivelos, S.A.; Danezis, G.P.; Halagarda, M.; Popek, S.; Georgiou, C.A. Geographical origin and botanical type honey authentication through elemental metabolomics via chemometrics. Food Chem. 2021, 338, 127936–127943. [Google Scholar] [CrossRef]
- Brugnerotto, P.; Silva, B.; Seraglio, S.K.T.; Schulz, M.; Blainski, E.; Dortzbach, D.; Gonzaga, L.V.; Fett, R.; Costa, A.C.O. Physicochemical characterization of honeys from Brazilian monitored beehives. Eur. Food Res. Technol. 2021, 247, 2709–2719. [Google Scholar] [CrossRef]
- Kaczmarek, A.M.; Muzolf-Panek, M.; Tomaszewska-Gras, J.; Konieczny, P. Predicting the botanical origin of honeys with chemometric analysis according to their antioxidant and physicochemical properties. Pol. J. Food Nutr. Sci. 2019, 69, 191–201. [Google Scholar] [CrossRef]
- Maione, C.; Barbosa Jr., F.; Barbosa, R.M. Predicting the botanical and geographical origin of honey with multivariate data analysis and machine learning techniques: A review. Comput. Electron. Agric. 2019, 157, 436–446. [Google Scholar] [CrossRef]
- Machado, A.M.; Antunes, M.; Miguel, M.G.; Vilas-Boas, M.; Figueiredo, A.C. Volatile profile of Portuguese monofloral honeys: Significance in botanical origin determination. Molecules 2021, 26, 4970. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Chen, L.; Liu, F.; Zhou, F.; Peng, J.; Sun, M. Fast classification of geographical origins of honey based on laser-induced breakdown spectroscopy and multivariate analysis. Sensors 2020, 20, 1878. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, M.S.; Seraglio, S.K.T.; Rocha, G.; Balderas, C.B.; Piovezan, M.; Gonzag, L.V.; de Barcellos Falkenberg, D.; Fett, R.; de Oliveira, M.A.L.; Costa, A.C.O. Free amino acid determination by GC-MS combined with a chemometric approach for geographical classification of bracatinga honeydew honey (Mimosa scabrella Bentham). Food Control 2017, 78, 383–392. [Google Scholar] [CrossRef]
- Ghidotti, M.; Fiamegos, Y.; Dumitrascu, C.; de la Calle, M.B. Use of elemental profiles to verify geographical origin and botanical variety of Spanish honeys with a protected denomination of origin. Food Chem. 2021, 342, 128350–128361. [Google Scholar] [CrossRef]
- Tananaki, C.; Thrasyvoulou, A.; Giraudel, J.L.; Montury, M. Determination of volatile characteristics of Greek and Turkish pine honey samples and their classification by using Kohonen self organising maps. Food Chem. 2007, 101, 1687–1693. [Google Scholar] [CrossRef]
- De Boishebert, V.; Giraudel, J.-L.; Montury, M. Characterization of strawberry varieties by SPME–GC–MS and Kohonen self-organizing map. Chemom. Intel. Lab. 2006, 80, 13–23. [Google Scholar] [CrossRef]
- Dinkov, D. Quality parameters of Bulgarian kinds of bee honey. Maced. Vet. Rev. 2014, 37, 35–41. [Google Scholar] [CrossRef][Green Version]
- Bulgarian State Standard—BDS 3050, Bee Honey. Rules for Sampling and Testing Methods, 1980. Available online: https://bds-bg.org/en/project/show/bds:proj:22905 (accessed on 26 November 2021).
- Bogdanov, S.; Martin, P.; Lullmann, C.; Borneck, R.; Flamini, C.; Morlot, M.; Lheritier, J.; Vorwohl, G.; Russmann, H.; Persano, L.; et al. Harmonized methods of the European Honey commission. Apidologie 1997, 28, 1–59. [Google Scholar]
- Chudzinska, M.; Baralkiewicz, D. Application of ICP-MS method of determination of 15 elements in honey with chemometric approach for the verification of their authenticity. Food Chem. Toxicol. 2011, 49, 2741–2749. [Google Scholar] [CrossRef]
- Kohonen, T. Self-Organization and Associative Memory; Springer: Berlin/Heidelberg, Germany, 1984; pp. 1464–1480. [Google Scholar]
- Voyslavov, T.; Tsakovski, S.; Simeonov, V. Surface water quality assessment using self-organizing maps and Hasse diagram technique. Chemom. Intel. Lab. 2012, 118, 280–286. [Google Scholar] [CrossRef]
- Vesanto, J. SOM-based data visualization methods. Intel. Data Anal. 1999, 3, 111–126. [Google Scholar] [CrossRef]


| Generator power | 1.00 kW |
| Type of nebulizer | V-groove |
| Plasma gas (argon) | 15 L/min |
| Cooling gas | 1.50 L/min |
| Gas of nebulizer | 1.40 kPa |
| Observation time | 5 s |
| Burner height | 5 mm |
| Pump speed | 30 rpm |
| Sample input time | 12 s |
| Time for stabilization | 15 s |
| Number of cues | 5 |
| Element | Spectral Line | Element | Spectral Line |
|---|---|---|---|
| Al | 396.152 nm | Mn | 257.610 nm |
| Ba | 455.403 nm | Na | 588.995 nm |
| Ca | 370.602 nm | P | 213.618 nm |
| Cu | 324.754 nm | S | 181.972 nm |
| Fe | 238.204 nm | Sr | 407.771 nm |
| K | 766.491 nm | Zn | 213.857 nm |
| Mg | 279.553 nm |
| Generator power | 1.20 kW |
| Type of nebulizer | Meinhardt (concentric) |
| Plasma gas (argon) | 9 L/min |
| Additional gas | 1.35 L/min |
| Nebulizer flow | 1.1 L/min |
| Pump speed | 20 rpm |
| Time for stabilization | 5 s |
| Number of cues | 5 |
| Number of scans for cues | 10 |
| Element | m/z | Collision Cell Included | Isobaric Interference | Parallel Analysis |
|---|---|---|---|---|
| Li | 7 | The concentration is confirmed by FAAS | ||
| Al | 27 | Yes | 12C15N+, 13C14N+, 1H12C14N+ | The concentration is confirmed by ICP-OES |
| V | 51 | Yes | 34S16O1H+, 35C16O+, 38Ar13C+, 36Ar15N+, 36Ar14N1H+, 37Cl14N+, 36S15N+, 33S18O+, 34S17O+ | |
| Fe | 57 | Yes | 40Ar16O1H+, 40Ca16O1H+, 40Ar17O+, 38Ar18O1H+, 38Ar19F+ | The concentration is confirmed by ICP-OES |
| Co | 59 | Yes | 43Ca16O+, 42Ca16O1H+, 24Mg35Cl+, 36Ar23Na+, 40Ar18O1H+ | |
| Cr | 52 | Yes | 35Cl16O1H+, 40Ar12C+, 36Ar16O+, 37Cl15N+ 34S18O+, 36S16O+, 38Ar14N+, 36Ar15N1H+, 35Cl17O+ | The concentration is confirmed by ICP-OES |
| Ni | 60 | Yes | 44Ca16O+, 23Na37Cl+, 43Ca16O1H+ | |
| Cu | 63 | Yes | 31P16O2+, 40Ar23Na+, 47Ti16O+, 23Na40Ca+, 46Ca16O1H+, 36Ar12C14N1H+, 14N12C37Cl+, 16O12C35Cl+ | The concentration is confirmed by ICP-OES |
| Ga | 71 | Yes | 35Cl18O2+, 37Cl16O18O+, 37Cl17O2+, 36Ar35Cl+, 38Ar33S+ | |
| As | 75 | Yes | 40Ar35Cl+, 59Co16O+, 36Ar38Ar1H+, 38Ar37Cl+, 36Ar39K, | |
| Se | 78 | Yes | 40Ar38Ar+, 38Ar40Ca+ | |
| Rb | 85 | The concentration is confirmed by FAAS | ||
| Cd | 111 | 95Mo16O+, 94Zr16O1H+, 39K216O21H+ | ||
| In | 115 | |||
| Cs | 133 | |||
| Ba | 137 | The concentration is confirmed by ICP-OES | ||
| Pb | 208 | |||
| Bi | 209 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Voyslavov, T.; Mladenova, E.; Balkanska, R. A New Approach for Determination of the Botanical Origin of Monofloral Bee Honey, Combining Mineral Content, Physicochemical Parameters, and Self-Organizing Maps. Molecules 2021, 26, 7219. https://doi.org/10.3390/molecules26237219
Voyslavov T, Mladenova E, Balkanska R. A New Approach for Determination of the Botanical Origin of Monofloral Bee Honey, Combining Mineral Content, Physicochemical Parameters, and Self-Organizing Maps. Molecules. 2021; 26(23):7219. https://doi.org/10.3390/molecules26237219
Chicago/Turabian StyleVoyslavov, Tsvetomil, Elisaveta Mladenova, and Ralitsa Balkanska. 2021. "A New Approach for Determination of the Botanical Origin of Monofloral Bee Honey, Combining Mineral Content, Physicochemical Parameters, and Self-Organizing Maps" Molecules 26, no. 23: 7219. https://doi.org/10.3390/molecules26237219
APA StyleVoyslavov, T., Mladenova, E., & Balkanska, R. (2021). A New Approach for Determination of the Botanical Origin of Monofloral Bee Honey, Combining Mineral Content, Physicochemical Parameters, and Self-Organizing Maps. Molecules, 26(23), 7219. https://doi.org/10.3390/molecules26237219







