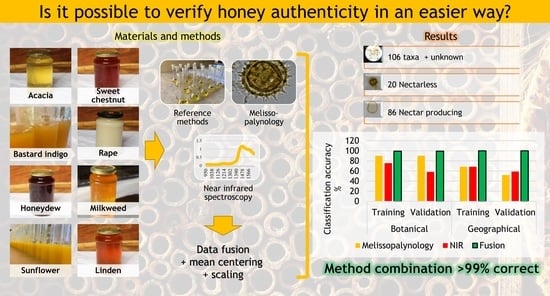
Origin Identification of Hungarian Honey Using Melissopalynology, Physicochemical Analysis, and Near Infrared Spectroscopy
Abstract
:1. Introduction
2. Results and Discussion
2.1. Results of the Physicochemical Analysis
2.2. Results of Melissopalinology
2.2.1. Results of the Cluster Analysis of the Melissopalynological Data
2.2.2. Results of the Botanical Origin Identification Models Using the Pollen Data
2.3. Results of the near Infrared Spectroscopy
2.4. Results of the Combined Data
3. Materials and Methods
3.1. Honey Samples
3.2. Determination of Physicochemical Parameters
3.3. Melissopalynology
3.4. Near Infrared Spectroscopy
3.5. Statistical Analysis
3.5.1. Physicochemical Data
3.5.2. Pollen Data
3.5.3. NIRS Data
3.5.4. Fusion of the Data
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- The European Council. Council Directive 2001/110/EC of 20 December 2001 relating to honey. Off. J. Eur. Communities 2001, 10, 47–52. [Google Scholar]
- Codex Alimentarius Commission. Codex standard for honey. CODEX STAN 12-1981 2001, 11, 7. [Google Scholar]
- Da Silva, P.M.; Gauche, C.; Gonzaga, L.V.; Costa, A.C.O.; Fett, R. Honey: Chemical composition, stability and authenticity. Food Chem. 2016, 196, 309–323. [Google Scholar] [CrossRef] [PubMed]
- European Council. European Parliament Directive 2014/63/EU of The European Parliament and of the Council of 15 May 2014 amending Council Directive 2001/110/EC relating to honey. Off. J. Eur. Communities 2014, L164, 1–5. [Google Scholar]
- Persano Oddo, L.; Piana, L.; Bogdanov, S.; Bentabol, A.; Gotsiou, P.; Kerkvliet, J.; Martin, P.; Morlot, M.; Ortiz Valbuena, A.; Von der Ohe, K. Botanical species giving unifloral honey in Europe. Apidologie 2006, 37, 452–461. [Google Scholar] [CrossRef]
- Ruoff, K.; Bogdanov, S. Authenticity of Honey and Other Bee Products. Apiacta 2004, 38, 317–327. [Google Scholar]
- Bogdanov, S. Honey Composition. In Book of Honey; Bee Hexagon Knowledge Network: Burnaby, BC, Canada, 2014; pp. 27–36. [Google Scholar]
- Persano Oddo, L.; Bogdanov, S. Determination of honey botanical origin: Problems and issues. Apidologie 2004, 35, S2–S3. [Google Scholar] [CrossRef] [Green Version]
- Anklam, E. A review of the analytical methods to determine the geographical and botanical origin of honey. Food Chem. 1998, 63, 549–562. [Google Scholar] [CrossRef]
- Hungarian Standards Institution. MSZ 6950-3:2017—Test of Honey. Part 3: Microscopical Examination; Hungarian Standards Institution: Budapest, Hungary, 2017. [Google Scholar]
- Persano Oddo, L.; Piro, R. Main European unifloral honeys: Descriptive sheets. Apidologie 2004, 35, S38–S81. [Google Scholar] [CrossRef]
- Thrasyvoulou, A.; Tananaki, C.; Goras, G.; Karazafiris, E.; Dimou, M.; Liolios, V.; Kanelis, D.; Gounari, S. Legislation of honey criteria and standards. J. Apic. Res. 2018, 57, 88–96. [Google Scholar] [CrossRef]
- Codex Alimentarius Hungaricus. 2–100 számú irányelv Megkülönböztető jelöléssel ellátott mézfélék (2–100 Honey bearing with a distinguishing quality mark). In Codex Alimentarius Hungaricus; Codex Alimentarius Hungaricus; Magyar Élelmiszerkönyv Bizottság: Budapest, Hungary, 2009; pp. 3–6. [Google Scholar]
- Yao, L.; Jiang, Y.; Singanusong, R.; D’Arcy, B.; Datta, N.; Caffin, N.; Raymont, K. Flavonoids in Australian Melaleuca, Guioa, Lophostemon, Banksia and Helianthus honeys and their potential for floral authentication. Food Res. Int. 2004, 37, 166–174. [Google Scholar] [CrossRef]
- Trifković, J.; Andrić, F.; Ristivojević, P.; Guzelmeric, E.; Yesilada, E. Analytical methods in tracing honey authenticity. J. AOAC Int. 2017, 100, 827–839. [Google Scholar] [CrossRef]
- Bogdanov, S.; Ruoff, K.; Oddo, L.P.; Persano Oddo, L. Physico-chemical methods for the characterisation of unifloral honeys: A review. Apidologie 2004, 35, S4–S17. [Google Scholar] [CrossRef] [Green Version]
- Pascual-Maté, A.; Osés, S.M.; Fernández-Muiño, M.A.; Sancho, M.T. Methods of analysis of honey. J. Apic. Res. 2018, 57, 38–74. [Google Scholar] [CrossRef]
- Aouadi, B.; Zaukuu, J.L.Z.; Vitális, F.; Bodor, Z.; Fehér, O.; Gillay, Z.; Bazar, G.; Kovacs, Z. Historical evolution and food control achievements of near infrared spectroscopy, electronic nose, and electronic tongue—critical overview. Sensors 2020, 20, 5479. [Google Scholar] [CrossRef]
- Chen, L.; Wang, J.; Ye, Z.; Zhao, J.; Xue, X.; Vander Heyden, Y.; Sun, Q. Classification of Chinese honeys according to their floral origin by near infrared spectroscopy. Food Chem. 2012, 135, 338–342. [Google Scholar] [CrossRef]
- Bodor, Z.; Koncz, F.A.; Rashed, M.S.; Kaszab, T.; Gillay, Z.; Benedek, C.; Kovacs, Z. Application of near infrared spectroscopy and classical analytical methods for the evaluation of Hungarian honey. Prog. Agric. Eng. Sci. 2018, 14, 11–23. [Google Scholar] [CrossRef]
- Di Rosa, A.R.; Leone, F.; Cheli, F.; Chiofalo, V. Fusion of electronic nose, electronic tongue and computer vision for animal source food authentication and quality assessment—A review. J. Food Eng. 2017, 210, 62–75. [Google Scholar] [CrossRef]
- Beć, K.B.; Grabska, J.; Plewka, N.; Huck, C.W. Insect Protein Content Analysis in Handcrafted Fitness Bars by NIR Spectroscopy. Gaussian Process Regression and Data Fusion for Performance Enhancement of Miniaturized Cost-Effective Consumer-Grade Sensors. Molecules 2021, 26, 6390. [Google Scholar] [CrossRef]
- Forshed, J.; Idborg, H.; Jacobsson, S.P. Evaluation of different techniques for data fusion of LC/MS and 1H-NMR. Chemom. Intell. Lab. Syst. 2007, 85, 102–109. [Google Scholar] [CrossRef]
- Sipos, L.; Végh, R.; Bodor, Z.; Zaukuu, J.L.Z.; Hitka, G.; Bázár, G.; Kovacs, Z. Classification of bee pollen and prediction of sensory and colorimetric attributes—a sensometric fusion approach by e-nose, e-tongue and NIR. Sensors 2020, 20, 6768. [Google Scholar] [CrossRef]
- Borràs, E.; Ferré, J.; Boqué, R.; Mestres, M.; Aceña, L.; Busto, O. Data fusion methodologies for food and beverage authentication and quality assessment—A review. Anal. Chim. Acta 2015, 891, 1–14. [Google Scholar] [CrossRef]
- Ríos-Reina, R.; Callejón, R.M.; Savorani, F.; Amigo, J.M.; Cocchi, M. Data fusion approaches in spectroscopic characterization and classification of PDO wine vinegars. Talanta 2019, 198, 560–572. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.; Zhao, H.; Wang, Q.; Wu, F.; Cao, W. A Novel Chinese Honey from Amorpha fruticosa L.: Nutritional Composition and Antioxidant Capacity In Vitro. Molecules 2020, 25, 5211. [Google Scholar] [CrossRef]
- Kasper-Szél, Z.; Amtmann, M.; Takáts, A.; Kardos-Neumann, Á. Comparative Analysis of Hungarian Robinia and Milkweed Honeys Based on Their Chemical and Physical Characteristics. Acta Aliment. 2003, 32, 395–403. [Google Scholar] [CrossRef]
- Institute of Apiculture and Bee Biology. Jellemző mézprofilok meghatározása (Determination of Characteristic honey profiles). Institute of Apiculture and Bee Biology: Gödöllő, Hungary, 2016. [Google Scholar]
- Farkas, Á.; Zajácz, E. Nectar production for the Hungarian honey industry. Eur. J. Plant Sci. Biotechnol. 2007, 1, 125–151. [Google Scholar]
- Bartha, D.; Bán, M.; Schmidt, D.; Tiborcz, V. OBMVascular plants of Hungary online database. Available online: http://floraatlasz.uni-sopron.hu (accessed on 28 June 2021).
- Qiu, P.Y.; Ding, H.B.; Tang, Y.K.; Xu, R.J. Determination of chemical composition of commercial honey by near-infrared spectroscopy. J. Agric. Food Chem. 1999, 47, 2760–2765. [Google Scholar] [CrossRef]
- Erdtman, G. The Acetolysis Method—A Revised Description. Svensk botanisk tidskrift 1960, 54, 561–564. [Google Scholar]
- Khansaritoreh, E.; Salmaki, Y.; Akbari Azirani, T.; Henareh, F.; Alizadeh, K.; Ramezani, E.; Zarre, S.; Beckh, G.; Behling, H. The sources and quality of Iranian honey. Heliyon 2021, 7, e06651. [Google Scholar] [CrossRef]
- Beug, H.-J. Leitfaden der Pollenbestimmung für Mitteleuropa und Angrenzende Gebiete; Friedrich Pfeil: München, Germany, 2004; ISBN 3-89937-043-0. [Google Scholar]
- Tabachnick, B.G.; Fidell, L.S. Using Multivariate Statistics, 6th ed.; Pearson Education: Harlow, UK, 2013. [Google Scholar]
- Grimm, E.C. Tilia and Tiliagraph; Illinois State Museum: Springfield, IL, USA, 1991; p. 101. [Google Scholar]
- Grimm, E. CONISS: A FORTRAN 77 program for stratigraphically constrained cluster analysis by the method of incremental sum of squares. Comput. Geosci. 1987, 13, 13–35. [Google Scholar] [CrossRef]
- Zhao, N.; Mio, W.; Liu, X. A hybrid PCA-LDA model for dimension reduction. In Proceedings of the 2011 International Joint Conference on Neural Networks, San Jose, CL, USA, 31 July–5 August 2011; pp. 2184–2190. [Google Scholar]
- Kovacs, Z.; Pollner, B. Aquaphotomics Course Data analysis and Software. In Proceedings of the Aquaphotomics: Understanding Water in Biology—2nd International Symposium, Kobe, Japan, 26–29 November 2016. [Google Scholar]
- Campos, M.P.; Reis, M.S. Data preprocessing for multiblock modelling—A systematization with new methods. Chemom. Intell. Lab. Syst. 2020, 199, 103959. [Google Scholar] [CrossRef]





| Botanical Origin | Moisture % | Electrical Conductivity µS/cm | pH |
|---|---|---|---|
| Acacia | 17.7 ± 1 a | 148.1 ± 20.4 a | 4.0 ± 0.2 bcd |
| Bastard indigo | 17.6 ± 1.1 a | 305.3 ± 167.7 abc | 3.9 ± 0.3 abd |
| Chestnut | 16.8 ± 1.8 a | 715.1 ± 120.6 c | 4.4 ± 0.2 e |
| Honeydew | 17.3 ± 1.2 a | 566.1 ± 205.2 bc | 4.2 ± 0.2 cde |
| Linden | 17.7 ± 1.4 a | 617.6 ± 134.1 bc | 4.3 ± 0.3 ce |
| Rape | 18 ± 1.1 a | 231.9 ± 75.2 a | 4.0 ± 0.1 bcde |
| Milkweed | 18.1 ± 1.4 a | 264.2 ± 106.3 a | 3.8 ± 0.1 a |
| Sunflower | 17.4 ± 1.1 a | 472.6 ± 96.4 b | 3.8 ± 0.4 ab |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bodor, Z.; Kovacs, Z.; Benedek, C.; Hitka, G.; Behling, H. Origin Identification of Hungarian Honey Using Melissopalynology, Physicochemical Analysis, and Near Infrared Spectroscopy. Molecules 2021, 26, 7274. https://doi.org/10.3390/molecules26237274
Bodor Z, Kovacs Z, Benedek C, Hitka G, Behling H. Origin Identification of Hungarian Honey Using Melissopalynology, Physicochemical Analysis, and Near Infrared Spectroscopy. Molecules. 2021; 26(23):7274. https://doi.org/10.3390/molecules26237274
Chicago/Turabian StyleBodor, Zsanett, Zoltan Kovacs, Csilla Benedek, Géza Hitka, and Hermann Behling. 2021. "Origin Identification of Hungarian Honey Using Melissopalynology, Physicochemical Analysis, and Near Infrared Spectroscopy" Molecules 26, no. 23: 7274. https://doi.org/10.3390/molecules26237274
APA StyleBodor, Z., Kovacs, Z., Benedek, C., Hitka, G., & Behling, H. (2021). Origin Identification of Hungarian Honey Using Melissopalynology, Physicochemical Analysis, and Near Infrared Spectroscopy. Molecules, 26(23), 7274. https://doi.org/10.3390/molecules26237274