Tris-(2-pyridyl)-pyrazolyl Borate Zinc(II) Complexes: Synthesis, DNA/Protein Binding and In Vitro Cytotoxicity Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Complex Synthesis
2.1.1. [TppyZnCl] (1)
2.1.2. [TppyZn(H2O)](BF4)] (2)
2.1.3. [TppyZnN3] (3)
2.2. CT DNA Interaction Study
2.3. Hydrodynamic Study
2.4. Interaction of the Complexes with BSA
2.5. Molecular Docking Study with DNA
2.6. In Vitro Cytotoxicity Study
3. Results and Discussion
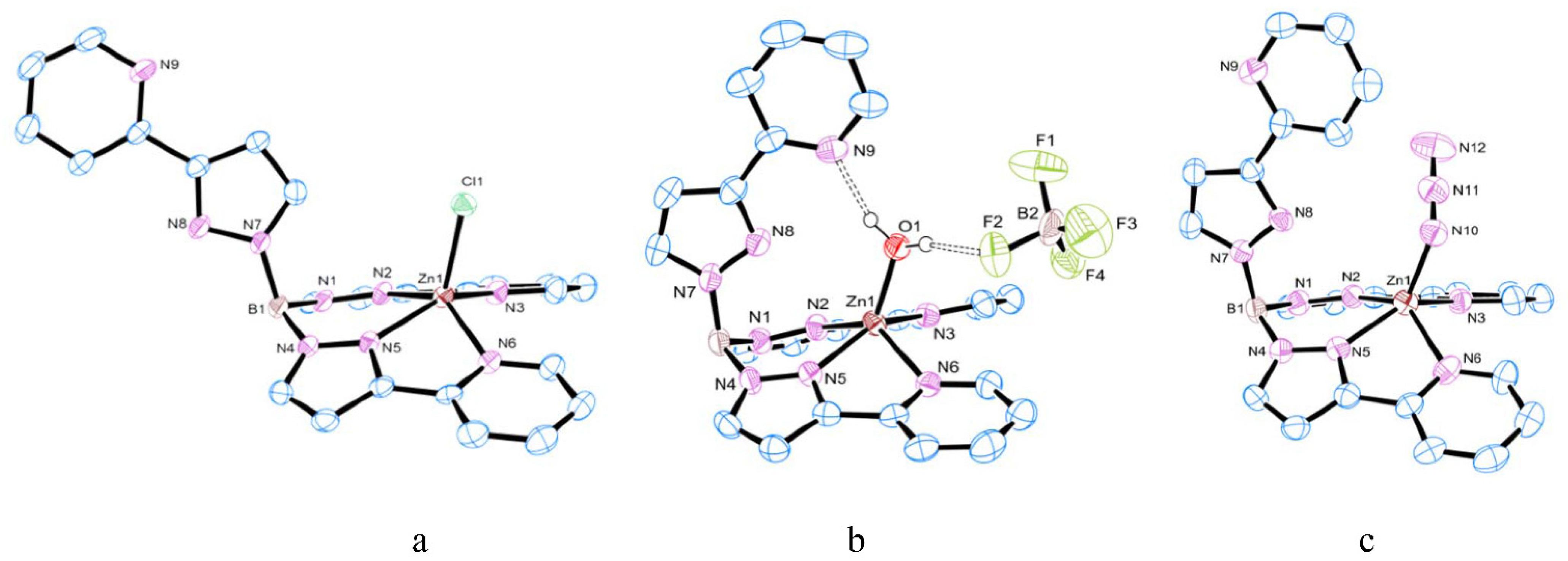
3.1. Synthesis and Structure of Zinc Complexes
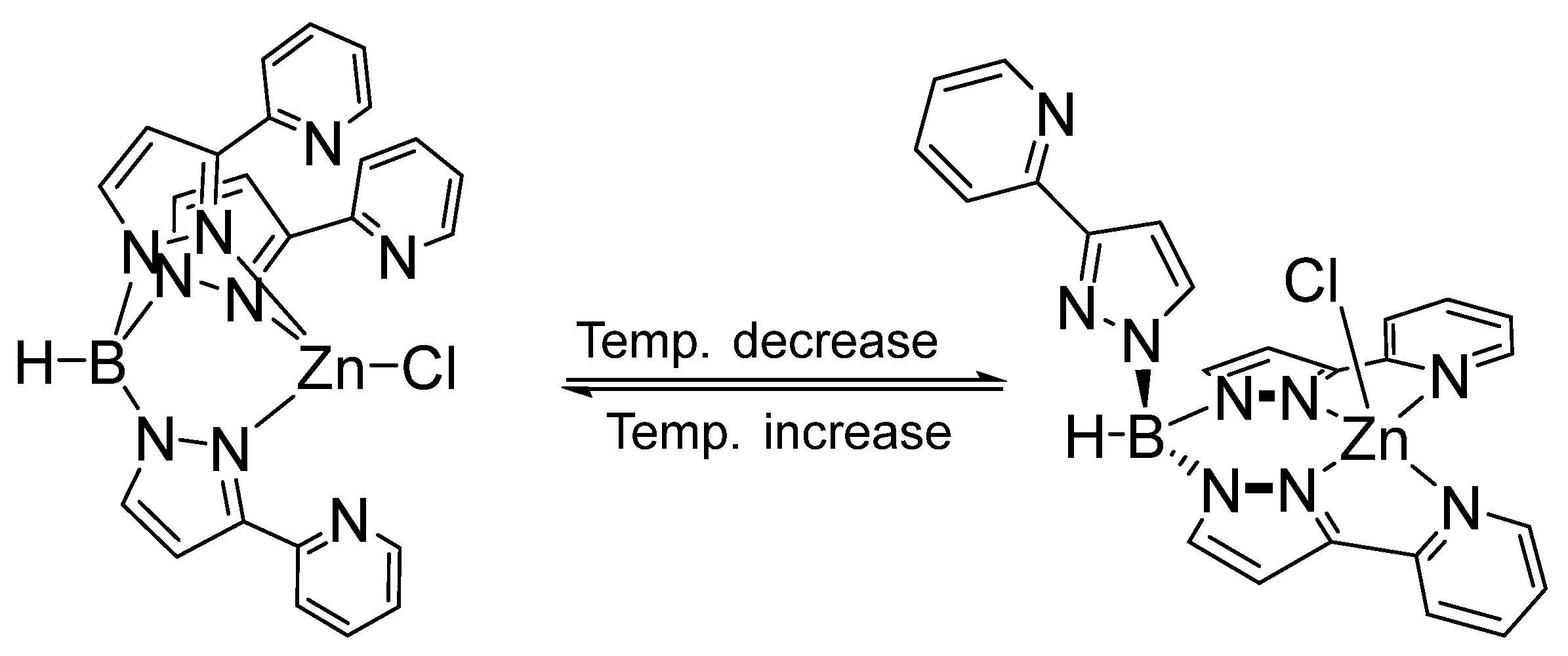
3.2. Substitution Reaction of Mononuclear Zinc Complexes
3.3. Solution Behavior of Zinc Complexes
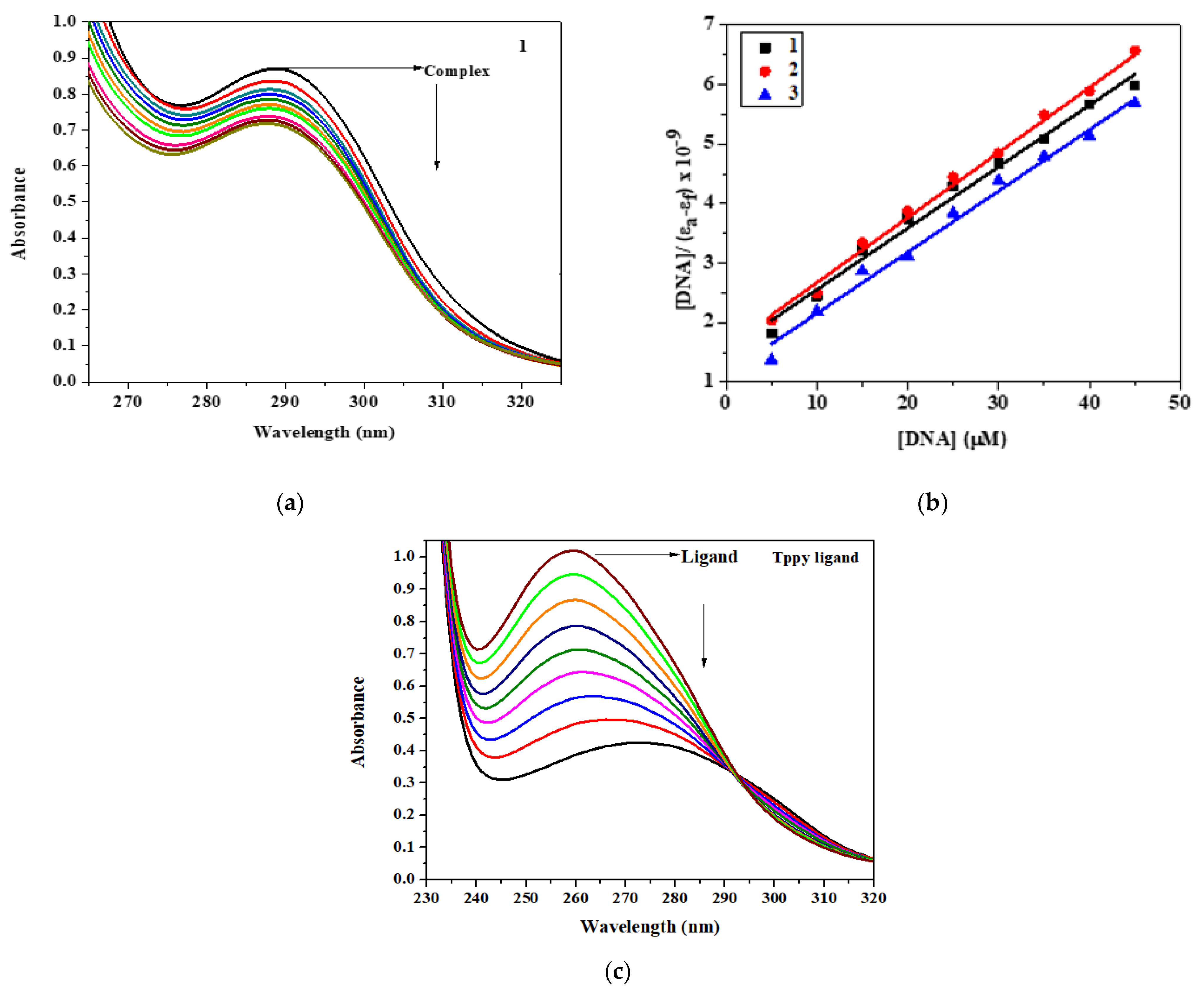
3.4. CT DNA Interaction Study
3.5. EB Displacement Study
3.6. Hydrodynamic Study
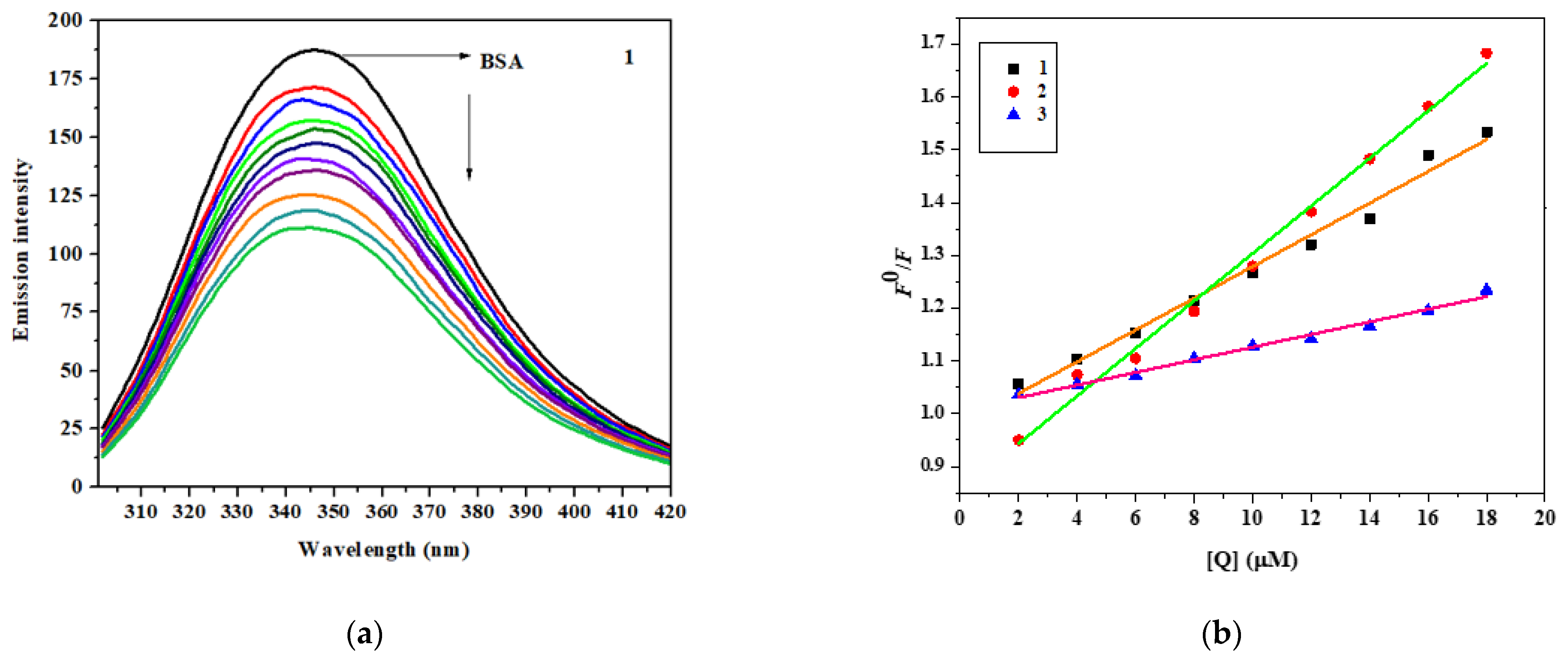
3.7. Protein Interaction Study of the Zn(II) Complexes with BSA
3.7.1. Absorption Study
3.7.2. Fluorescence Measurement
3.8. Molecular Docking Investigation with DNA
3.9. In Vitro Cytotoxicity Evaluation of the Zn(II) Complexes: Cell Viability Assay Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Atar, D.; Backx, P.H.; Appel, M.M.; Gao, W.D.; Marban, E. Excitation-transcription coupling mediated by zinc influx through voltage-dependent calcium channels. J. Biol. Chem. 1995, 270, 2473–2477. [Google Scholar] [CrossRef]
- Vallee, B.L.; Falchuk, K.H. The biochemical basis of zinc physiology. Physiol. Rev. 1993, 73, 79–118. [Google Scholar] [CrossRef]
- Li, X.; Hayik, S.A.; Merz, K.M. QM/MM X-ray refinement of zinc metalloenzymes. J. Inorg. Biochem. 2010, 104, 512–522. [Google Scholar] [CrossRef][Green Version]
- Falchuk, K.H. The molecular basis for the role of zinc in developmental biology. In Molecular and Cellular Effects of Nutrition on Disease Processes; Springer: Boston, MA, USA, 1998; pp. 41–48. [Google Scholar]
- Zalewski, P.D.; Forbes, I.J.; Betts, W.H. Correlation of apoptosis with change in intracellular labile Zn(II) using zinquin [(2-methyl-8-p-toluenesulphonamido-6-quinolyloxy)acetic acid], a new specific fluorescent probe for Zn(II). Biochem. J. 1993, 296, 403–408. [Google Scholar] [CrossRef]
- Wilson, M.; Hogstrand, C.; Maret, W. Picomolar concentrations of free zinc(II) ions regulate receptor protein-tyrosine phosphatase β activity. J. Biol. Chem. 2012, 287, 9322–9326. [Google Scholar] [CrossRef]
- Rusch, P.; Hirner, A.V.; Schmitz, O.; Kimmig, R.; Hoffmann, O.; Diel, M. Zinc distribution within breast cancer tissue of different intrinsic subtypes. Arch. Gynecol. Obstet. 2021, 303, 195–205. [Google Scholar] [CrossRef]
- Krężel, A.; Maret, W. The biological inorganic chemistry of zinc ions. Arch. Biochem. Biophys. 2016, 611, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Maret, W. Metallothionein redox biology in the cytoprotective and cytotoxic functions of zinc. Exp. Gerontol. 2008, 43, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Marino, S.M.; Gladyshev, V.N. Redox biology: Computational approaches to the investigation of functional cysteine residues. Antioxid. Redox Signal. 2011, 15, 135–146. [Google Scholar] [CrossRef]
- Aizenman, E.; Loring, R.H.; Reynolds, I.J.; Rosenberg, P.A. The redox biology of excitotoxic processes: The NMDA receptor, TOPA quinone, and the oxidative liberation of intracellular zinc. Front. Neurosci. 2020, 14, 778. [Google Scholar] [CrossRef] [PubMed]
- Maret, W. The redox biology of redox-inert zinc ions. Free Radic. Biol. Med. 2019, 134, 311–326. [Google Scholar] [CrossRef]
- Kadhim, M.I.; Husein, I. Pharmaceutical and biological application of new synthetic compounds of pyranone, pyridine, pyrmidine, pyrazole and isoxazole incoporating on 2-flouroquinoline moieties. Syst. Rev. Pharm. 2020, 11, 679–684. [Google Scholar]
- Mallikarjuna Rao, R.; Sreeramulu, J.; Ravindranath, L.; Nagaraja Reddy, G.; Hanumanthurayudu, K.; Nageswara Reddy, G.; Jayaraju, A.; Madhusudhan, P. Synthesis and biological screening of some pyridine and pyrrole derivatives of pyrazolo [3, 4-c] pyrazoles. J. Chem. Pharm. Res. 2012, 4, 272–278. [Google Scholar]
- Illán-Cabeza, N.A.; Jiménez-Pulido, S.B.; Hueso-Ureña, F.; Ramírez-Expósito, M.J.; Sánchez-Sánchez, P.; Martínez-Martos, J.M.; Moreno-Carretero, M.N. Effects on estrogen-dependent and triple negative breast cancer cells growth of Ni(II), Zn(II) and Cd(II) complexes with the schiff base derived from pyridine-2-carboxaldehyde and 5,6-diamino-1,3-dimethyluracil explored through the renin-angiotensin system (RAS)-regulating aminopeptidases. J. Inorg. Biochem. 2018, 185, 52–62. [Google Scholar]
- Porchia, M.; Pellei, M.; Del Bello, F.; Santini, C. Zinc complexes with nitrogen donor ligands as anticancer agents. Molecules 2020, 25, 5814. [Google Scholar] [CrossRef] [PubMed]
- Huggins, D.J.; Sherman, W.; Tidor, B. Rational approaches to improving selectivity in drug design. J. Med. Chem. 2012, 55, 1424–1444. [Google Scholar] [CrossRef]
- Gao, C.-Y.; Qiao, X.; Ma, Z.-Y.; Wang, Z.-G.; Lu, J.; Tian, J.-L.; Xu, J.-Y.; Yan, S.-P. Synthesis, characterization, DNA binding and cleavage, BSA interaction and anticancer activity of dinuclear zinc complexes. Dalton Trans. 2012, 41, 12220–12232. [Google Scholar] [CrossRef]
- Schmidt, A.; Casini, A.; Kühn, F.E. Self-assembled M2L4 coordination cages: Synthesis and potential applications. Coord. Chem. Rev. 2014, 275, 19–36. [Google Scholar] [CrossRef]
- Therrien, B. Drug delivery by water-soluble organometallic cages. Top Curr. Chem. 2011, 319, 35–55. [Google Scholar]
- Bardwell, D.A.; Jeffery, J.C.; Jones, P.L.; McCleverty, J.A.; Psillakis, E.; Reeves, Z.; Ward, M.D. Lanthanide complexes of the tetradentate N-donor ligand dihydrobis[3-(2-pyridyl)pyrazolyl]borate and the terdentate N-donor ligand 2,6-bis(1H-pyrazol-3-yl)pyridine: Syntheses, crystal structures and solution structures based on luminescence lifetime studies. Dalton Trans. 1997, 12, 2079–2086. [Google Scholar]
- Amoroso, A.J.; Jeffery, J.C.; Jones, P.L.; McCleverty, J.A.; Rees, L.; Rheingold, A.L.; Sun, Y.; Takats, J.; Trofimenko, S.; Ward, M.D.; et al. Complexes of the podand ligand tris[3-(2-pyridyl)-pyrazol-1-yl]borate with lanthanoids and actinoids: Rare examples of icosahedral N12 coordination. Chem. Comm. 1995, 18, 1881–1882. [Google Scholar] [CrossRef]
- Paul, R.L.; Amoroso, A.J.; Jones, P.L.; Couchman, S.M.; Reeves, Z.R.; Rees, L.H.; Jeffery, J.C.; McCleverty, J.A.; Ward, M.D. Effects of metal coordination geometry on self-assembly: A monomeric complex with trigonal prismatic metal coordination vs. tetrameric complexes with octahedral metal coordination. Dalton Trans. 1999, 10, 1563–1568. [Google Scholar] [CrossRef]
- Narwane, M.; Chang, Y.-L.; Ching, W.-M.; Tsai, M.-L.; Hsu, S.C.N. Investigation on the coordination behaviors of tris(2-pyridyl)pyrazolyl borates iron(II) complexes. Inorg. Chim. Acta 2019, 495, 118966. [Google Scholar] [CrossRef]
- Reichmann, M.; Rice, S.; Thomas, C.; Doty, P. A further examination of the molecular weight and size of desoxypentose nucleic acid. J. Am. Chem. Soc. 1954, 76, 3047–3053. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, H.; Chen, C.; Deng, H.; Lu, T.; Ji, L. Interaction of macrocyclic copper(II) complexes with calf thymus DNA: Effects of the side chains of the ligands on the DNA binding behaviors. Dalton Trans. 2003, 1, 114–119. [Google Scholar] [CrossRef]
- Ou-Yang, F.; Tsai, I.H.; Tang, J.Y.; Yen, C.Y.; Cheng, Y.B.; Farooqi, A.A.; Chen, S.R.; Yu, S.Y.; Kao, J.K.; Chang, H.W. Antiproliferation for breast cancer cells by ethyl acetate extract of nepenthes thorellii x (ventricosa x maxima). Int. J. Mol. Sci. 2019, 20, 3238. [Google Scholar] [CrossRef] [PubMed]
- Kavithaa, K.; Paulpandi, M.; Padma, P.R.; Sumathi, S. Induction of intrinsic apoptotic pathway and cell cycle arrest via baicalein loaded iron oxide nanoparticles as a competent nano-mediated system for triple negative breast cancer therapy. RSC Adv. 2016, 6, 64531–64543. [Google Scholar] [CrossRef]
- Bardwell, D.A.; Jeffery, J.C.; Jones, P.L.; McCleverty, J.A.; Ward, M.D. A reversible intramolecular hydrogen-bonding interaction involving second-sphere coordination of a water ligand. Dalton Trans. 1995, 17, 2921–2922. [Google Scholar] [CrossRef]
- Halcrow, M.A.; McInnes, E.J.L.; Mabbs, F.E.; Scowen, I.J.; McPartlin, M.; Powell, H.R.; Davies, J.E. Syntheses, structures and electrochemistry of [CuL1(LR)]BF4 [L1 = 3-{2,5-dimethoxyphenyl)-1-(2-pyridyl)pyrazole; LR = tris(3-arylpyrazolyl)hydroborate] and [CuL12][BF4]2. Effects of graphitic interactions on the stability of an aryl radical cation. Dalton Trans. 1997, 21, 4025–4036. [Google Scholar] [CrossRef]
- Amoroso, A.J.; Jeffery, J.C.; Jones, P.L.; McCleverty, J.A.; Psillakis, E.; Ward, M.D. Crystal structures of silver(I) and thallium(I) complexes of tris[3-(2-pyridyl)-pyrazol-1-yl]borate; Encapsulation of either a single thallium(I) ion or a trinuclear silver(I) cluster by a hexadentate podand. Chem. Comm. 1995, 11, 1175–1176. [Google Scholar] [CrossRef]
- Trofimenko, S.; Calabrese, J.C.; Thompson, J.S. Novel polypyrazolylborate ligands-Coordination control through 3-substituents of the pyrazole ring. Inorg. Chem. 1987, 26, 1507–1514. [Google Scholar] [CrossRef]
- Trofimenko, S. Recent advances in poly(pyrazolyl)borate(scorpionate) chemistry. Chem. Rev. 1993, 93, 943–980. [Google Scholar] [CrossRef]
- Bergquist, C.; Fillebeen, T.; Morlok, M.M.; Parkin, G. Protonation and reactivity towards carbon dioxide of the mononuclear tetrahedral zinc and cobalt hydroxide complexes, [TpBut,Me]ZnOH and [TpBut,Me]CoOH: Comparison of the reactivity of the metal hydroxide function in synthetic analogues of carbonic anhydrase. J. Am. Chem. Soc. 2003, 125, 6189–6199. [Google Scholar] [PubMed]
- Ruf, M.; Weis, K.; Vahrenkamp, H. Zn−O2H3−Zn: A coordination mode of the hydrolytic zinc−aqua function and a possible structural motif for oligozinc enzymes. J. Am. Chem. Soc. 1996, 118, 9288–9294. [Google Scholar] [CrossRef]
- Ibrahim, M.M.; Olmo, C.P.; Tekeste, T.; Seebacher, J.; He, G.; Maldonado Calvo, J.A.; Böhmerle, K.; Steinfeld, G.; Brombacher, H.; Vahrenkamp, H. Zn−OH2 and Zn−OH complexes with hydroborate derived tripod ligands: A comprehensive study. Inorg. Chem. 2006, 45, 7493–7502. [Google Scholar] [CrossRef] [PubMed]
- Sénèque, O.; Rager, M.-N.; Giorgi, M.; Reinaud, O. Supramolecular stabilization of a tris(imidazolyl) Zn−aqua complex evidenced by x-ray analysis: A structural model for mono-zinc active sites of enzymes. J. Am. Chem. Soc. 2001, 123, 8442–8443. [Google Scholar] [CrossRef]
- Bergquist, C.; Parkin, G. Protonation of the hydroxide ligand in a synthetic analogue of carbonic anhydrase, [TpBut,Me]ZnOH: Inhibition of reactivity towards CO2. J. Am. Chem. Soc. 1999, 121, 6322–6323. [Google Scholar] [CrossRef]
- Bergquist, C.; Bridgewater, B.M.; Harlan, C.J.; Norton, J.R.; Friesner, R.A.; Parkin, G. Aqua, alcohol, and acetonitrile adducts of tris(perfluorophenyl)borane: Evaluation of bronsted acidity and ligand lability with experimental and computational methods. J. Am. Chem. Soc. 2000, 122, 10581–10590. [Google Scholar] [CrossRef]
- Denisov, V.P.; Jonsson, B.-H.; Halle, B. Dynamics of functional water in the active site of native carbonic anhydrase from 17O magnetic relaxation dispersion. J. Am. Chem. Soc. 1999, 121, 2327–2328. [Google Scholar] [CrossRef]
- Toba, S.; Colombo, G.; Merz, K.M. Solvent dynamics and mechanism of proton transfer in human carbonic anhydrase II. J. Am. Chem. Soc. 1999, 121, 2290–2302. [Google Scholar] [CrossRef]
- Christianson, D.W.; Fierke, C.A. Carbonic anhydrase: Evolution of the zinc binding site by nature and by design. Acc. Chem. Res. 1996, 29, 331–339. [Google Scholar] [CrossRef]
- Das, D.; Chand, B.G.; Sarker, K.K.; Dinda, J.; Sinha, C. Zn(II) azide complexes of diimine and azoimine functions: Synthesis, spectra and X-ray structures. Polyhedron 2006, 25, 2333–2340. [Google Scholar] [CrossRef]
- Dapporto, P.; Formica, M.; Fusi, V.; Giorgi, L.; Micheloni, M.; Paoli, P.; Pontellini, R.; Rossi, P. Addition of small molecules by Zn(II) and Cu(II) dinuclear complexes obtained by an amino-phenolic ligand. Crystal Structures of the dinuclear zinc complex assembling butanolate and azide anions. Inorg. Chem. 2001, 40, 6186–6192. [Google Scholar] [CrossRef] [PubMed]
- Pérez Olmo, C.; Böhmerle, K.; Steinfeld, G.; Vahrenkamp, H. New polar pyrazolylborate ligands and their basic zinc complex chemistry. Eur. J. Inorg. Chem. 2006, 2006, 3869–3877. [Google Scholar] [CrossRef]
- Huang, Y.-C.; Haribabu, J.; Chien, C.-M.; Sabapathi, G.; Chou, C.-K.; Karvembu, R.; Venuvanalingam, P.; Ching, W.-M.; Tsai, M.-L.; Hsu, S.C. Half-sandwich Ru(η6-p-cymene) complexes featuring pyrazole appended ligands: Synthesis, DNA binding and in vitro cytotoxicity. J. Inorg. Biochem. 2019, 194, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Dorairaj, D.P.; Haribabu, J.; Chang, Y.-L.; Hsu, S.C.; Echeverria, C.; Echeverria, J.; Karvembu, R. Effect of new Pd (II)-aroylthiourea complex on pancreatic cancer cells. Inorg. Chem. Commun. 2021, 134, 109018. [Google Scholar] [CrossRef]
- Dorairaj, D.P.; Lin, Y.-F.; Haribabu, J.; Murugan, T.; Narwane, M.; Karvembu, R.; Neelakantan, M.A.; Kao, C.-L.; Chiu, C.-C.; Hsu, S.C. Binding mode transformation and biological activity on the Ru(II)-DMSO complexes bearing heterocyclic pyrazolyl ligands. J. Inorg. Biochem. 2021, 223, 111545. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.; Shimer Jr, G.H.; Meehan, T. Polycyclic aromatic hydrocarbons physically intercalate into duplex regions of denatured DNA. Biochemistry 1987, 26, 6392–6396. [Google Scholar] [CrossRef]
- Saha, S.; De, A.; Ghosh, A.; Ghosh, A.; Bera, K.; Das, K.S.; Akhtar, S.; Maiti, N.C.; Das, A.K.; Das, B.B. Pyridine-pyrazole based Al(III) ‘turn on’ sensor for MCF7 cancer cell imaging and detection of picric acid. RSC Adv. 2021, 11, 10094–10109. [Google Scholar] [CrossRef]
- Desai, N.C.; Vaja, D.V.; Jadeja, K.A.; Joshi, S.B.; Khedkar, V.M. Synthesis, biological evaluation and molecular docking study of pyrazole, pyrazoline clubbed pyridine as potential antimicrobial agents. Anti-Infect. Agents 2020, 18, 306–314. [Google Scholar] [CrossRef]
- Lakowicz, J.R.; Weber, G. Quenching of fluorescence by oxygen. Probe for structural fluctuations in macromolecules. Biochemistry 1973, 12, 4161–4170. [Google Scholar] [CrossRef]
- Dorairaj, D.P.; Haribabu, J.; Chithravel, V.; Vennila, K.N.; Bhuvanesh, N.; Echeverria, C.; Hsu, S.C.; Karvembu, R. Spectroscopic, anticancer and antioxidant studies of fluxional trans-[PdCl2(S-acylthiourea)2] complexes. Results Chem. 2021, 3, 100157. [Google Scholar] [CrossRef]
- Haribabu, J.; Balachandran, C.; Tamizh, M.M.; Arun, Y.; Bhuvanesh, N.S.; Aoki, S.; Karvembu, R. Unprecedented formation of palladium(II)-pyrazole based thiourea from chromone thiosemicarbazone and [PdCl2(PPh3)2]: Interaction with biomolecules and apoptosis through mitochondrial signaling pathway. J. Inorg. Biochem. 2020, 205, 110988. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.M.; Zhao, H.; Lee, J.I.; Reynolds, K.; Zhang, L.; Temple, R.; Lesko, L. Therapeutic protein–drug interactions and implications for drug development. Clin. Pharmacol. Ther. 2010, 87, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wan, R.; Liu, H.; Cheng, C.-M.; Zhao, Y.-F. Cleavage of BSA by a dipeptide seryl-histidine. Lett. Sci. 2000, 7, 325–329. [Google Scholar] [CrossRef]
- Ramachandran, E.; Senthil Raja, D.; Rath, N.P.; Natarajan, K. Role of substitution at terminal nitrogen of 2-oxo-1, 2-dihydroquinoline-3-carbaldehyde thiosemicarbazones on the coordination behavior and structure and biological properties of their palladium(II) complexes. Inorg. Chem. 2013, 52, 1504–1514. [Google Scholar] [CrossRef] [PubMed]
- Jeyalakshmi, K.; Haribabu, J.; Bhuvanesh, N.S.; Karvembu, R. Half-sandwich RuCl2(η6-p-cymene) core complexes containing sulfur donor aroylthiourea ligands: DNA and protein binding, DNA cleavage and cytotoxic studies. Dalton Trans. 2016, 45, 12518–12531. [Google Scholar] [CrossRef]
- Wei, X.L.; Xiao, J.B.; Wang, Y.; Bai, Y. Which model based on fluorescence quenching is suitable to study the interaction between trans-resveratrol and BSA? Spectrochim. Acta A Mol. Biomol. Spectrosc. 2010, 75, 299–304. [Google Scholar] [CrossRef]
- Suganthi, M.; Elango, K.P. Synthesis, characterization and serum albumin binding studies of vitamin K3 derivatives. J. Photochem. Photobiol. B Biol. 2017, 166, 126–135. [Google Scholar] [CrossRef]
- Huerta-Aguilar, C.A.; Pandiyan, T.; Raj, P.; Singh, N.; Zanella, R. Fluorescent organic nanoparticles (FONs) for the selective recognition of Zn2+: Applications to multi-vitamin formulations in aqueous medium. Sens. Actuators B Chem. 2016, 223, 59–67. [Google Scholar] [CrossRef]
- Chakraborty, B.; Basu, S. Interaction of BSA with proflavin: A spectroscopic approach. J. Lumin. 2009, 129, 34–39. [Google Scholar] [CrossRef]
- Ismail-Khan, R.; Bui, M.M. A review of triple-negative breast cancer. Cancer Control 2010, 17, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.D.; Sun, Y.; Zhou, W.J.; Xie, X.Z.; Zhou, Q.M.; Lu, Y.Y.; Su, S.B. Resveratrol enhances inhibition effects of cisplatin on cell migration and invasion and tumor growth in breast cancer MDA-MB-231 cell models in vivo and in vitro. Molecules 2021, 26, 2204. [Google Scholar] [CrossRef] [PubMed]
- Gambini, V.; Tilio, M.; Maina, E.W.; Andreani, C.; Bartolacci, C.; Wang, J.; Iezzi, M.; Ferraro, S.; Ramadori, A.T.; Simon, O.C.; et al. In vitro and in vivo studies of gold(I) azolate/phosphane complexes for the treatment of basal like breast cancer. Eur. J. Med. Chem. 2018, 155, 418–427. [Google Scholar] [CrossRef] [PubMed]




| Cancer Cell Lines | 1 | 2 |
|---|---|---|
| MDA-MB-231 | 6.81 0.98 | 8.85 1.05 |
| MDA-MB-468 | 16.56 1.32 | 10.85 1.72 |
| HCC1937 | 13.54 1.77 | 10.60 1.04 |
| Hs578T | 12.51 1.84 | 6.68 1.16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Narwane, M.; Dorairaj, D.P.; Chang, Y.-L.; Karvembu, R.; Huang, Y.-H.; Chang, H.-W.; Hsu, S.C.N. Tris-(2-pyridyl)-pyrazolyl Borate Zinc(II) Complexes: Synthesis, DNA/Protein Binding and In Vitro Cytotoxicity Studies. Molecules 2021, 26, 7341. https://doi.org/10.3390/molecules26237341
Narwane M, Dorairaj DP, Chang Y-L, Karvembu R, Huang Y-H, Chang H-W, Hsu SCN. Tris-(2-pyridyl)-pyrazolyl Borate Zinc(II) Complexes: Synthesis, DNA/Protein Binding and In Vitro Cytotoxicity Studies. Molecules. 2021; 26(23):7341. https://doi.org/10.3390/molecules26237341
Chicago/Turabian StyleNarwane, Manmath, Dorothy Priyanka Dorairaj, Yu-Lun Chang, Ramasamy Karvembu, Yu-Han Huang, Hsueh-Wei Chang, and Sodio C. N. Hsu. 2021. "Tris-(2-pyridyl)-pyrazolyl Borate Zinc(II) Complexes: Synthesis, DNA/Protein Binding and In Vitro Cytotoxicity Studies" Molecules 26, no. 23: 7341. https://doi.org/10.3390/molecules26237341
APA StyleNarwane, M., Dorairaj, D. P., Chang, Y.-L., Karvembu, R., Huang, Y.-H., Chang, H.-W., & Hsu, S. C. N. (2021). Tris-(2-pyridyl)-pyrazolyl Borate Zinc(II) Complexes: Synthesis, DNA/Protein Binding and In Vitro Cytotoxicity Studies. Molecules, 26(23), 7341. https://doi.org/10.3390/molecules26237341






